Abstract
We report the transient response of a patient with theETV6-ABL fusion gene to imatinib mesylate (STI571). A 38-year-old man was referred with an erroneous diagnosis of Philadelphia-positive chronic myeloid leukemia in blastic transformation for treatment with the ABL tyrosine kinase inhibitor, STI571. Further investigation indicated that the patient in fact had acute myeloid leukemia; no evidence of the Philadelphia translocation or BCR-ABL was found using fluorescence in situ hybridization (FISH) or reverse transcription–polymerase chain reaction. Detailed FISH analysis identified a cryptic t(9;12) translocation, and molecular studies confirmed the presence of the ETV6-ABL fusion transcript. Because the patient was gravely ill at presentation, treatment was commenced immediately with STI571 monotherapy, resulting in considerable initial improvement. However within 10 days the patient's condition again deteriorated, and he required conventional chemotherapy. This case has implications for the design of future studies using STI571 in leukemias involving ABL-encoded fusion proteins other than BCR-ABL.
Introduction
ETV6-ABL (also known asTEL-ABL) is a rare translocation previously reported in only 5 patients—2 with acute lymphoblastic leukemia,1,2 one with acute myeloid leukemia,3 and 2 withBCR-ABL–negative chronic myeloid leukemia (CML).2,4 This abnormality is rarely visible cytogenetically, and it is also rare to detect occult ETV6-ABL translocations using fluorescence in situ hybridization (FISH) and reverse transcription–polymerase chain reaction (RT-PCR).1,5 In vitro studies have shown thatETV6 dysregulates the tyrosine kinase activity ofABL by inducing oligomerization of ETV6-ABLprotein molecules through its pointed amino-terminal domain, in a process similar to that involving the BCR dimerization domain.3,6 At least some of the signal transduction pathways activated by the ETV6-ABL protein are similar to those activated by p210BCR-ABL.7,8 The functional similarity of the BCR-ABL and ETV6-ABLoncoproteins is further supported by the fact that STI571 can inhibit the growth of ETV6-ABL-positive cells in vitro.9
Imatinib mesylate (STI571, Gleevec) is a 2′ phenylaminopyrimidine compound that inhibits the tyrosine kinase activity of BCR-ABL and ABL and of thePDGF and KIT receptor tyrosine kinases.10,11 In vitro studies confirmed its ability to inhibit the growth of CML cells,12,13 and the results of phase 1 and 2 studies show STI571 to be a highly effective agent in Philadelphia (Ph)-positive leukemias with minimal toxicity.14
Study design
In February 2000, a 38-year-old man was referred with, in retrospect, an erroneous diagnosis of Philadelphia-positive CML in blast transformation for treatment with STI571. His local hematologist examined him in May 1999 for flulike illness and generalized lymphadenopathy. He had leukocytosis with 10% blasts, and his white blood cell (WBC) count was initially controlled with intermittent hydroxyurea. At the time of referral to our center, the patient was gravely ill with massive generalized lymphadenopathy, 3-cm splenomegaly, and the following laboratory values: WBC, 77.6 × 109/L with 92% blasts; hemoglobin, 103 g/L; platelets, 90 × 109/L. Screening investigations, including bone marrow cytogenetic analysis, were performed. Bone marrow was hypercellular, and 57% of the cells had the morphologic appearance of myeloblasts; immunophenotyping was consistent with acute myeloid leukemia or myeloid blast transformation of CML.
Because the patient was seriously ill, therapy with STI571 at 600 mg daily by mouth was started immediately. The patient made a considerable clinical recovery over the next 48 hours, with reduction in lymphadenopathy, improvement in the WBC, and no tumor lysis syndrome. At this stage the locally analyzed cytogenetic data and the historic data from the referring hospital became available. Successive cytogenetic analyses showed a 46, XY karyotype in May 1999, a 49, XY,+11,+12,+19 karyotype in January 2000, and a 49, XY, add(9)(q34),+11,+12,+19, der(22)t(1;22)(q21;q11) karyotype in February 2000. The Philadelphia translocation was not detected, nor was the BCR-ABL mRNA fusion transcript detected by RT-PCR. FISH studies were then performed to elucidate the nature of the cytogenetic abnormalities.
Cytogenetic data suggested that the ABL gene might be involved, though not in the context of BCR-ABL. Additional molecular studies were therefore conducted. Total RNA extraction and cDNA synthesis with random hexamers were performed as previously described.15 RT-PCR amplifications of ABL,ETV6, and ETV6-ABL transcripts were primed with the following oligonucleotides: NIb+ (ABL sense), 5′-CACGAATTCTGGAAAGGGGTACCTATTA; Jc- (ABLantisense), 5′-GGAGTGTTTCTCCAGACTGTTG; CA3-(ABLantisense), 5′-TGTTGACTGGCGTGATGTAGTTGCTTGG; TelE+(ETV6 sense), 5′- GATGACGTAGCCCAGTGGCTC; TelA+(ETV6 sense), 5′-TCCGTGGATTTCAAACAGTCCAG; and TelC-(ETV6 antisense), 5′-GTCAGCTCGGTCAGGCAACCCTA. Thermocycling conditions were 35 cycles of denaturation at 96°C for 30 seconds, annealing at 60°C for 50 seconds, and extension at 72°C for 1 minute, followed by a final 10-minute extension at 72°C. Rapid amplification of cDNA ends (RACE-PCR) was performed on cDNA primed with the ABL Jc- oligonucleotide using a 5′RACE kit (Gibco-BRL, Paisley, United Kingdom), according to the manufacturer's protocol. The ABL nested 3′ primersCA3- and A2e- (5′-CGACAAGCTTTAGTTATGCTTAGAGTGTTA) were used for the first- and second-step RACE amplifications, respectively. Final PCR products were cloned into the TOPO-TA vector (Invitrogen, NV Leek, The Netherlands), and mini-preparations from selected clones were made. The inserts were then PCR amplified from the plasmids with M13 standard primers, resolved on an agarose gel, and blotted onto a nylon membrane. Hybridization with an ABL oligonucleotide probe (A2+,5′-TTCAGCGGCCAGTAGCATCTGACTT) internal to the second-step primer was used to verify positive clones. Sequencing was performed by the chain-termination method using fluorescence primers.
Results and discussion
In view of the 9q34 and 22q11 breakpoints, we used FISH and Vysis BCR-ABL probes to investigate the possibility of a variant Ph rearrangement. This showed no fusion signal—that is, no evidence of cryptic Ph rearrangement—but one ABL signal was split between 9q34 and the short (p) arms of 2 of the 3 copies of chromosome 12 (Figure 1A). Initial hybridization with an ETV6 probe suggested that it was not involved. When probing for BCR-ABL andETV6-AML was carried out in a combined hybridization procedure, 2 red–green fusion signals were observed on the short arms of 2 of the 3 copies of chromosome 12, confirming a cryptic translocation between 9q34 and 12p1, which would be consistent with fusion of the ETV6 andABL genes. The revised karyotype was therefore 49, XY,+11, t(9;12)(q34;p1?), +der(12)t(9;12), +19, der(22)t(1;22)(q21;q11).
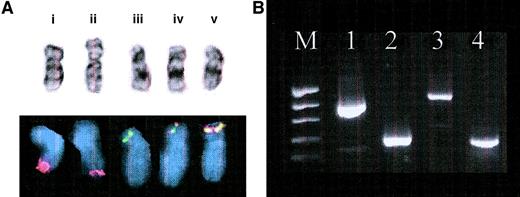
Confirmation of ETVG-ABL t(9;12)translocation.
(A) Cytogenetic and FISH results. (i) Normal chromosome 9 with normal ABL signal (red), (ii) add(9)(q34) with diminished ABL FISH signal, (iii) normal chromosome (12) with normal ETV6 signal (green), and (iv, v) 2 copies of der(12) showing yellow fusion signal in simultaneousETV6-ABL FISH. (B) Agarose gel electrophoresis of RT-PCR products. Lanes: M, molecular weight marker (pEMBL18 digested with TaqI); 1, ETV6 (TelE+ ↔ TelC-); 2,ABL (NIb+ ↔ Jc-); 3,ETV6-ABL (TelE+ ↔ Jc-); 4,ETV6-ABL (TelA+ ↔ CA3-). The larger and stronger band on lane 3 corresponds to the type BETV6-ABL transcript, whereas the smaller and weaker band represents the type A transcript. The latter can be more clearly detected with the TelA+ primer, which is closer to theETV6 exon 4–ABL exon 2 junction, as shown on lane 4.
Confirmation of ETVG-ABL t(9;12)translocation.
(A) Cytogenetic and FISH results. (i) Normal chromosome 9 with normal ABL signal (red), (ii) add(9)(q34) with diminished ABL FISH signal, (iii) normal chromosome (12) with normal ETV6 signal (green), and (iv, v) 2 copies of der(12) showing yellow fusion signal in simultaneousETV6-ABL FISH. (B) Agarose gel electrophoresis of RT-PCR products. Lanes: M, molecular weight marker (pEMBL18 digested with TaqI); 1, ETV6 (TelE+ ↔ TelC-); 2,ABL (NIb+ ↔ Jc-); 3,ETV6-ABL (TelE+ ↔ Jc-); 4,ETV6-ABL (TelA+ ↔ CA3-). The larger and stronger band on lane 3 corresponds to the type BETV6-ABL transcript, whereas the smaller and weaker band represents the type A transcript. The latter can be more clearly detected with the TelA+ primer, which is closer to theETV6 exon 4–ABL exon 2 junction, as shown on lane 4.
Initial ETV6-ABL RT-PCR analysis of the random hexamer-primed cDNA with primers TelE+ and Jc-was negative. Although a positive ETV6-ABL control was not available, correct amplification of ABL andETV6 transcripts from that cDNA suggested that the absence of ETV6-ABL message was a genuine finding, which was in agreement with the initial FISH results. However, when the 5′RACE-PCR method was used to identify the 5′partner of ABL, it was revealed to be ETV6, as also revealed by the subsequently modified FISH analysis. Repeat of the initial RT-PCR amplifications using the ABL primer-specific cDNA template used for RACE and alternative ETV6 primers confirmed anETV6-ABL fusion. The hybrid gene in this case produced 2 alternative transcripts, with the type B (ETV6 exon 5–ABL exon 2 junction) and type A (ETV6 exon 4–ABL exon 2 junction) structures,6respectively (Figure 1B).
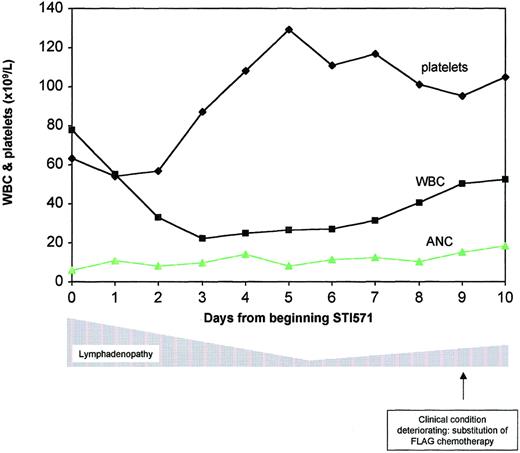
The patient continued to respond to STI571 monotherapy for 10 days (Figure 2), at which point the WBC count began to escalate again; conventional chemotherapy with fludarabine, Ara-C, and G-CSF (FLAG) was instituted; and STI571 therapy was discontinued. He underwent several additional courses of conventional chemotherapy but ultimately died of relapsed disease. Although not by design, our patient appears to be the only known person with the t(9;12) ETV6-ABL translocation to be treated thus far with STI571. There was an impressive but transient response to the drug, suggesting that inhibition of ABL kinase in such patients may be therapeutically useful but not sufficient to induce remission. Although the mechanism of resistance in this case is unknown, the possibility that high-dose STI571 may avert the development of resistance in such leukemias deserves further consideration.16
Clinical progress.
Significant resolution of the patient's lymphadenopathy was seen as the WBC count initially dropped. STI571 was substituted with FLAG chemotherapy because the WBC count failed to respond, and the patient's clinical condition deteriorated.
Clinical progress.
Significant resolution of the patient's lymphadenopathy was seen as the WBC count initially dropped. STI571 was substituted with FLAG chemotherapy because the WBC count failed to respond, and the patient's clinical condition deteriorated.
This case has implications for the design of future studies in leukemias involving the ABL gene, but not necessarily in the context of BCR-ABL. Furthermore, because STI571 is known to also inhibit the kinase activity associated with the KIT andPDGF receptors,10 11 it is possible that inhibition of these pathways as well as, or instead of, inhibition of ABL kinase may be therapeutically important.
We thank Michael Deininger (Leipzig, Germany) for help in tracking down a valuable cDNA sample and Nick Telford (Christie Hospital, Manchester, United Kingdom) for help with retrospective cytogenetic data.
Supported by the Tyneside Leukaemia Research Association (S.G.O.), Novartis Pharma AG (S.G.O.), and Fundação para a Ciencia e Tecnologia (Praxis XXI BD/15769/98).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen G. O'Brien, Department of Haematology, University of Newcastle, Royal Victoria Infirmary, NE1 4LP, United Kingdom; e-mail: s.g.o'brien@ncl.ac.uk

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal