Key Points
FRD is a well-tolerated oral triplet regimen with durable responses in myeloma.
Correlative analysis identified MAGEA1 as a functional biomarker of resistance.
Abstract
Phase 3 studies combining histone deacetylase inhibitors with bortezomib were hampered by gastrointestinal (GI) intolerance, which was not observed when combined with immunomodulatory drugs. This study is a single-center phase 2 study of panobinostat with lenalidomide and dexamethasone (FRD). Twenty-seven relapsed multiple myeloma patients were enrolled. Twenty-two patients (81%) were lenalidomide refractory and 9 (33%), 14 (52%), and 7 (26%) were refractory to pomalidomide, bortezomib, and carfilzomib, respectively. High-risk molecular findings were present in 17 (63%) patients. Responses included 2 complete responses (CRs), 4 very good partial responses (VGPRs), 5 partial responses (PRs), and 9 minimal responses (MRs) for an overall response rate of 41%, clinical benefit rate of 74%, and a disease control rate of 96%. The median progression-free survival (PFS) was 7.1 months. In the 22 lenalidomide-refractory patients, there were 1 CR, 4 VGPRs, 3 PRs, and 7 MRs, with a median PFS of 6.5 months. Median overall survival was not reached. Grade 3/4 toxicities were primarily hematologic. Gene expression profiling of enrollment tumor samples revealed a set of 1989 genes associated with short (<90 days) PFS to therapy. MAGEA1 RNA and protein expression were correlated with short PFS, and laboratory studies demonstrated a role for MAGE-A in resistance to panobinostat-induced cell death. FRD demonstrates durable responses, even in high-risk, lenalidomide-refractory patients, indicating the essential role of panobinostat in attaining responses. MAGEA1 expression may represent a functional biomarker for resistance to panobinostat. In contrast to PANORAMA 1, there were no significant GI toxicities and primarily expected hematologic toxicities. This trial was registered at www.clinicaltrials.gov as #NCT00742027.
Introduction
Treatment options for patients with multiple myeloma (MM) refractory to immunomodulatory drugs (IMID) and proteasome inhibitors (PIs) are urgently needed. A promising strategy is the use of epigenetic agents, such as the histone deacetylase inhibitor (HDACi) panobinostat, which inhibits class I, II, and IV HDACs and is therefore a pan-HDAC inhibitor, to modulate the acetylation of histones and proteins, such as p53, α-tubulin, HIF-1α, and hsp90, that are involved in oncogenesis. Treatment with HDACi has been shown to induce histone acetylation and activation of tumor suppressor genes that have been linked to MM progression.1 Preclinical studies with panobinostat demonstrate increased gene dysregulation and synergy in inhibition of MM xenograft tumor growth when combined with dexamethasone, lenalidomide, and bortezomib.1
Clinically, data from the phase 3 PANORAMA 1 study of 768 patients randomized to receive intravenous bortezomib and dexamethasone with either panobinostat or placebo revealed a 3.9-month increase in progression-free survival (PFS) with panobinostat along with an increase in complete response (CR) rates.2 However, this was accompanied by 25% grade 3/4 diarrhea vs 8% in the placebo arm. Importantly, PANORAMA 2 also demonstrated that the addition of panobinostat to bortezomib in bortezomib-refractory patients resulted in a response rate (RR) of 34.5% and PFS of 5.4 months.3 On February 23, 2015, panobinostat was approved by the US Food and Drug Administration for the treatment of MM patients who have received at least 2 prior regimens, including bortezomib and an immunomodulatory agent.
To date, despite the preclinical data demonstrating synergy between panobinostat and lenalidomide, there are very limited clinical data available regarding this combination. The safety and preliminary efficacy of the panobinostat-lenalidomide-dexamethasone (FRD) triplet regimen (Table 1) were assessed in a phase 1b study of 46 relapsed or relapsed/refractory MM patients4 but was complicated by 7 deaths due to respiratory failure (n = 3), myocardial infarction, bronchial disorder, intestinal perforation, and multiorgan failure (1 patient each) toxicities, likely due to the high-dose dexamethasone toxicities. The maximum tolerated dose of panobinostat and dexamethasone in that study is the dose selected for this phase 2 study. However, we investigated a modified schedule (Table 1) of this triplet regimen. Here, panobinostat is given thrice weekly only every other week (instead of weekly), given data that 2 weeks on/1 week off schedule was not well tolerated with bortezomib-based regimens,3,4 and dexamethasone is given weekly (instead of three 4-day pulses).
Methods
This study was an open-label, single-arm, and single-center phase 2 study and was approved by the program for the protection of human subjects at the Icahn School of Medicine at Mt. Sinai in accordance with the Declaration of Helsinki. The primary objective was to evaluate the best overall response rate (ORR). Secondary objectives were to evaluate the safety, response duration, overall survival, (OS) and PFS. Each drug was administered at the doses and schedule shown in Table 1.
Inclusion criteria were patients with relapsed or relapsed/refractory MM (including IMID and PI refractory), measurable disease (m spike ≥1.0, Bence Jones protein >200 mg per 24 hours, serum free light chain ≥10 mg/dL with abnormal serum free light chain ratio), adequate performance status, organ function (creatinine clearance ≥50), normal hepatic function, normal electrolytes, and hematologic parameters (absolute neutrophil count ≥1.5 vs >1.0, platelet ≥100 000 vs ≥75 000 if bone marrow [BM] plasma cell % < or ≥50%, respectively).
Patients previously treated with an HDACi or currently receiving medications with a risk of prolonging the QTc interval were excluded. Patients with QT Fridericia's correction formula >480, significant cardiac disease, impaired gastrointestinal (GI) functions, and diarrhea > grade 2 (National Cancer Institute Common Terminology Criteria for Adverse Events 3.0) were also excluded.
ORR was calculated using International Myeloma Working Group criteria,5 and PFS and OS were calculated by Kaplan-Meier curve analysis. Safety was characterized using Common Terminology Criteria for Adverse Events criteria.
RNA sequencing and data analysis
BM aspirates were obtained from MM patients in the study. Mononuclear cells were isolated from aspirates by density gradient centrifugation using Ficoll-Paque media following the Miltenyi MidiMACS BM mononuclear cell protocol. CD138+ cells were then selected using a RoboSep instrument (Stemcell Technologies) for fully automated cell separation. DNA, RNA, and protein were isolated simultaneously with the help of the Allprep DNA/RNA/Protein mini kit (Qiagen). RNA-Seq libraries were constructed from ribosomal RNA–depleted RNA using the TrueSeq RNA sample preparation guide (Illumina) and submitted for the paired-end 100-base-pairs sequencing on a HiSeq2500 (Illumina) following the manufacturer's instructions. RNAseq fastq files were aligned to the human genome (GRCh37) using the tool STAR.6 Reads were quantified at the gene level using the tool featureCounts from the Subread package.7 Raw counts were log2 batch-corrected and normalized using the function Voom from the Limma package.8,9 Patients were split in 2 groups based on PFS rate at 90 days, and then differentially expressed genes were identified by Limma. Further pathways and functional analyses for differentially expressed genes were carried out using the Ingenuity Pathway Analysis (Ingenuity Systems; http://www.ingenuity.com/products/pathways_analysis.html).
Immunoblotting
Protein lysates from CD138+ BM tumor cells prior to treatment were prepared using Allprep DNA/RNA/Protein mini kit (Qiagen). Protein extracts were then resolved by sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis followed by immunoblotting with MAGE-A1 antibody (MA454, mouse monoclonal antibody; Santa Cruz Biotechnologies, sc-20033), CRBN (Ab68763, mouse polyclonal; Abcam), Aiolos (D1C1E, rabbit polyclonal; Cell Signaling Technologies, 12720), Ikaros (rabbit polyclonal; Cell Signaling Technologies, 5443), and actin antibody (C-11, horseradish peroxidase, goat polyclonal antibody; Santa Cruz Biotechnologies, sc-1615HPR), and detected by enhanced chemiluminescence (BioRad). Blot patterns were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/), providing a quantitative measure of protein expression. Cell pellets were harvested at the start of panobinostat treatment to check for decrease of MAGE-A via western blot. Cells were lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% SDS, 1% sodium deoxycholate) containing 1× protease and phosphatase inhibitor (Pierce, 78447). Protein concentration was determined by Bradford assay (Bio-Rad, 500-0205). Ten micrograms of lysate was run on an SDS–polyacrylamide gel electrophoresis gel and transferred to a PVDF membrane. Blocking and antibody dilutions were done in 5% nonfat dry milk in Tris-buffered saline with Tween 20 (25 mM Tris, 140 mM NaCl, 2.7 mM KCl, 0.1% Tween 20, pH 7.5). The antibodies used were MAGE-A (Santa Cruz, sc-20034), glyceraldehyde-3-phosphate dehydrogenase (Abcam, ab8245), and goat anti-mouse immunoglobulin G (IgG) horseradish peroxidase conjugate (Thermo Scientific, 32430). Blots were visualized with Supersignal West Pico Chemiluminescent Substrate (Thermo Scientific, 34087) or Supersignal West Femto Chemiluminescent Substrate (Thermo Scientific, 34096).
Cell lines
Human myeloma cell lines (HMCLs), H929 and MM.1r, were purchased from ATCC and cultured at 37°C, 5% CO2 in RPMI-1640 (Invitrogen, A1049101) supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich, F0926).
shRNA lentiviral particles
Lentiviral short hairpin RNA (shRNA) particles targeting MAGE-A3 (shMA3) and a scrambled nontarget sequence (shNT) were produced following the lentiviral packaging protocol from Sigma-Aldrich (SHP001). Low-passage HEK293T cells were transfected with plasmid constructs containing the shRNA targeting MAGE-A3 (TRCN0000128375; Sigma-Aldrich) or a nontarget sequence (SHC002; Sigma-Aldrich). Viral supernatants were harvested and concentrated using Lenti-X concentrator (Clontech, 631231), and titer was obtained by HIV-1 p24 enzyme-linked immunosorbent assay (Zeptometrix, 0801111). Optimal multiplicity of infection was determined for each cell line on a lot-to-lot basis.
Lentiviral transduction and in vitro drug treatment
Overnight cultures were harvested and resuspended in R10 media supplemented with 8 μg/mL polybrene (Sigma-Aldrich, H9268). Cells were then plated at 30 000 cells or 40 000 cells per well into a 96-well round-bottom plate for MM.1r and H929, respectively. Lentiviral particles were added at optimal multiplicity of infection for each cell line. Cells were incubated for 18 hours at 37°C; then, the cells were harvested and washed twice with R10 media to remove excess virus and polybrene. Washed cells were resuspended in R10 media at 60 000 cells/mL or 150 000 cells/mL for MM.1r and H929, respectively. Resuspended cells were then aliquoted at 100 μL per well into a 96-well flat-bottom white plate. Plates were incubated at 37°C, 5% CO2, until panobinostat (SelleckChem, S1030) or lenalidomide (Santa Cruz Biotechnology, sc-218656) treatment time, which was at 24 hours after transduction for MM.1r and at 48 hours after transduction for H929. Cells were treated in triplicates for each concentration. Viability was assayed for after 24 hours of panobinostat treatment with the CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7572). Viability curves were generated with Prism 6.0 and normalized to vehicle-treated controls.
Results
A total of 27 patients completed cycle 1 of all 3 agents and therefore were considered evaluable for efficacy and safety. The baseline characteristics of these patients, all of whom had progressive disease (PD) at screening, are presented in Table 2. The median age was 64 (52% > 65 years old) with a median of 3 lines of prior therapy (range 1-10) over 4 years since diagnosis. High-risk molecular findings were present in 17 patients (63%), including 12 with gain of 1q21 by fluorescence in situ hybridization and 5 with del p53. Four of the patients with a gain of 1q21 also had t(4;14). Twenty-two (81%) patients were lenalidomide refractory, and 1 other patient only had a minimal response (MR) to bortezomib, lenalidomide, and dexamethasone induction. Refractory to pomalidomide, bortezomib, and carfilzomib were 33%, 52%, and 26%, respectively; 40% were refractory to both an IMID and PI. Twelve (44%) patients had only achieved an MR or less as the best response during the last line of treatment.
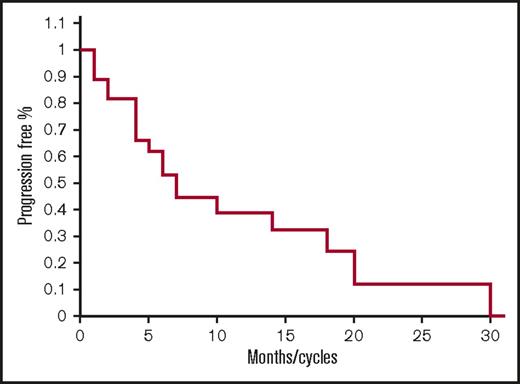
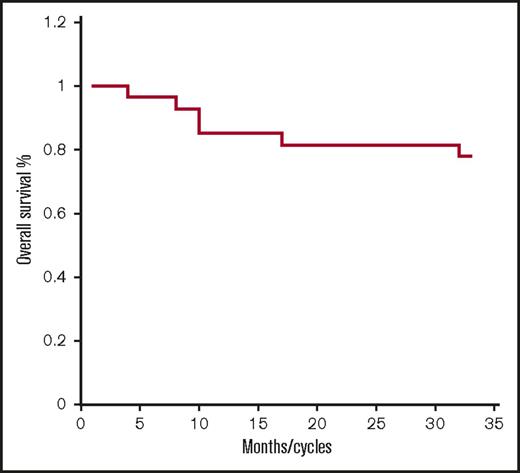
As shown in Table 3, responses include 2 CR, 4 very good partial responses (VGPR), 5 partial responses (PR), 9 MR, 6 stable diseases (SDs), and 1 PD for an ORR of 41%, and clinical benefit rate (CBR; ie, MR or greater) of 74%, and disease control rate (DCR; ie, SD or greater for >2 months) of 96%. The median PFS was 7.1 months (range 1-31) (Figure 1). The median overall survival has not been reached (Figure 3). Twenty-one out of 27 (77%) patients remain alive after 33 months.
Progression-free survival for all patients. Kaplan-Meier PFS curve for all patients treated with FRD.
Progression-free survival for all patients. Kaplan-Meier PFS curve for all patients treated with FRD.
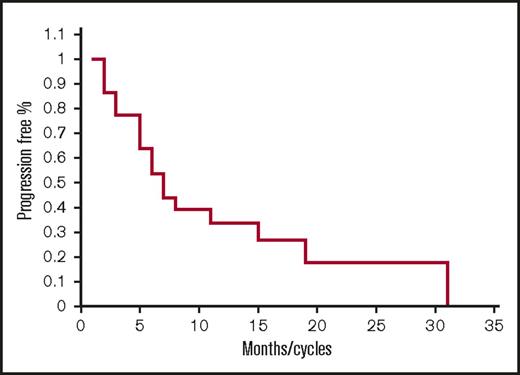
Of the 22 lenalidomide-refractory patients, there was 1 CR, 4 VGPRs, 3 PRs, 7 MRs, 6 SDs, and 1 PD for a 36% ORR, 68% CBR, and 95% DCR with a median PFS of 6.5 months (Figure 2) (range 1-31). Of note, responses were seen even in patients who were previously refractory to lenalidomide 25 mg with 1 CR, 2 VGPRs, 1 PR, and 1 MR and also pomalidomide-refractory patients with 1, CR, 1 VPGR, 1 PR, and 3 MRs (Table 4).
An additional 5 patients did not complete cycle 1 and therefore were inevaluable for efficacy but were considered evaluable for safety. Of these 5 patients, 1 had rapidly PD, 1 had a mechanical fall unrelated to study therapy, and 1 had deterioration in performance status in the setting of pneumonia and then developed a pulmonary embolism. Of the remaining 2, 1 had grade 3 QTc prolongation and the other had grade 4 thrombocytopenia.
Including all 32 patients, grade 3/4 toxicities (regardless of drug attribution) were primarily hematologic, with neutropenia (59%), thrombocytopenia (31%), and anemia (5%), respectively, but were generally not associated with sequelae of infections or bleeding and manageable with granulocyte colony-stimulating factor and/or dose reductions. Grade 3/4 nonhematologic adverse events included infections in 5 (1 while neutropenic), 3 diarrhea (transient), 4 fatigue, and 1 pulmonary embolus, and 1 patient each with neck pain, QTc prolongation, and weight loss (Figure 2).
Progression-free survival for lenalidomide-refractory patients. Kaplan-Meier PFS curve for lenalidomide-refractory patients treated with FRD (n = 22).
Progression-free survival for lenalidomide-refractory patients. Kaplan-Meier PFS curve for lenalidomide-refractory patients treated with FRD (n = 22).
The numbers of patients requiring dose reductions of lenalidomide/panobinostat were 4/5 for neutropenia, 6/2 for thrombocytopenia, 1/1 for febrile neutropenia, 5/2 for fatigue, and 1 panobinostat for asymptomatic T-wave inversions, respectively. Three patients required dose reduction of dexamethasone: 2 due to insomnia and 1 due to mood alteration. No doses of any of the study medications were held or reduced for GI toxicities.
Overall survival. Kaplan-Meier overall survival curve for all patients treated with FRD (n = 27).
Overall survival. Kaplan-Meier overall survival curve for all patients treated with FRD (n = 27).
Correlative studies
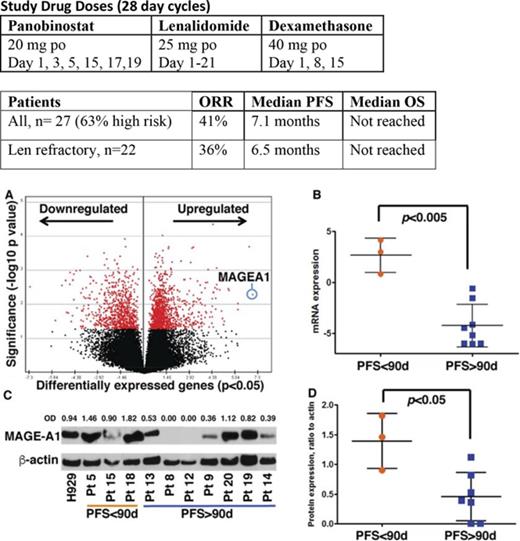
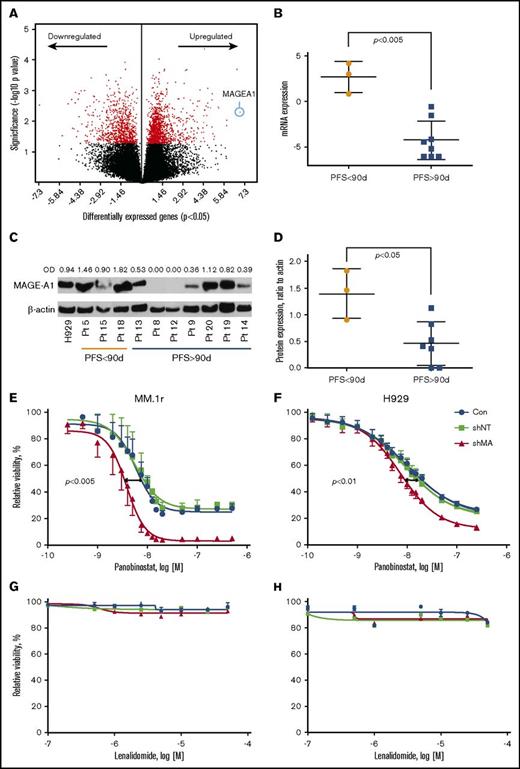
Gene expression profiling by RNA sequencing was performed on CD138+ myeloma cells from BM aspirates acquired at screening on all subjects. Supervised clustering analysis of gene expression based on PFS <90 days (short, n = 3 subjects) vs PFS > 90 days (long, n = 8) revealed a set of 1989 genes that were differentially expressed in the short PFS group (P < .05) (Figure 4A; supplemental Table 1). The top canonical pathways enriched by these genes included Cell Cycle Control of Chromosomal Replication and G2/M DNA Damage Checkpoint Regulation (supplemental Table 2). Top molecular functions of differentially expressed genes were Cell Cycle, DNA Replication, Recombination, and Repair, Cell Death, and Survival (supplemental Table 3). DNA damage-related genes, ATR, CCNB1, CCNB2, CDK4, CDK6, and PLK1, and survival-promoting genes, MAGE-A1 and BRCA1, were seen upregulated in the tumor BM samples from the patients with worse PFS (supplemental Table 1). MAGEA1, a type I Melanoma Antigen Gene, was the most overexpressed gene in the short PFS group (Figure 4A-B; supplemental Table 1), and other members of this family, notably MAGE-A3 and C2, were among the top scoring overexpressed genes (supplemental Figure 1A-B). Whole cell extracts from the same specimens were probed by western blot for MAGE-A1 protein expression, which was also correlated with short PFS (Figure 4C-D). The classes of prior therapy and refractory vs sensitivity status (eg, to lenalidomide) did not appear to have any impact on MAGE expression; however, the numbers in these subgroups were too small to draw statistically significant inference. Importantly, high MAGEA1 expression did not correlate with patients being refractory to lenalidomide or pomalidomide upon study entry (Table 4). These results suggested the hypothesis that MAGE-A may promote resistance to FRD chemotherapy and short PFS.
Correlation of MAGE-A1 expression with PFS in patients treated with FRD and knockdown of MAGE-A in HMCLs. MAGE-A1 expression is correlated with resistance to FRD chemotherapy. (A) Analysis of gene expression profiling data by RNASeq from CD138+ myeloma cells obtained at screening based on PFS < 90 days (short PFS) vs PFS > 90 days (long PFS) enriched a set of 1898 differentially expressed genes (down- or upregulated relative to comparator). MAGEA1 was the most highly upregulated gene associated with short PFS (blue circle). (B) Quantitative analysis of MAGEA1 transcript abundance in short vs long PFS subjects demonstrated significantly higher expression associated with short PFS (P < .005). (C) Western blot for MAGE-A1 protein in lysates from screening specimens. OD, relative optical density = OD (MAGE-A1)/OD β-actin load control. (D) Quantitative analysis of MAGE-A1 relative OD in short vs long PFS subjects demonstrated significantly higher protein expression associated with short PFS (P < .05). (E-H) MM.1r (E,G) or H929 (F,H) HMCL were treated with MAGE-A shRNA lenti or controls for 24 hours (MM.1r) or 48 hours (H929) and then incubated with increasing concentrations of panobinostat (E-F) or lenalidomide (G-H). Cell viability was assessed 24 hours later by Cell TiterGlo assay (ProMega). Knockdown of MAGE-A was correlated with a significant increase in sensitivity to panobinostat-induced cell death. MM.1r (E), 50% inhibitory concentration (IC50) of shMA group = 3.8 nM vs IC50 of shNT group 5.6 nM, P < .005. H929 (F), IC50 (shMA) = 7.4 nM vs IC50 (shNT) = 9.2 nM, P < .01. Error bars indicate standard error of the mean; data pooled from 3 experiments. Con, control; mRNA, messenger RNA.
Correlation of MAGE-A1 expression with PFS in patients treated with FRD and knockdown of MAGE-A in HMCLs. MAGE-A1 expression is correlated with resistance to FRD chemotherapy. (A) Analysis of gene expression profiling data by RNASeq from CD138+ myeloma cells obtained at screening based on PFS < 90 days (short PFS) vs PFS > 90 days (long PFS) enriched a set of 1898 differentially expressed genes (down- or upregulated relative to comparator). MAGEA1 was the most highly upregulated gene associated with short PFS (blue circle). (B) Quantitative analysis of MAGEA1 transcript abundance in short vs long PFS subjects demonstrated significantly higher expression associated with short PFS (P < .005). (C) Western blot for MAGE-A1 protein in lysates from screening specimens. OD, relative optical density = OD (MAGE-A1)/OD β-actin load control. (D) Quantitative analysis of MAGE-A1 relative OD in short vs long PFS subjects demonstrated significantly higher protein expression associated with short PFS (P < .05). (E-H) MM.1r (E,G) or H929 (F,H) HMCL were treated with MAGE-A shRNA lenti or controls for 24 hours (MM.1r) or 48 hours (H929) and then incubated with increasing concentrations of panobinostat (E-F) or lenalidomide (G-H). Cell viability was assessed 24 hours later by Cell TiterGlo assay (ProMega). Knockdown of MAGE-A was correlated with a significant increase in sensitivity to panobinostat-induced cell death. MM.1r (E), 50% inhibitory concentration (IC50) of shMA group = 3.8 nM vs IC50 of shNT group 5.6 nM, P < .005. H929 (F), IC50 (shMA) = 7.4 nM vs IC50 (shNT) = 9.2 nM, P < .01. Error bars indicate standard error of the mean; data pooled from 3 experiments. Con, control; mRNA, messenger RNA.
To investigate the impact of type I MAGE expression in resistance to FRD chemotherapy, we transduced HMCLs MM.1r and H929 with lentiviral shRNA constructs that silence MAGE-A expression, including A1, A3/6, and A4,10 or with a nontarget shRNA lenti construct. Knockdown of MAGE-A proteins was confirmed by western blot (supplemental Figure 1C). Lenti-transduced cells or untreated controls were incubated with increasing concentrations of panobinostat or lenalidomide, and cell viability was assessed after 24 hours of incubation with drug. As shown in Figure 4E-F, knockdown of MAGE-A significantly increased sensitivity to panobinostat-induced cell death. In contrast, MM.1r and H929 HMCL did not demonstrate loss of viability at any of the tested concentrations of lenalidomide, and this resistance was not affected by knockdown of MAGE-A (Figure 4G-H). These results strongly suggest that in the context of these in vitro studies, MAGE-A plays a role in resistance to chemotherapy-induced cell death that is relatively specific to panobinostat.
To determine whether expression of the IMID target Cereblon (CRBN) could predict response to the therapy in MM patients, we first compared CRBN protein levels in MM patient samples collected prior to the treatment. In addition, we evaluated the expression level of the Ikaros and Aiolos transcription factors, known substrates of CRBN. We found that the level of CRBN at baseline as well as Ikaros and Aiolos and the ratio of CRBN:Ikaros or CRBN:Aiolos were not associated with PFS in patients on panobinostat lenalidomide dexamethasone therapy (supplemental Figure 2).
Discussion
In relapsed/refractory MM patients, the completely oral FRD regimen demonstrates encouraging ORR and PFS in lenalidomide and pomalidomide–refractory patients with high-risk molecular findings, indicating the essential role of panobinostat in attaining responses. These results suggest that panobinostat modulates expression of genes to restore sensitivity to lenalidomide. In notable contrast to PANORAMA 1 results, this completely oral regimen was well tolerated with no grade 3/4 GI toxicities and primarily expected hematologic toxicities.
Our correlative analysis revealed a novel association between short PFS and expression of MAGE family genes, particularly MAGE-A1, and demonstrated that loss of MAGE-A expression sensitized myeloma cells to panobinostat-induced cell death. MAGE proteins partner with RING domain proteins such as Kap1 to form ubiquitin ligase complexes,11 and they appear to have a range of functions that may affect transcriptional regulation, cell cycle control, and survival in laboratory models of cancer.10-12 MAGE-A proteins are commonly detected in MM, and their expression correlates with progression of disease after chemotherapy.10,13 They inhibit p53-dependent and -independent mechanisms of apoptosis in MM cells.10 Interestingly, MAGE-A1 interacts with the adapter protein Ski Interacting Protein to recruit HDAC1 to chromatin and repress transcription.14 MAGE-A2/p53 complexes recruit HDAC3 to p53 transcription sites, and this activity is associated with resistance to etoposide-induced apoptosis in melanoma cell lines.15 MAGE may protect against panobinostat through direct interactions with HDACs that prevent access to target sites through allosteric inhibition, or through posttranslational modifications, such as ubiquitinylation, that render the HDAC insensitive to panobinostat. In contrast, knockdown of MAGE-A did not appear to affect resistance to lenalidomide in the tested HMCL. Because MAGEA expression also did not appear to correlate with refractoriness to immunomodulatory agents, these results suggest that the functional mechanism of resistance was relatively specific to panobinostat. We did not examine MAGE-A activity in relation to dexamethasone in these correlative experiments. This was not done in part due to the report that MM.1r are resistant to dexamethasone due to transcriptional repression of the glucocorticoid receptor,16 and therefore unlikely to demonstrate any changes in sensitivity regardless of MAGE status. These results support MAGE-A expression as a functional biomarker of resistance to panobinostat. These findings illustrate the significant potential for GEP and biochemical analysis to reveal functional biomarkers of resistance or response in the setting of a clinical trial.
Recently, the E3-ubiquitin ligase CRBN has been identified to be a biomarker for Len function.17 In addition, the Ikaros and Aiolos transcription factors have recently been identified as substrates of CRBN18,19 and may be proximal biomarkers for its clinical activity. Analysis of protein expression indicates that the clinical response to FRD therapy is independent of CRBN and IMIDs–related markers Ikaros and Aiolos, suggesting that panobinostat has an independent contribution to duration of response to FRD.
The addition of only 6 tablets of panobinostat to a standard 28-day cycle lenalidomide and dexamethasone regimen resulted in a completely oral, well-tolerated, and efficacious regimen even in lenalidomide and pomalidomide–refractory patients. FRD represents a novel use for the US Food and Drug Administration–approved agent panobinostat in relapsed refractory MM and is worthy of further investigation.
The full-text version of this article contains a data supplement.
Acknowledgment
This work was funded and supported by Novartis Pharmaceuticals.
Authorship
Contribution: A.C. provided investigation, conceptualization, methodology, analysis, resources, original draft, supervision review, and edits of the manuscript; H.J.C. provided analysis, investigation, resources, review, and edits for the correlative studies; V.L., A.L., D.P., A.H.-C.M., K.T., and J.F. provided data analysis and creation of tables and figures for the correlative studies; A.D., L.L., K.Z., and E.C. provided formal analysis of the data, provision of study materials, research data management, and creation of tables and figures, review and edits of the manuscript, and preparation of the data presentation; G.M., E.F., D.C., D.V., and N.S. provided research and investigation process, resources, review and edits of the manuscript, and visualization of the project; S.J. and S.P. provided conceptualization, methodology, research and investigation process, review and edits of the manuscript, supervision, and funding acquisition of the project; and all authors provided final approval of the published version.
Conflict-of-interest disclosure: The following authors have served as paid consultants for the identified companies: A.C. for Novartis and Celgene; S.J. for Novartis and Celgene; and D.C. and D.V. for Celgene speakers bureau. The remaining authors declare no competing financial interests.
Correspondence: Ajai Chari, Mount Sinai Hospital, Icahn School of Medicine, 1 Gustave L. Levy Pl, Box 1185, New York, NY 10029; e-mail: ajai.chari@mssm.edu.