Key Points
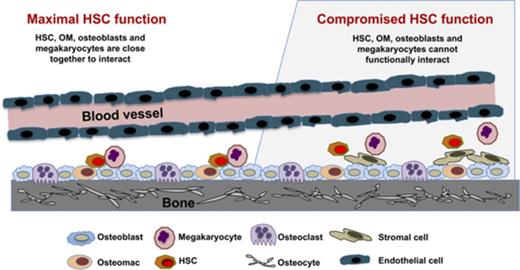
OM, osteoblast, and megakaryocyte interactions regulate HSC function in the niche.
OMs differ functionally and phenotypically from BM-derived macrophages.
Abstract
Networking between hematopoietic stem cells (HSCs) and cells of the hematopoietic niche is critical for stem cell function and maintenance of the stem cell pool. We characterized calvariae-resident osteomacs (OMs) and their interaction with megakaryocytes to sustain HSC function and identified distinguishing properties between OMs and bone marrow (BM)–derived macrophages. OMs, identified as CD45+F4/80+ cells, were easily detectable (3%-5%) in neonatal calvarial cells. Coculture of neonatal calvarial cells with megakaryocytes for 7 days increased OM three- to sixfold, demonstrating that megakaryocytes regulate OM proliferation. OMs were required for the hematopoiesis-enhancing activity of osteoblasts, and this activity was augmented by megakaryocytes. Serial transplantation demonstrated that HSC repopulating potential was best maintained by in vitro cultures containing osteoblasts, OMs, and megakaryocytes. With or without megakaryocytes, BM-derived macrophages were unable to functionally substitute for neonatal calvarial cell–associated OMs. In addition, OMs differentiated into multinucleated, tartrate resistant acid phosphatase–positive osteoclasts capable of bone resorption. Nine-color flow cytometric analysis revealed that although BM-derived macrophages and OMs share many cell surface phenotypic similarities (CD45, F4/80, CD68, CD11b, Mac2, and Gr-1), only a subgroup of OMs coexpressed M-CSFR and CD166, thus providing a unique profile for OMs. CD169 was expressed by both OMs and BM-derived macrophages and therefore was not a distinguishing marker between these 2 cell types. These results demonstrate that OMs support HSC function and illustrate that megakaryocytes significantly augment the synergistic activity of osteoblasts and OMs. Furthermore, this report establishes for the first time that the crosstalk between OMs, osteoblasts, and megakaryocytes is a novel network supporting HSC function.
Introduction
Maintenance of hematopoietic stem cell (HSC) function is an orchestrated event between multiple cell types within the hematopoietic niche.1,2 We previously demonstrated that osteoblasts play a major role in sustaining HSC function and that immature osteoblasts from neonatal calvariae of 2-day-old pups mediate a robust in vitro hematopoiesis-enhancing activity.3,4 We also showed that this activity is inversely correlated with osteoblast maturation and the level of CD166 expression4,5 which is a functional marker of murine and human HSCs and niche competency.6 Our group also demonstrated that megakaryocytes can increase osteoblast proliferation in vitro and in vivo,7-9 thus implicating megakaryocytes, albeit indirectly, in HSC maintenance. These observations identified critical interactions between osteoblasts and megakaryocytes and offered a unique experimental model to assess how megakaryocytes impact HSC function via osteoblasts.
Recently, independent groups have illustrated that megakaryocytes potentially regulate HSCs through the expression of platelet factor 4, transforming growth factor–β, or nuclear factor erythroid 2,10-12 although alternative mechanisms have been reported.13 Other cell types are also involved in the competence of the hematopoietic niche, including a unique class of CD45+F4/80+ macrophages known as osteomacs (OMs) that line the endosteum.14 The loss of OMs led to the mobilization of HSCs into the periphery, and the depletion of macrophages was detrimental for the maintenance of the niche.14 Furthermore, several groups have shown that macrophages are involved in HSC regulatory networks15-17 and the production of reactive oxygen species.18 Although bone marrow (BM)–derived macrophages express F4/80 (in addition to other markers, including CD45, CD68 [microsialin], CD169, and variable amounts of CD11b), it is unknown whether BM-derived macrophages and OMs (defined as bone-associated CD45+F4/80+ cells) are similar in phenotype and functional properties.
Recently, we confirmed that freshly isolated neonatal calvarial cells (NCCs) from 2-day-old pups,3,4 which are a common source of osteoblasts and osteoblast progenitors (but devoid of measurable hematopoietic progenitors), also contain a small group of CD45+F4/80+ OMs.19 In this study, we show that these OMs increase in number after culture with megakaryocytes. Interestingly, we also show that without CD45+F4/80+ cells, the hematopoietic-enhancing activity of NCCs is substantially reduced. The presence of OMs in NCC preparations gave us the opportunity to closely investigate how OMs cooperate with osteoblasts and megakaryocytes to impact HSC function. Our studies document that CD45+F4/80+ cells in NCC preparations are identical to the previously described OMs14,19,20 and that these cells are critical for the maintenance of HSC function. Furthermore, we illustrate that although BM-derived macrophages share many phenotypic properties with NCC-derived CD45+F4/80+ cells, these 2 cell types are phenotypically and functionally different.
Materials and methods
Preparation of fresh NCCs
Fresh NCCs were prepared as previously described.21 Calvariae from 2- to 3-day-old neonatal mice were pretreated with 10 mM EDTA in phosphate-buffered saline (PBS) for 30 minutes. Calvariae were then subjected to sequential collagenase digestions (200 U/mL; Collagenase, Type 2, Worthington Biochemicals, Lakewood NJ). Fractions 3 to 5 (collected between 45-60, 60-75, and 75-90 minutes during the digestion) were collected as fresh NCCs and are referred to as NCCs unless otherwise specified. These cells are >95% osteoblasts or osteoblast precursors as previously demonstrated.22
Preparation of fetal liver-derived megakaryocytes
Murine megakaryocytes were prepared from fetal livers as previously described.21 Fetuses were dissected from pregnant mice at embryonic day 13-15. The livers were removed, and single-cell suspensions were made by forcing cells through sequentially smaller gauge needles (18, 20, and 23 gauge). Cells were washed twice with Dulbecco’s modified Eagle’s medium (DMEM), 10% fetal calf serum and then seeded in 100-mL culture dishes (5 fetal livers per 100-mm dish), in DMEM 10% fetal calf serum containing 1% murine thrombopoietin. Megakaryocytes were obtained 3 to 5 days later by separating them from lymphocytes and other cells using a 1-step albumin gradient. The bottom layer was 3% albumin in PBS (bovine albumin, protease free and fatty acid poor, Serological Proteins, Inc., Kankakee, IL); the middle layer was 1.5% albumin in PBS, and the top layer was media containing the cells to be separated. All of the cells were sedimented through the layers at 1 g for 40 minutes at room temperature. The megakaryocyte-rich fraction was collected from the bottom of the tube.
Cell cultures
The cultures described in this article were established in 12-well culture plates. Cell types were mixed together (as indicated) in each individual well without the use of transwells or separating membranes. When indicated, fractions of freshly prepared NCCs were separated by cell sorting into CD45+F4/80+ cells (OMs) and CD45−F4/80− cells, referred to hereafter as osteoblasts. In some experiments, as indicated in the figure legends, total NCCs or OMs or osteoblasts sorted from total NCCs were first cultured with or without megakaryocytes for 1 week in α-minimum essential medium (α-MEM). At the end of 1 week, megakaryocytes were removed by gentle agitation23 and 1000 freshly sorted Lin− Sca1+ Kit+ (LSK) cells were added. Cultures were then maintained in medium consisting of a 1:1 mix of Iscove modified Dulbecco medium and α-MEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% L-glutamine and were supplemented with murine stem cell factor and interleukin-3 (10 ng/mL, Peprotech), IGF1 and TPO (20 ng/mL, Peprotech), and interleukin-6 and Flt3 (25 ng/mL, Peprotech) as previously reported.3 Cells were harvested 1 week later for colony-forming unit (CFU) assay and the CFU fold increase was calculated relative to that obtained from 250 fresh LSK cells.
BM repopulating assay
BoyJ, C57BL/6J, and C57BL/6JXBoyJ F1 mice were bred and housed at Indiana University School of Medicine. Eight- to 12-week-old mice were used. Recipients (F1 mice) received 1000 cGy (650 and 350 cGy split dose, 4 hours apart) before IV injection of test cells in 200 μl PBS. All procedures were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine and followed National Institutes of Health guidelines. Freshly isolated BoyJ (CD45.1) LSK cells (1000 cells) or the progeny of 1000 LSK cells cultured for 5 days with NCCs or different fractions of NCCs (as defined above) were co-transplanted with 100 000 C57/B6 (CD45.2) competitor cells. Chimerism was assessed monthly. Four months after the primary transplantation, the BM content of half of a femur from each primary recipient was transplanted into a lethally irradiated CD45.2 × CD45.1 F1 secondary recipient without competitor cells, and engraftment was assessed monthly. We have previously demonstrated3 that total NCCs do not contain any detectable BM-repopulating or clonogenic cells.
Cell staining, flow cytometry, and cell sorting
Cells were washed with stain wash (PBS, 1% bovine calf serum, and 1% penicillin/streptomycin) and stained for 15 minutes on ice. Freshly isolated cells (NCCs, neonatal BM cells, or BM cells from 8-week-old mice) were stained with CD45 and F4/80 and sorted (FACSAria or SORP FACSAria, BD Biosciences) into CD45+F4/80+ (OMs) and CD45−F4/80− (osteoblasts). Low-density BM cells were stained with fluorescein isothiocyanate–conjugated lineage anti-mouse antibodies CD3, CD4, CD45R, Ter119, and Gr1 (BioLegend), allophycocyanin (APC) c-Kit (CD117, BioLegend), and phycoerythrin (PE) Sca-1 (BioLegend) to collect LSK cells by cell sorting. For the multicolor analysis, cells were stained with a cocktail of Pacific Blue anti-CD45, APC anti-F4/80, APC/Cy7 anti-CD11b, PerCP/Cy5.5 anti-CD68, PE/Dazzle 594 anti-Ly-6G, Alexa Fluor 488 anti-Mac-2, PE/Cy7 anti-macrophage colony-stimulating factor (M-CSF) receptor, PE anti-CD166, and Brilliant Violent 605 anti-CD169. All antibodies except anti-CD166 were obtained from BioLegend. Anti-CD166 was obtained from Invitrogen. Cells were analyzed on a FACS Fortessa cell analyzer.
Osteoclast differentiation
Murine OMs or BM monocytes (20 000 cells per well) were seeded in a 96-well plate in α-MEM with 10% fetal bovine serum in the presence of M-CSF (20 ng/mL) and RANK ligand (RANKL) (80 ng/mL). The medium was changed every other day for 5 to 6 days. Once mature osteoclasts were formed, cells were fixed with 3.7% formaldehyde in PBS for 15 minutes at room temperature. Cells were stained for tartrate resistant acid phosphatase (TRAP) using a kit from Sigma (387A). Osteoclasts, defined as TRAP+ cells with ≥3 nuclei were counted. Mature osteoclasts were trypsinized and cultured on dentin with M-CSF and RANKL. After 3 days, osteoclasts were removed by mechanical agitation. Dentin slices were incubated with 20 μg/mL of peroxidase-conjugated wheat germ agglutinin for 90 minutes. After washing with PBS, 3,3′-diaminobenzidine (Sigma-Aldrich) was added to stain osteoclast resorption pits. Osteoclasts were imaged using Image-Pro version 7.0 software on a Leica DM1400 inverted epifluorscent microscope fitted with an automated stage and a Retiga EXi digital camera.
Statistical analysis
Experiments were repeated ≥3 times. Data are presented as the mean ± standard deviation unless otherwise stated. Statistical differences were determined by Student t test, 1-way analysis of variance (ANOVA) or 2-way ANOVA with Tukey post-hoc analysis. Significance was set at P < .05
Results
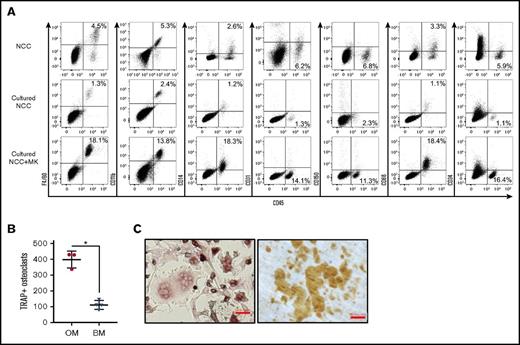
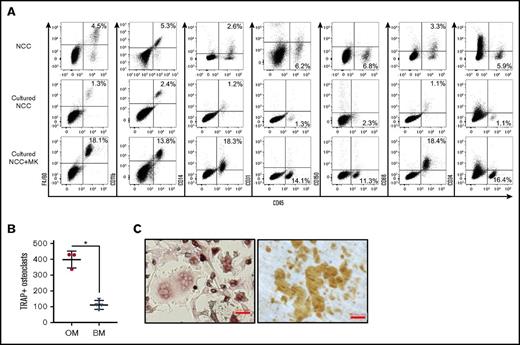
Consistent with a previous report,24 we identified in freshly isolated NCCs (which are predominantly CD45−) a population of CD45+ F4/80+ cells that share multiple characteristics with OMs14,19 (Figure 1A; NCC, left dot plot). CD45+ cells in NCCs expressed some classical macrophage markers, but were mostly negative for others (Figure 1A, NCC). Because megakaryocytes promote in vitro and in vivo osteoblast expansion,7,9,25 we examined the impact of megakaryocyte stimulation on NCCs. When cultured alone for 7 days, NCCs increased in number, but the percentage of CD45+ cells remained relatively unchanged or declined (Figure 1A; cultured NCC, left dot plot). However, NCCs cocultured with megakaryocytes displayed a significant increase in the percentage of CD45+ cells (Figure 1A; cultured NCC+MK, left dot plot), indicating that megakaryocytes promote CD45+ cell proliferation in vitro.
Phenotypic analysis of fresh and cultured OMs. (A) Representative data from the flow cytometric analysis of freshly isolated NCCs, NCCs cultured alone for 7 days (Cultured NCC), or cultured for 7 days with megakaryocytes (Cultured NCC+MK). In the example shown in the figure, cultures were initiated with 2 × 104 NCCs. On day 7, cultures contained 7 × 104 cells without and 1.8 × 105 cells with megakaryocytes, indicating that the number of CD45+F4/80+ cells in these cultures was ∼900 and 32 500 on day 7, respectively. Staining included CD41 to ensure that only CD41− cells were analyzed. (B) CD45+F/48+ OMs from NCCs or BM cells from long bones were cultured with RANKL and M-CSF to induce osteoclast formation. TRAP+ osteoclasts with ≥3 nuclei were quantified. *P < .05. (C) Osteoclasts formed from the calvariae-derived (left panel) OMs were replated on dentin, and resorption pits were stained after 3 days (right panel). Representative images of TRAP+ osteoclasts and resorption pits formed by these cells are shown. Slides were stained as described in “Materials and methods.” Scale bar indicates 50 μm.
Phenotypic analysis of fresh and cultured OMs. (A) Representative data from the flow cytometric analysis of freshly isolated NCCs, NCCs cultured alone for 7 days (Cultured NCC), or cultured for 7 days with megakaryocytes (Cultured NCC+MK). In the example shown in the figure, cultures were initiated with 2 × 104 NCCs. On day 7, cultures contained 7 × 104 cells without and 1.8 × 105 cells with megakaryocytes, indicating that the number of CD45+F4/80+ cells in these cultures was ∼900 and 32 500 on day 7, respectively. Staining included CD41 to ensure that only CD41− cells were analyzed. (B) CD45+F/48+ OMs from NCCs or BM cells from long bones were cultured with RANKL and M-CSF to induce osteoclast formation. TRAP+ osteoclasts with ≥3 nuclei were quantified. *P < .05. (C) Osteoclasts formed from the calvariae-derived (left panel) OMs were replated on dentin, and resorption pits were stained after 3 days (right panel). Representative images of TRAP+ osteoclasts and resorption pits formed by these cells are shown. Slides were stained as described in “Materials and methods.” Scale bar indicates 50 μm.
We performed a series of experiments to examine previously reported14,19,24,26 functional properties of OMs. OMs were characterized as endosteal macrophages that contribute to osteoblast mineralization26 and do not differentiate into osteoclasts.24 NCC-derived CD45+F4/80+ cells did not differentiate into osteoblasts (data not shown). However, the removal of CD45+F4/80+ cells from NCCs significantly reduced the ability of the remaining cells (CD45−F4/80− fraction of NCC) to mineralize (calcium levels of 3.03 ± 0.04 µg/mL for CD45−F4/80− cells alone vs 5.59 ± 0.01 µg/mL for CD45−F4/80− cells with OMs; P < .05), thus confirming previously published observations.20,26 On the other hand, RANKL and M-CSF induced CD45+F4/80+ cells to differentiate into multinucleated osteoclasts that express TRAP (Figure 1B-C, left micrograph) and were capable of forming dentin resorption pits (Figure 1C, right micrograph) suggesting that, contrary to published data,24 NCC-derived CD45+F4/80+ cells contain osteoclast progenitors. Furthermore, using our culture conditions, we established that osteoclast formation was increased in calvariae-derived OMs compared with BM CD45+F4/80+ cells (Figure 1B).
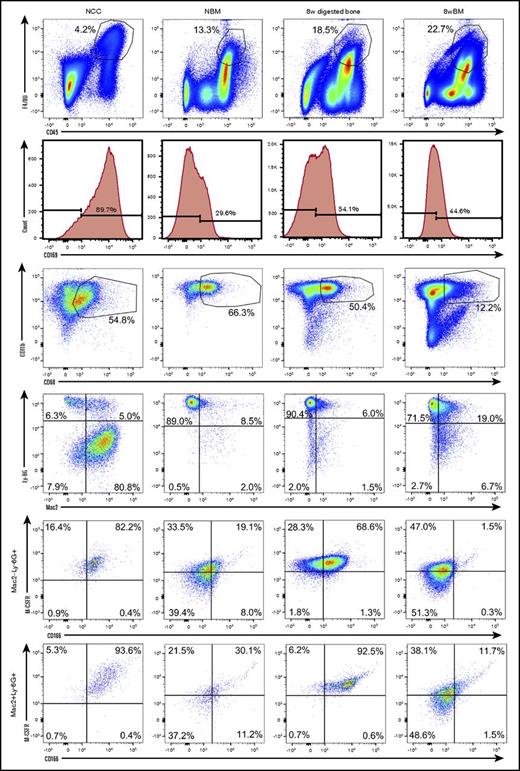
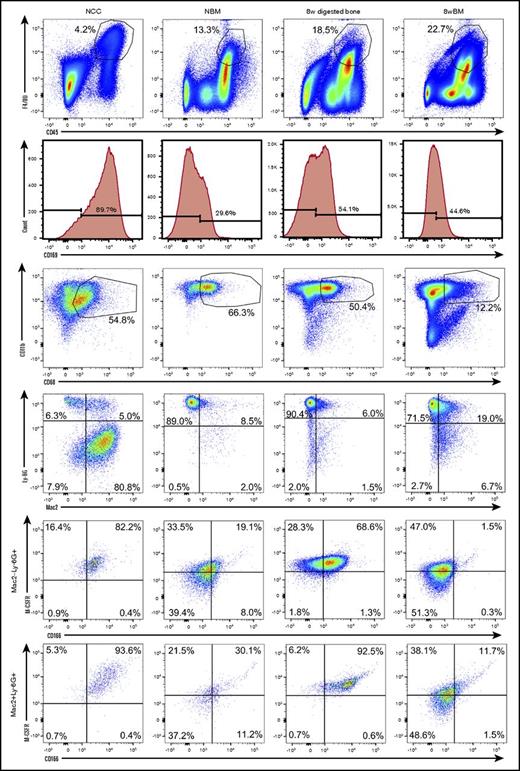
We next examined the phenotypic and functional properties of OMs contained in NCCs and their CD45+F4/80+ BM-derived counterparts collected from the long bones of neonatal pups. To identify possible developmental changes in the phenotypic makeup of BM-derived macrophages, we also analyzed BM cells collected from the long bones of 8-week-old mice. To confirm that OMs are osteolineage-associated macrophages (present in the bones of neonatal and adult mice) with a unique identity, we also analyzed digested bones (previously flushed) from 8-week-old mice. Figure 2 demonstrates that cells from all 4 sources shared similar profiles for multiple markers. However, only NCC-derived CD45+F4/80+ cells and the enzymatically released CD45+F4/80+ cells from flushed 8-week long bones contained a distinct subgroup of cells that coexpressed CD166 and M-CSFR. Double expression of CD166 and M-CSFR was evident among Ly-6G+Mac2− and Ly-6G+Mac2+ cells (Figure 2). Based on fluorescence minus one data (supplemental Figure 2), BM-derived macrophages and NCCs are Mac2low (Galectin-3low) as has been reported by others17,26,27 suggesting that Ly-6G+Mac2+ cells may in fact be considered Ly-6G+Mac2low cells. This is in agreement with reports indicating that tissue resident macrophages (such as OM) are Mac2− or low as opposed to Mac2high inflammatory macrophages.17 Expression of other surface markers by OMs and BM-derived macrophages is shown in supplemental Figure 3. Interestingly, the expression of CD169 did not discriminate between OMs and BM-derived macrophages because both groups of cells expressed this marker (Figure 2). In fact, in the example shown in Figure 2, a higher percentage of NCCs expressed CD169 than that detected among neonatal BM cells.
Phenotypic analysis of OMs and BM-derived macrophages from neonatal and adult donors. Flow cytometric analysis (representative data) of freshly isolated NCCs, neonatal BM (NBM) cells, digested (after flushing BM) long bones (8-week digested bone) and flushed BM cells (8-week BM) from 8-week-old mice. An analysis of digested bones and BM from 2 age groups was done to ensure that the identified phenotypes were not confined to a single developmental stage. Dot plots displaying similarly gated events (top to bottom) containing phenotypically similar cells are shown. Fluorescence minus one (FMO) controls collected for these assays are shown in supplemental Figure 1. Additional phenotypic data from these analyses are shown in supplemental Figure 3. Please note that BM-derived macrophages express M-CSFR as expected. However, 2-day– and 8-week–derived BM macrophages do not contain the subgroup of CD166+M-CSFR+ cells.
Phenotypic analysis of OMs and BM-derived macrophages from neonatal and adult donors. Flow cytometric analysis (representative data) of freshly isolated NCCs, neonatal BM (NBM) cells, digested (after flushing BM) long bones (8-week digested bone) and flushed BM cells (8-week BM) from 8-week-old mice. An analysis of digested bones and BM from 2 age groups was done to ensure that the identified phenotypes were not confined to a single developmental stage. Dot plots displaying similarly gated events (top to bottom) containing phenotypically similar cells are shown. Fluorescence minus one (FMO) controls collected for these assays are shown in supplemental Figure 1. Additional phenotypic data from these analyses are shown in supplemental Figure 3. Please note that BM-derived macrophages express M-CSFR as expected. However, 2-day– and 8-week–derived BM macrophages do not contain the subgroup of CD166+M-CSFR+ cells.
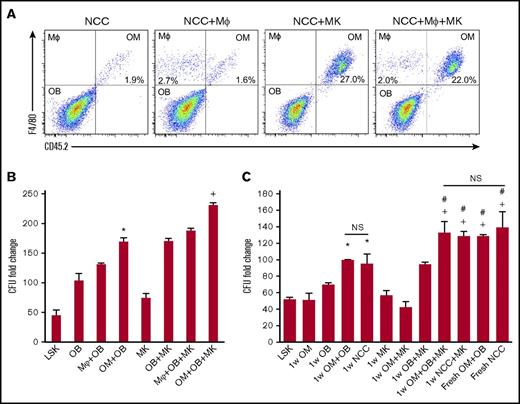
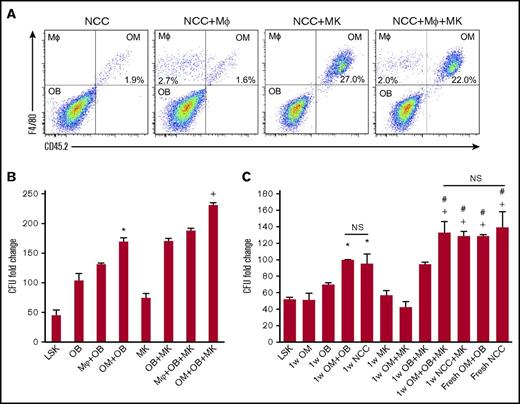
We examined whether BM-derived macrophages can functionally substitute for OMs to deliver the well-established hematopoiesis-enhancing activity.3,5 First, we assessed if megakaryocytes can promote BM-derived CD45+F4/80+ cells proliferation in vitro as we demonstrated for OMs (Figure 3A). We cultured NCCs and megakaryocytes from C57Bl/6J mice (CD45.2) for 1 week with BM-derived macrophages from BoyJ mice (CD45.1). Megakaryocytes promoted the expansion of OMs, but had no effect on BM CD45+F4/80+ macrophages (Figure 3A). Similar results were obtained when OMs in NCCs were substituted by BM-derived macrophages (data not shown). These results illustrate that NCC-derived OMs are a unique class of tissue-resident macrophages that display a distinctive response to megakaryocyte stimulation. We next examined if OMs are essential for the maximal expansion of CFUs by NCCs. As shown in Figure 3B, megakaryocytes promoted a significant enhancement of CFU fold increase. However, regardless of whether megakaryocytes were added to the cocultures, BM-derived macrophages did not support CFU fold increase to the same level sustained by OMs, demonstrating that BM-derived macrophages cannot functionally substitute for OMs. Osteoblasts alone were capable of expanding CFUs, but not to the same level as that observed when osteoblasts were mixed with OMs (Figure 3B). These data demonstrate that although osteoblasts can mediate the previously reported hematopoiesis-enhancing activity,3 OMs are required for the maximal hematopoiesis-enhancing activity promoted by NCCs. Promotion of the hematopoiesis-enhancing activity was not restricted to OMs isolated from neonatal calvariae because OMs collected from the long bones of adult mice mediated the same activity (supplemental Figure 4). In addition, megakaryocytes were also able to promote the proliferation of OMs harvested from the long bones of adult mice (data not shown), illustrating that the megakaryocyte-mediated enhancement of OM proliferation is not restricted to calvarial OMs only.
Effect of megakaryocytes on OM function. (A) Representative flow cytometric analysis of OMs and macrophages (MΦ) cultured for 1 week with megakaryocytes. NCCs and megakaryocytes were isolated from CD45.2 mice, and BM-derived macrophages and CD45+F4/80+ cells were collected from CD45.1 mice. (B) Cocultures of NCC-derived OBs (CD45−F4/80− cells) mixed with either the original number of OMs contained in freshly isolated NCCs (OM+OB) or a complimentary number (equivalent to the original number of OMs in NCCs) of BM-derived CD45+F4/80+ macrophages (MΦ+OB) were established. LSK cells were added with or without MKs, and LSK progeny were assessed for CFU content on day 7. CFU fold increase was calculated relative to that obtained from 250 fresh LSK cells. *P < .001 vs LSK, OB, MΦ+OB; +P < .001 vs MK, OB+MK, MΦ+OB+MK; 1-way ANOVA). (C) Fresh NCCs or fractions of NCCs (OBs or OMs) cultured with or without MKs (as indicated on the x-axis) were seeded with LSK cells then assayed 7 days later for their CFU content. CFU fold increase was calculated relative to that obtained from 250 fresh LSK cells. *P < .05 vs LSK, OM, and OB groups; +P < .005 vs MK, OM+MK, and OB+MK groups; #P < .05 vs 1w OM+OB and 1wNCC groups; one-way ANOVA). Please note that data in panels B and C are reported as fold increase rather than absolute numbers of CFUs to highlight the magnitude 1wNCC of CFU increase in these cultures. MK, megakaryocyte; OB, osteoblast.
Effect of megakaryocytes on OM function. (A) Representative flow cytometric analysis of OMs and macrophages (MΦ) cultured for 1 week with megakaryocytes. NCCs and megakaryocytes were isolated from CD45.2 mice, and BM-derived macrophages and CD45+F4/80+ cells were collected from CD45.1 mice. (B) Cocultures of NCC-derived OBs (CD45−F4/80− cells) mixed with either the original number of OMs contained in freshly isolated NCCs (OM+OB) or a complimentary number (equivalent to the original number of OMs in NCCs) of BM-derived CD45+F4/80+ macrophages (MΦ+OB) were established. LSK cells were added with or without MKs, and LSK progeny were assessed for CFU content on day 7. CFU fold increase was calculated relative to that obtained from 250 fresh LSK cells. *P < .001 vs LSK, OB, MΦ+OB; +P < .001 vs MK, OB+MK, MΦ+OB+MK; 1-way ANOVA). (C) Fresh NCCs or fractions of NCCs (OBs or OMs) cultured with or without MKs (as indicated on the x-axis) were seeded with LSK cells then assayed 7 days later for their CFU content. CFU fold increase was calculated relative to that obtained from 250 fresh LSK cells. *P < .05 vs LSK, OM, and OB groups; +P < .005 vs MK, OM+MK, and OB+MK groups; #P < .05 vs 1w OM+OB and 1wNCC groups; one-way ANOVA). Please note that data in panels B and C are reported as fold increase rather than absolute numbers of CFUs to highlight the magnitude 1wNCC of CFU increase in these cultures. MK, megakaryocyte; OB, osteoblast.
Based on the established ability of immature osteoblasts to support the most robust hematopoiesis-enhancing activity4,5 and the potential of megakaryocytes to suppress osteoblast differentiation,7 we examined if megakaryocytes can augment the osteoblast-mediated hematopoiesis-enhancing activity via their interactions with OMs. Unsorted NCCs, sorted osteoblasts (CD45−F4/80−), or OMs from C57BL/6J mice (CD45.2) were used for these assays. NCCs were cultured fresh (NCC) or were maintained for 1 week without (1w NCC) or with megakaryocytes (1w NCC+MK) before their use in culture (Figure 3C). Similarly, we established cultures of osteoblasts (OB in Figure 3C) and OMs with megakaryocytes (1w OB+MK and 1w OM+MK, respectively). At the 1-week time point, cultures were seeded with freshly sorted BoyJ-derived (CD45.1) LSK cells. Cells were harvested 7 days later and assayed for CFUs (Figure 3C). Consistent with our previous findings, NCCs maintained in culture for 1 week (1w NCC) did not provide a comparable hematopoiesis-enhancing activity relative to fresh NCCs. Megakaryocytes promoted the hematopoiesis-enhancing activity of 1w NCC (1w NCC+MK) as evidenced by comparable CFU counts compared with fresh NCC cultures (Figure 3C). Interestingly, the megakaryocyte-enhanced activity was lost when OMs were removed from NCCs by cell sorting (1w OB+MK), suggesting that osteoblasts, OMs, and megakaryocytes cooperate to promote hematopoietic progenitor and HSC function. Data depicting the breakdown of the CFU subtypes in these cultures are shown in supplemental Figure 5.
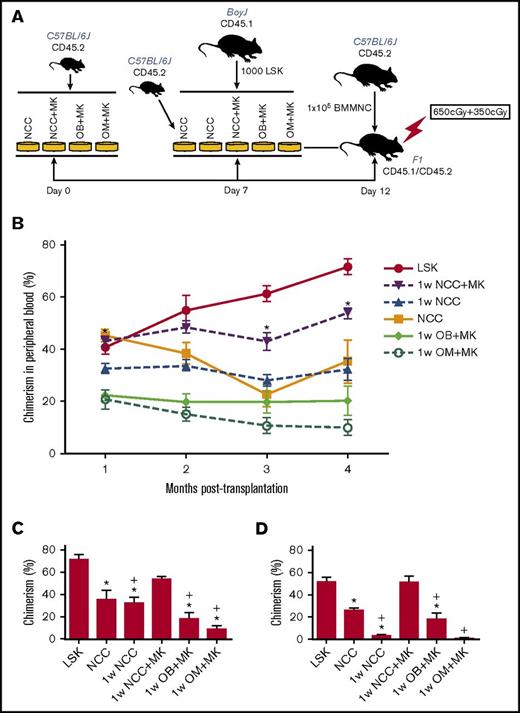
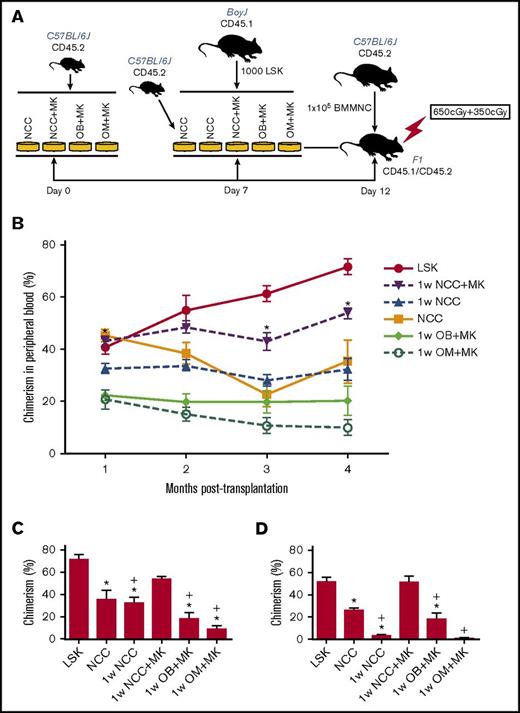
In vitro observations were validated in competitive repopulation assays (Figure 4). An illustration depicting the experimental set-up of these studies is shown in Figure 4A. The kinetics of chimerism in the peripheral blood of primary recipients over a period of 4 months are shown in Figure 4B. LSK cells maintained for 1 week in NCC+MK cultures retained the highest level of repopulating activity relative to fresh LSK cells (Figure 4B-C), suggesting that megakaryocytes were able to reverse the decline in the hematopoiesis-enhancing activity of NCCs cultured alone without megakaryocytes (1w NCC). Secondary transplantation data (Figure 4D) demonstrated again that 1w NCC cultured with megakaryocytes (1w NCC+MK) retained a robust hematopoiesis-enhancing activity relative to LSK cells cultured with NCCs maintained alone for 1 week. Neither osteoblasts nor OMs alone sustained the BM repopulating potential of LSK cells, suggesting that the previously documented hematopoiesis-enhancing activity of NCCs3 is synergistically mediated by osteoblasts and OMs. Reconstitution was multilineage (supplemental Figure 6).
In vivo assays of cells maintained in vitro with NCCs or sorted fractions of NCCs. (A) Schema summarizing the protocol for maintaining LSK cells in cocultures followed by transplantation of progeny cells into CD45.1/CD45.2 recipients. (B) Progeny of 1000 LSK cells from C57BL/6 (CD45.2) mice cocultured for 5 days with each group of cells identified in panel A were transplanted in a competitive repopulation assay with 100 000 BoyJ (CD45.1) cells via tail vein injection in lethally irradiated (split dose of 650 and 350 cGy) CD45.1 × CD45.2 F1 recipients. Freshly isolated LSK cells from BoyJ mice were used as control. Engraftment was assessed in the peripheral blood monthly and plotted for each group (n = 8–9 mice per group from 2 independent experiments). *P < .05 vs 1w NCC, 1-way ANOVA. (C) Chimerism in the BM of primary recipients (shown in panel B) at 4 months posttransplantation. (D) At 4 months post–primary transplantation, halves of femurs from primary recipients were transplanted into lethally irradiated secondary recipients. Chimerism in secondary recipients at 3 months posttransplantation is shown (4 mice per group) *P < .05 vs LSK group, +P < .05 vs 1w NCC+MK group, 1-way ANOVA.
In vivo assays of cells maintained in vitro with NCCs or sorted fractions of NCCs. (A) Schema summarizing the protocol for maintaining LSK cells in cocultures followed by transplantation of progeny cells into CD45.1/CD45.2 recipients. (B) Progeny of 1000 LSK cells from C57BL/6 (CD45.2) mice cocultured for 5 days with each group of cells identified in panel A were transplanted in a competitive repopulation assay with 100 000 BoyJ (CD45.1) cells via tail vein injection in lethally irradiated (split dose of 650 and 350 cGy) CD45.1 × CD45.2 F1 recipients. Freshly isolated LSK cells from BoyJ mice were used as control. Engraftment was assessed in the peripheral blood monthly and plotted for each group (n = 8–9 mice per group from 2 independent experiments). *P < .05 vs 1w NCC, 1-way ANOVA. (C) Chimerism in the BM of primary recipients (shown in panel B) at 4 months posttransplantation. (D) At 4 months post–primary transplantation, halves of femurs from primary recipients were transplanted into lethally irradiated secondary recipients. Chimerism in secondary recipients at 3 months posttransplantation is shown (4 mice per group) *P < .05 vs LSK group, +P < .05 vs 1w NCC+MK group, 1-way ANOVA.
Discussion
In this article, we further characterized the osteolineage-associated macrophages known as OMs and showed that, phenotypically and functionally, OMs are different than BM-derived macrophages, defined as CD45+F4/80+ cells. In performing these studies, it was crucial to design methodologies that can physically separate BM-derived macrophages from osteolineage-associated OMs in long bones from adult mice. This approach was important for 2 reasons. First, it was critical to demonstrate that OMs exist in the long bones and that they are not a unique developmental cell type present in only neonatal calvariae. Our studies expand the phenotypic definition previously attributed to these cells and confirm that OMs are not a developmentally or anatomically unique cell population (Figure 2). More importantly, our studies also confirm that OMs collected from the hematopoietic niche of adult mice possess the same functional properties attributed to neonatal OMs (supplemental Figure 4). Second, a reliable and reproducible isolation protocol that consistently yielded separate preparations of BM-derived macrophages and OMs from long bones was required to accurately assess the phenotypic and functional differences between the 2 cell types. Our protocol described in “Materials and methods” provided these necessary isolation parameters.
Using this approach coupled with multicolor phenotypic analysis, we were able to identify key expression differences that separated BM-associated macrophages from bone-associated OMs. Only OMs and collagenase-digested long bones contained a group of OMs that coexpressed M-CSFR and CD166. This group of cells constituted a very small fraction of OMs (Figure 2), which was not present among BM-derived macrophages from either neonatal or adult animals. Whether these cells (or a different subfraction of OMs) are responsible for the functional properties of OM requires additional investigations. Because we previously demonstrated that CD166 is expressed on HSCs and is critical to the competence of the hematopoietic niche,6 it is possible that OMs contribute to the fitness of the niche through CD166-CD166 homotypic interactions. It is not clear at present if these homotypic interactions actually take place or whether they involve OMs and HSCs.
Our studies suggest that CD169, which was previously shown to identify a niche macrophage critical to HSC function,15,16 was not a distinguishing marker between OMs and BM-derived macrophages. Interestingly, our data demonstrate that BM macrophages, identified by Chow et al.15 as CD169+ cells, may have also been OMs, because CD169 is not a discriminatory marker between OMs and BM-derived macrophages. Therefore, our findings suggest that previous studies15 in which clodronate was used to eliminate macrophages may have also inadvertently removed OMs (in addition to macrophages and osteoclasts) and incorrectly attributed the role of supporting erythropoiesis only to BM-resident macrophages instead of both macrophages and OMs. Our present findings do not necessarily contradict those of Chow et al.,15 but clarify the origin and source of these CD169 cells and their anatomic association in the hematopoietic niche. It is important, however, to keep in mind that, at least in the functional assays reported in this article, BM-derived macrophages which are predominantly CD169+ could not substitute for osteolineage OMs that are also predominantly CD169+, suggesting that these 2 cell types are functionally distinct.
Our current studies confirm many of the previously reported functional properties of OMs,20,24,26 such as their ability to enhance osteoblast mineralization. However, in our hands, highly purified CD45+F4/80+ OMs from NCCs differentiated into TRAP+ osteoclasts that were able to resorb dentin (Figure 1C). Previously, Chang et al24 reported that OMs did not differentiate into TRAP+ osteoclasts in vitro. Although it remains possible that different OM subpopulations were used by the different laboratories, carefully controlled and optimized culture conditions are also necessary to induce osteoclast differentiation, which could possibly explain the differing results. Alexander et al26 and Chang et al24 also reported the absence of TRAP+ osteoclasts in the immediate microenvironment of OMs using the macrophage Fas-induced apoptosis mouse model or clodronate liposome delivery system. In their assessment, these authors indicated that bone modeling, rather than remodeling, was occurring at these sites. However, our findings that OMs contain a population that can form osteoclasts, provides an alternative explanation, that is, that the reported strategies to inhibit OMs also blocked osteoclast formation.
In conclusion, this report presents evidence that OMs are bone-associated macrophages that are phenotypically and functionally distinct from their BM-derived CD45+F4/80+ counterparts. These investigations extended the definition of OMs to osteal macrophages that have a unique CD166+M-CSFR+ subgroup that is not present among BM-derived macrophages. Furthermore, we illustrate that OMs are critical for the hematopoiesis-enhancing activity previously attributed to NCCs3 and that megakaryocytes augment this activity. These findings describe a unique and new network between osteoblasts, OMs, and megakaryocytes that maintain HSC function and demonstrate the importance of OMs in bone health, in sustaining HSC function, and in maintaining the competence of the hematopoietic niche. Further detailed analyses are required to molecularly define how these cell types interact together.
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the members of the Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility for their outstanding technical support. The authors also thank the In Vivo Therapeutics Core.
This work was supported in part by National Institutes of Health (NIH), National Institute of Arthritis and Musculoskeletal and Skin Disease grant R01 AR060332. The Indiana University Melvin and Bren Simon Cancer Center Flow Cytometry Resource Facility and the In Vivo Therapeutics Core are funded in part by NIH, National Cancer Institute grant P30 CA082709 and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant U54 DK106846. The Flow Cytometry Resource Facility is supported in part by NIH instrumentation grant 1S10D012270. This work was also supported in part by a postdoctoral NIH NIDDK T32 training grant in hematopoiesis, T32 DK007519 (P.J.C.).
Authorship
Contribution: S.F.M., L.X., J.G., P.J.C., I.A., E.R.H., H.W., M.B.A., K.M.D., A.A.-P., and J.M.H. performed the experiments; S.F.M., L.X., M.A.K., A.B., and E.F.S. designed the experiments and interpreted the data; L.X. and E.F.S. wrote the manuscript; S.F.M. J.G., M.A.K., and A.B. provided critical suggestions and edited the manuscript; E.F.S. conceived and supervised the study; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edward F. Srour, Indiana University School of Medicine, 980 West Walnut St, R3-C312, Indianapolis, IN 46202; e-mail: esrour@iu.edu.
References
Author notes
S.F.M. and L.X. contributed equally to this work.