Abstract
Decisions about cell survival or death are central components of adaptive immunity and occur at several levels in immune system development and function. The Bcl-2 family of homologous proteins plays an important role in these decisions in lymphoid cells. Bcl-2, Bcl-xL, and A1 are differentially expressed during B- and T-cell development, and they have shared and distinct roles in regulating cell death. We sought to gain insight into the role of A1 in immune system development and function. A murine A1-a transgene was expressed under the control of the Eμ enhancer, and mice with A1 overexpression in B- and T-cell lineages were derived. Thymocytes and early B cells in Eμ-A1 mice showed extended survival. B-lineage development was altered, with expansion of the pro–B cell subset at the expense of pre–B cells, suggesting an impairment of the pro– to pre–B-cell transition. This early B-cell phenotype resembled Eμ–Bcl-xL mice but did not preferentially rescue cells with completed V(D)J rearrangements of the immunoglobulin heavy chain. In contrast to Eμ–Bcl-2 transgenes, A1 expression in pro–B cells did not rescue pre–B-cell development in SCID mice. These studies indicate that A1 protects lymphocytes from apoptosis in vitro but that it has lineage- and stage-specific effects on lymphoid development. Comparison with the effects of Bcl-2 and Bcl-xL expressed under similar control elements supports the model that antiapoptotic Bcl-2 homologs interact differentially with intracellular pathways affecting development and apoptosis in lymphoid cells.
Introduction
Regulation of cell survival is a critical process in the development and function of multicellular organisms.1,2 The family of proteins sharing homology with Bcl-2 plays key roles in regulating cell fate and includes members that antagonize and others that promote cell death.3,4Available studies suggest a role for the antiapoptotic proteins Bcl-2, Bcl-xL, and A1 in the development, function, or both of the immune system. In mice, Bcl-2 plays a critical role in the survival of mature T and B cells.5-7 Variations in its expression may enhance specific immunity or may limit the extent of antigen responses.4,8 Bcl-xL is developmentally regulated in the thymus and is required for the survival of immature thymocytes, suggesting a potential role in the generation of central tolerance.9-11 In the bone marrow, Bcl-xL is expressed in pre–B cells and is required for the survival of early B-lineage cells.10,12 Bcl-xL is down-regulated at later stages of B-cell development, but it is induced in activated lymphocytes and may play a role in the selection of beneficial clones.12-14Less is known regarding A1, which is inducible as an early-response gene to a variety of stimuli in myeloid, lymphoid, and endothelial cells.15-20 Like Bcl-2 and Bcl-xL, A1 is developmentally regulated in the immune system and is induced on cellular activation, suggesting a role in development and immune responses.18A1 is unusual in that it is encoded by 4 distinct genes in mice, termedA1-a through A1-d.21 Aside from A1-c, which bears a frameshift insertion,A1-a, A1-b, and A1-d are 97% identical, and in neutrophils all 3 isoforms are expressed. Increased apoptosis of neutrophils has been demonstrated in mice with targeted inactivation of the A1-a isoform, but this subtle phenotype likely reflects functional compensation by the other genes.
Although Bcl-2, Bcl-xL, and A1 all have the property of enhancing cell survival, it remains unclear whether the individual functions of these proteins represent the same fundamental activity targeted to particular developmental and physiologic circumstances. Alternatively, these homologs could have important individual activities within the cell, such as regulating different categories of death signals or other types of cellular stimuli. To study the functional properties of A1 in lymphoid cells in vivo, we have constructed transgenic mice that overexpress A1-a under the control of the immunoglobulin intronic enhancer Eμ. This control element has been well characterized in transgenic systems. Moreover, numerous studies have been performed using Bcl-2 and Bcl-xL overexpressed under Eμ control. By comparing the similarities and differences in the phenotypes of Eμ–Bcl-2, Eμ–Bcl-xL, and Eμ-A1 mice, we sought to gain insight into the distinct cellular functions of these homologs.
Materials and methods
Eμ-A1 transgenic mice
Murine cDNA corresponding to positions 1 to 676 of theA1-a gene15 was amplified from mouse spleen RNA by reverse transcription–polymerase chain reaction (RT-PCR), subcloned into the pT7BlueR vector (Novagen, Madison, WI), and verified by DNA sequencing. The A1-a cDNA sequence was recloned into theBamH1 site in the pHSE3′ expression cassette22(Figure 1A). This vector includes the H-2Kb promoter and the Eμ enhancer, and it directs expression to the B- and T-lymphoid lineages.22 The Eμ-A1 transgene construct was liberated from the plasmid vector using a XhoI digest and was introduced into C57BL/6 eggs by pronuclear injection using standard techniques. Founder mice were detected by Southern blot analysis of tail DNA and were bred with C57Bl/6 mice to generate individual mouse lines. Offspring were typed using PCR for wild-type and transgene-specific A1-asequences using the following primers: 5′-GCG ATC ACC AAG AAC CAA TC-3′ (5′ H-2Kb primer); 5′-GCC ATC TTC CCT GGC AGA GC-3′ (5′ A1 primer); and 5′-GCCATCTTCCCTGGCAGAGC-3′ (3′ A1 primer). All mice were housed in a specific pathogen-free facility. Eμ-A1 line 8 mice were bred with SCID mice maintained on a C57Bl/6 background in our facility. Transgene-positive offspring were interbred, and SCID/SCID offspring were identified by screening peripheral blood for T cells using flow cytometry.
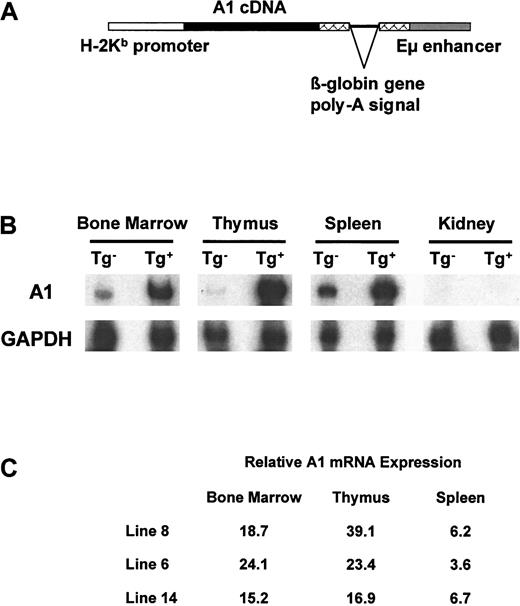
Eμ-A1 transgenic mice.
(A) Eμ-A1 transgene construct, containing full-length A1-acDNA, Eμ enhancer, and H-2Kb promoter. (B) A1 mRNA expression in transgenic mice and wild-type littermates. Ribonuclease protection assays for A1-a and GAPDH control are shown for the indicated tissues for line 8 mice. (C) Overexpression ofA1-a mRNA in 3 Eμ-A1 transgenic mouse lines. Figures represent -fold increases in A1-a mRNA in the indicated tissues over those of wild-type littermates. Results are the mean from 2 animals of each genotype and are normalized to the GAPDH signal.
Eμ-A1 transgenic mice.
(A) Eμ-A1 transgene construct, containing full-length A1-acDNA, Eμ enhancer, and H-2Kb promoter. (B) A1 mRNA expression in transgenic mice and wild-type littermates. Ribonuclease protection assays for A1-a and GAPDH control are shown for the indicated tissues for line 8 mice. (C) Overexpression ofA1-a mRNA in 3 Eμ-A1 transgenic mouse lines. Figures represent -fold increases in A1-a mRNA in the indicated tissues over those of wild-type littermates. Results are the mean from 2 animals of each genotype and are normalized to the GAPDH signal.
To assess transgene expression, total RNA was harvested from thymus, spleen, bone marrow, and kidney using Tri-reagent (Sigma, St Louis, MO), according to the manufacturer's instructions. RNA (1.5 μg per sample) was used in a ribonuclease protection assay (Multi-probe RPA; BD PharMingen, San Diego, CA) as previously described.17 23 The probe used in this assay protected nucleotides 140 to 503 in the A1-a cDNA. Within the range of this A1-a probe, there were 6 mismatches for theA1-b mRNA subtype, 9 mismatches for A1-c, and 4 mismatches for A1-d. As a result, only the A1-asubtype mRNA was expected to yield the longest (364 base pair [bp]) protected fragment. Quantitation of protected bands was performed using densitometric analysis of autoradiograms (National Institutes of Health Image software, version 1.62).
Polymerase chain reaction assays
Semiquantitative PCR assays for DJ and V(D)J rearrangement were performed using purified genomic DNA as described.24Equivalence of sample DNA was assessed in parallel PCR amplifications performed to amplify CD14 and was analyzed on ethidium bromide–stained agarose gels. RT-PCR was performed on RNA extracted from 107 bone marrow cells with the RNA-Easy mini kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Four microliters RNA preparation was treated with 10 U RNase-Free DNase I (Roche Molecular Biochemicals, Mannheim, Germany) in Superscript-II RT buffer (Life Technologies/Invitrogen, Carlsbad, CA) for 10 minutes at 25°C, followed by inactivation at 65°C for 10 minutes. One half of the reaction mixture was immediately reverse transcribed with Superscript-II, according to the manufacturer's standard protocol. The other half was carried through identical reactions, except that the reverse transcriptase was omitted to verify the lack of a signal derived from genomic DNA. For PCR, 1 μL reverse-transcribed reaction mixture was used under conditions shown to be semiquantitative, with primer pairs (30 μM each) specific for β-actin, λ5, RAG-1, and RAG-2, as described by Li et al.25 In similar PCR reactions, the murine cμ primer sequences used were TAA GAA TCT GGT GGC CAT GG (5′ cμ primer) and TTG TTC TCG ATG GTC ACC G (3′ cμ primer) and yielded a single 453-bp product. The Eμ-A1 primers used were the 5′ H-2Kb primer and the 3′ A1 primer detailed above; it yielded a single 250-bp product.
Analysis of lymphoid development
Serum immunoglobulin levels were determined using enzyme-linked immunosorbent assays as described.26 Flow cytometry was performed using antibodies conjugated to fluorescein isothiocyanate, phycoerythrin, biotin, Cychrome, or allophycocyanin obtained from BD PharMingen. Biotin-conjugated antibodies were developed with streptavidin–Cychrome (PharMingen). Cell suspensions were stained as described27 and were analyzed using a FACScalibur instrument equipped with dual lasers (Becton Dickinson, San Jose, CA). Data were analyzed using FlowJo software (Treestar, San Carlos, CA). Cell-sorting experiments were performed on bone marrow cells pooled from 2 to 4 animals and were stained with fluorescein isothiocyanate–anti-IgM, phycoerythrin–anti-CD43, and Cychrome–anti-B220 (BD PharMingen). Using a FACSVantage cell sorter (Becton Dickinson), pre–B (B220+IgM−CD43−) and pro–B (B220+ IgM−CD43+) subsets were sorted. Reanalysis of sorted cells indicated greater than 94% purity. Cell-cycle analysis was performed using simultaneous surface staining with anti-B220 antibodies and DAPI (4,6-diamidino-2-phenylindole) staining for DNA content. Antibody-stained bone marrow cells were resuspended in Tris-buffered saline containing 10 mg/mL DAPI and 0.1% Nonidet-P40 (both from Accurate Chemical, Westbury, NY). Samples were analyzed on a dual-laser LSR instrument (Becton Dickinson) with UV and 488-nm excitation. Data were analyzed using FlowJo software and were gated to exclude doublets. Cycling cells were calculated using the Watson pragmatic algorithm.
Cell survival assays
Thymocyte survival in vitro was assessed after disrupting thymic lobes over nylon mesh and plating cells at a density of 1 × 106/mL in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM L-glutamine, nonessential amino acids, and antibiotics. Cells were cultured in 24-well plates at 37% in a humidified 6% CO2 incubator in medium or in dexamethasone (0.2 or 1 μM) or were exposed to γ-irradiation (2.5 or 10 Gy) and were studied for the ensuing 4 days. Cells were counted using a hemacytometer, and viability was determined by trypan blue exclusion. Bone marrow survival was studied as described.28
Results
Increased A1 mRNA expression in lymphoid tissues of Eμ-A1 transgenic mice
To study the in vivo effects of A1 overexpression on immune system development, we inserted cDNA representing the A1-a isoform into an expression system based on the IgH intronic enhancer Eμ and the H-2Kb promoter22 (Figure 1). Steady-state levels of A1-a mRNA were studied using RNase protection assays in 3 independent transgenic lines. We found that A1-amRNA levels were substantially increased in thymus, bone marrow, and spleen but not in kidney in 3 independent transgenic lines. Expression of transgenic mRNA was subsequently confirmed using PCR to detect A1 transcripts containing sequences from the heterologous control element (see below and Figure 7). The observed expression pattern was consistent with previous studies using this transgene regulatory system.22
Effects of A1 overexpression on immune system development
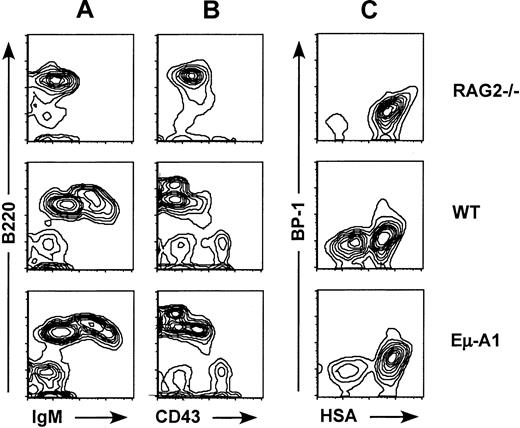
Four independent lines of Eμ-A1 transgenic mice were analyzed for effects on primary and secondary lymphoid tissues using flow cytometry. In all lines, no discernible effects on thymic cellularity or subsets defined by CD4 and CD8 expression were observed (data not shown). B lymphopoiesis was analyzed using 3-color flow cytometry with markers for IgM, B220, and CD43. In all 4 lines, an increase in the percentage of B220+IgM−CD43+pro–B cells was observed (Figure 2, Table 1). This effect was most pronounced in line 8, which was selected for more detailed studies. Analysis of absolute cell numbers contained in 2 femurs from line 8 or control mice indicated that the proportional increase in the ratio of pro– to pre–B cells was the result of an increase in pro–B-cell numbers and a decrease in pre–B-cell numbers. B-cell developmental subsets were further analyzed using 4-color flow cytometry according to the scheme of Li et al25 and Hardy et al29(Figures 2, 3). This analysis showed that the expanded population of B220+IgM−CD43+ pro–B cells was accounted for primarily by an increase in fraction B (heat-stable antigen [HSA+] BP1−), a population that precedes delivery of the pre–B-cell receptor signal (Figure 3). Taken together, these data indicate that the Eμ-A1 transgene led to an expansion of pro–B cells, and they suggest that this was in part caused by impaired progression from the pro–B-cell to the pre–B-cell stage.
Flow cytometric analysis of bone marrow B-lineage cells in Eμ-A1 mice.
Comparison with Rag-2−/− mice and wild-type (WT) littermates. (A, B) Bone marrow cells stained with antibodies to B220, IgM, and CD43 and gated on lymphocytes by scatter characteristics. (C) Four-color analysis of bone marrow cells stained with antibodies to B220, CD43, BP-1, and HSA and gated on B220+CD43+ pro–B cells.
Flow cytometric analysis of bone marrow B-lineage cells in Eμ-A1 mice.
Comparison with Rag-2−/− mice and wild-type (WT) littermates. (A, B) Bone marrow cells stained with antibodies to B220, IgM, and CD43 and gated on lymphocytes by scatter characteristics. (C) Four-color analysis of bone marrow cells stained with antibodies to B220, CD43, BP-1, and HSA and gated on B220+CD43+ pro–B cells.
Expansion of bone marrow pro–B cells in 4 independently derived mouse lines bearing an Eμ-A1 transgene
| Line . | N . | IgM+B . | Pre-B . | Pro-B . | Pre-B/Pro-B . | |||
|---|---|---|---|---|---|---|---|---|
| % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | |||
| Wild-type | 9 | 47 ± 2.0 | 2.0 ± 0.93 | 46 ± 13 | 2.5 ± 0.15 | 6.7 ± 2.4 | 0.41 ± 0.21 | 7.4 |
| 8 | 8 | 41 ± 8 | 1.5 ± 0.61 | 34 ± 10 | 1.3 ± 0.57 | 25 ± 3.4 | 0.94 ± 0.44 | 1.4 |
| 6 | 2 | 39 | ND | 42 | ND | 19 | ND | 2.3 |
| 14 | 2 | 58 | ND | 28 | ND | 10 | ND | 2.8 |
| 95 | 5 | 40 ± 5.3 | ND | 47 ± 7.2 | ND | 9.8 ± 3.1 | ND | 5.5 |
| Line . | N . | IgM+B . | Pre-B . | Pro-B . | Pre-B/Pro-B . | |||
|---|---|---|---|---|---|---|---|---|
| % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | |||
| Wild-type | 9 | 47 ± 2.0 | 2.0 ± 0.93 | 46 ± 13 | 2.5 ± 0.15 | 6.7 ± 2.4 | 0.41 ± 0.21 | 7.4 |
| 8 | 8 | 41 ± 8 | 1.5 ± 0.61 | 34 ± 10 | 1.3 ± 0.57 | 25 ± 3.4 | 0.94 ± 0.44 | 1.4 |
| 6 | 2 | 39 | ND | 42 | ND | 19 | ND | 2.3 |
| 14 | 2 | 58 | ND | 28 | ND | 10 | ND | 2.8 |
| 95 | 5 | 40 ± 5.3 | ND | 47 ± 7.2 | ND | 9.8 ± 3.1 | ND | 5.5 |
Bone marrow cells from mice 4 to 8 weeks of age were analyzed using 3-color flow cytometry for the expression of B220, IgM, and CD43. Populations were expressed as a percentage of B220+ cells, including IgM+ B cells, IgM−CD43−pre–B cells, and IgM-CD43+ pro–B cells. In WT and line 8 mice, absolute cell numbers contained in 2 femurs are also given. Ratios of pre– to pro–B cells are also given. Mean ± SD are given for the indicated number of mice analyzed, except for lines 6 and 14, in which only 2 mice were studied. ND indicates not done.
Analysis of bone marrow B-cell lineage development.
Flow cytometric analysis in Eμ-A1 transgenic mice (shaded bars) and wild-type littermates (open bars) using the convention devised by Hardy et al.29 Flow cytometry data were gated as described “Materials and methods.” Data are presented as the mean (± SD) percentage of the B220+ lymphocyte population for 4 mice of each genotype. Statistically significant differences (P < .05) were found in fraction B (subset of pro–B cells) and fraction D (pre–B cells), signified by asterisks.
Analysis of bone marrow B-cell lineage development.
Flow cytometric analysis in Eμ-A1 transgenic mice (shaded bars) and wild-type littermates (open bars) using the convention devised by Hardy et al.29 Flow cytometry data were gated as described “Materials and methods.” Data are presented as the mean (± SD) percentage of the B220+ lymphocyte population for 4 mice of each genotype. Statistically significant differences (P < .05) were found in fraction B (subset of pro–B cells) and fraction D (pre–B cells), signified by asterisks.
The pro–B-cell stage is characterized by ordered D to J, followed by V to DJ rearrangement of the IgH locus. Successful in-frame IgH rearrangement leads to expression of the μH protein and signals the transition to the pre–B-cell stage.30 We assessed IgH DJ and V(D)J rearrangement in the expanded pro–B-cell population in Eμ-A1 transgenic mice. B220+ bone marrow cells representing pro– and pre–B-cell subsets were purified by cell sorting (Figure 4). Genomic DNA from equivalent cell numbers was extracted and subjected to PCR as previously described24 using a primer downstream of JH3, paired with a degenerate primer complementary to 5′ DH segments or to VH segments. Amplification of polyclonal B-lineage populations gives a 3-part ladder representing rearrangements to JH1, JH2, and JH3. The CD14 gene was amplified from each sample as a control. As expected, the V(D)J signal was lower in pro–B cells than in pre–B cells, indicating that only a fraction of IgH alleles in the former population had completed rearrangement. There were no significant differences in DJ or V(D)J rearrangements when pro–B cells from wild-type and Eμ-A1 transgenic mice were compared, suggesting that rearrangement of the IgH locus was not blocked by the transgene and that pro–B-cell expansion did not represent selective accumulation of cells with completed V(D)J rearrangements of the IgH locus.
IgH locus rearrangements in pre– and pro–B-cell subsets from Eμ-A1 transgenic mice.
DNA from IgM−B220+CD43+ and IgM−B220+CD43− bone marrow populations purified by cell sorting was amplified using PCR for DJ and V(D)J rearrangements under semiquantitative conditions. Southern blots of PCR products were hybridized with internal oligonucleotide probes (see “Materials and methods”). Parallel amplification of CD14 served as a control and was assessed by ethidium bromide staining.
IgH locus rearrangements in pre– and pro–B-cell subsets from Eμ-A1 transgenic mice.
DNA from IgM−B220+CD43+ and IgM−B220+CD43− bone marrow populations purified by cell sorting was amplified using PCR for DJ and V(D)J rearrangements under semiquantitative conditions. Southern blots of PCR products were hybridized with internal oligonucleotide probes (see “Materials and methods”). Parallel amplification of CD14 served as a control and was assessed by ethidium bromide staining.
Peripheral lymphoid populations were analyzed in line 8 mice (Table2). We noted a modest but statistically significant decrease in splenic B cells in transgene-positive mice. Similar analyses performed on the other Eμ-A1 lines showed no definable difference in peripheral B cell number compared with littermate controls (data not shown). Serum immunoglobulin levels were also analyzed in 6- to 8-week-old line 8 mice and showed a correspondingly modest decrease in mean levels of serum IgG1 and IgG2a but not of IgM (Table 3).
Peripheral lymphoid subsets in Eμ-A1 line 8 mice compared with wild-type littermates
| . | B220+IgM+ . | CD4+ . | CD8+ . | |||
|---|---|---|---|---|---|---|
| % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | |
| Wild-type spleen | 56.0 ± 6.9 | 23.0 ± 9.4 | 24.0 ± 7.2 | 9.0 ± 5.6 | 15.0 ± 5.3 | 5.4 ± 2.9 |
| Eμ-A1 spleen | 49.0 ± 7.4* | 15.0 ± 5.1* | 23.0 ± 14 | 6.3 ± 4.3 | 17.0 ± 7.5 | 4.9 ± 3.2 |
| Wild-type lymph node | 25.0 ± 5.0 | 2.2 ± 1.1 | 36.0 ± 13 | 3.0 ± 1.5 | 28.0 ± 8.9 | 2.3 ± 1.2 |
| Eμ-A1 lymph node | 18.0 ± 5.5 | 1.6 ± 0.71 | 38.0 ± 11 | 3.2 ± 1.7 | 33.0 ± 7.1 | 2.6 ± 1.2 |
| . | B220+IgM+ . | CD4+ . | CD8+ . | |||
|---|---|---|---|---|---|---|
| % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | % . | Cells (× 10−6) . | |
| Wild-type spleen | 56.0 ± 6.9 | 23.0 ± 9.4 | 24.0 ± 7.2 | 9.0 ± 5.6 | 15.0 ± 5.3 | 5.4 ± 2.9 |
| Eμ-A1 spleen | 49.0 ± 7.4* | 15.0 ± 5.1* | 23.0 ± 14 | 6.3 ± 4.3 | 17.0 ± 7.5 | 4.9 ± 3.2 |
| Wild-type lymph node | 25.0 ± 5.0 | 2.2 ± 1.1 | 36.0 ± 13 | 3.0 ± 1.5 | 28.0 ± 8.9 | 2.3 ± 1.2 |
| Eμ-A1 lymph node | 18.0 ± 5.5 | 1.6 ± 0.71 | 38.0 ± 11 | 3.2 ± 1.7 | 33.0 ± 7.1 | 2.6 ± 1.2 |
Lymph node and spleen cells from 4- to 8-week-old mice were counted and analyzed for the indicated subsets using flow cytometry. Values indicate the average percentage or absolute numbers of cells for the indicated subset (± SD) for 6 mice analyzed in each group. For splenic B-cell numbers, the difference is statistically significant.
P < .02, compared to wild-type.
Serum immunoglobulin levels in 6- to 8-week-old Eμ-A1 transgenic mice and wild-type littermates
| . | Wild-type (μg/mL) . | Eμ-A1 (μg/mL) . |
|---|---|---|
| IgM | 128 ± 100 | 116 ± 34 |
| IgG1 | 427 ± 417 | 223 ± 216 |
| IgG2a | 114 ± 72 | 75 ± 49 |
| . | Wild-type (μg/mL) . | Eμ-A1 (μg/mL) . |
|---|---|---|
| IgM | 128 ± 100 | 116 ± 34 |
| IgG1 | 427 ± 417 | 223 ± 216 |
| IgG2a | 114 ± 72 | 75 ± 49 |
Mean ± SD for 5 mice tested in each group.
Increased survival of Eμ-A1 transgenic thymocytes and B-cell precursors in vitro
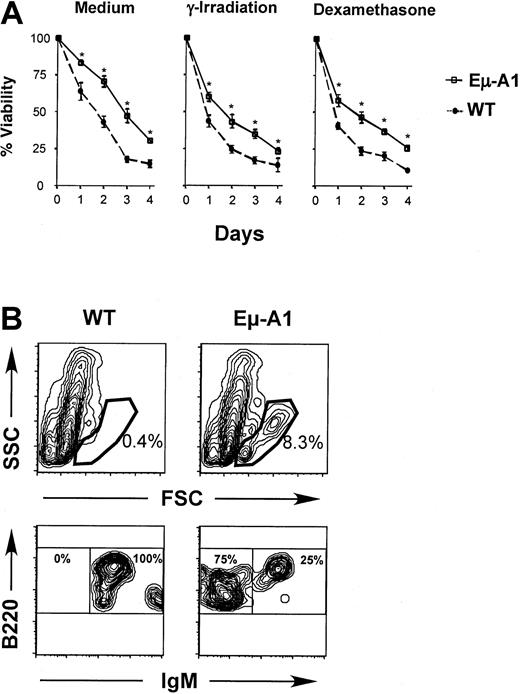
Enforced expression of A1 in cell lines leads to enhanced survival in the absence of growth signals or in response to apoptotic stimuli.31 Similar effects are noted when other antiapoptotic Bcl-2 homologs are overexpressed in lymphoid cells in vivo.32-34 To verify the expression of A1 protein in Eμ-A1 transgenic lymphoid cells, we studied the survival of thymocytes and bone marrow B-lineage cells. Compared with nontransgenic controls, thymocytes from transgenic animals survived better ex vivo in tissue culture medium and were more tolerant toward death-inducing stimuli, including γ-irradiation (2.5 Gy) and 0.2 μM dexamethasone (Figure 5A). Similar protective effects were observed with higher doses of γ-irradiation (10 Gy) and dexamethasone (1 μM) over the same culture period (data not shown). Survival of bone marrow B-lineage cells was studied by in vitro culture, followed by flow cytometric analysis of surviving cells defined by light-scatter characteristics (Figure 5B). In 2 independent experiments, after 11 and 12 days, respectively, substantially more cells survived in the lymphoid gate in transgenic mice. When these events were analyzed, essentially all cells were B220+. In wild-type mice, rare surviving B-lineage cells were almost exclusively IgM+, whereas in Eμ-A1 mice, surface IgM−B-precursor cells and IgM+ B cells survived. Short-term in vitro survival assays were also performed on lymph node and spleen cells in which no significant differences were found between wild-type and Eμ-A1 transgenic mice (data not shown). These data demonstrate that Eμ-A1 transgenic thymocytes and bone marrow B-lineage cells exhibit a survival advantage in vitro, and they suggest that the transgene leads to overexpression of functional A1 protein.
Survival analysis of developing lymphoid cells in Eμ-A1 transgenic mice.
(A) Thymocytes from Eμ-A1 transgenic or wild-type (WT) littermates (3 mice aged 5 weeks in each group) were cultured in vitro with medium alone, after γ-irradiation (2.5 Gy), or in the presence of 0.2 μM dexamethasone. Viability on the indicated days was ascertained by trypan blue exclusion. Representative results from 1 of 2 independent experiments are shown. (B) Survival analysis of B-lineage bone marrow cells. Bone marrow was cultured for 11 days and subjected to flow cytometric analysis for the indicated markers. Analysis is based on 100 000 events. Upper panels show light-scatter parameters on B220+ events and gate used to classify viable lymphoid cells. Lower panels show B220 and IgM expression on gated data. Percentages within each gate are shown. Data are representative of 2 experiments (the second was assayed after 12 days), each containing 2 or 3 mice of each genotype.
Survival analysis of developing lymphoid cells in Eμ-A1 transgenic mice.
(A) Thymocytes from Eμ-A1 transgenic or wild-type (WT) littermates (3 mice aged 5 weeks in each group) were cultured in vitro with medium alone, after γ-irradiation (2.5 Gy), or in the presence of 0.2 μM dexamethasone. Viability on the indicated days was ascertained by trypan blue exclusion. Representative results from 1 of 2 independent experiments are shown. (B) Survival analysis of B-lineage bone marrow cells. Bone marrow was cultured for 11 days and subjected to flow cytometric analysis for the indicated markers. Analysis is based on 100 000 events. Upper panels show light-scatter parameters on B220+ events and gate used to classify viable lymphoid cells. Lower panels show B220 and IgM expression on gated data. Percentages within each gate are shown. Data are representative of 2 experiments (the second was assayed after 12 days), each containing 2 or 3 mice of each genotype.
Failure of the Eμ-A1 transgene to rescue V(D)J recombination and pre–B cells in SCID mice
In SCID mice, mutation of the Prkdc gene, encoding the DNA-dependent protein kinase catalytic subunit, leads to impairment of V(D)J recombination and failure of T- and B-cell development.30 The defect is leaky, and T and B cells accumulate in these mice as they age. Eμ–Bcl-2 transgenes expressed in the SCID background lead to rescue of pre–B-cell development, a phenotype that is dependent on functional IgH rearrangement.35,36 To determine whether the expression of A1 in SCID B-lineage cells had a similar effect on developmental progression, we introduced the Eμ-A1 transgene into the SCID background. The number of B220+ cells recovered from both femurs in Eμ-A1 SCID mice was comparable to that in SCID littermates (Table 4). Surprisingly, we found no effect on the phenotype of early B-lineage cells in the bone marrow of transgenic SCID mice (Figure 6A). We next assayed bone marrow cells from SCID and Eμ-A1 SCID mice for DJ and V(D)J rearrangements (Figure 6B). Faint DJ signals were seen more strongly in Eμ-A1 SCID than in SCID bone marrow, suggesting that there might have been a small difference in the number of cells undergoing the first step of IgH rearrangement in the presence of the Eμ-A1 transgene. However, no V(D)J rearrangements were detected in either Eμ-A1 SCID or SCID mice. These findings indicate that A1 overexpression does not lead to successful V(D)J rearrangement in SCID pro–B cells or to rescue of pre–B cells as seen in Eμ-Bcl-2 SCID mice. Bcl-2 expression in either the SCID or the Rag-2−/−background leads to the accumulation of B220+ cells in the spleen.35 36 SCID mice expressing the A1 transgene had no such accumulation of B-lineage cells (Table4).
Expression of Eμ-A1 transgene in the SCID background
| . | B220+ cells (× 10−3) . | |
|---|---|---|
| Bone marrow . | Spleen . | |
| SCID | 570 ± 32 | 940 ± 940 |
| Eμ-A1 SCID | 510 ± 30 | 570 ± 210 |
| . | B220+ cells (× 10−3) . | |
|---|---|---|
| Bone marrow . | Spleen . | |
| SCID | 570 ± 32 | 940 ± 940 |
| Eμ-A1 SCID | 510 ± 30 | 570 ± 210 |
Bone marrow and spleen cells were analyzed for B220 expression using flow cytometry. Mean absolute cell numbers (± SD) are given for 2 femurs or whole spleen from 7 mice of each genotype. Differences between SCID and Eμ-A1 SCID groups were not statistically significant.
B-lineage development in Eμ-A1 SCID mice.
(A) Flow cytometric analysis of bone marrow of SCID mice with and without the Eμ-A1 transgene. B-lineage cells were IgM−and CD43+ and were indistinguishable from nontransgenic SCID littermates. Results are representative of 7 mice analyzed for each genotype. (B) Analysis of IgHI DJ and V(D)J rearrangements in SCID and Eμ-A1 SCID bone marrow using PCR. A faint DJ rearrangement signal is seen in the Eμ-A1 SCID samples, but no V(D)J rearrangements were detected. Wild-type bone marrow and kidney are shown for comparison. The V(D)J signal in the kidney sample likely reflects low levels of lymphoid cells in the organ.
B-lineage development in Eμ-A1 SCID mice.
(A) Flow cytometric analysis of bone marrow of SCID mice with and without the Eμ-A1 transgene. B-lineage cells were IgM−and CD43+ and were indistinguishable from nontransgenic SCID littermates. Results are representative of 7 mice analyzed for each genotype. (B) Analysis of IgHI DJ and V(D)J rearrangements in SCID and Eμ-A1 SCID bone marrow using PCR. A faint DJ rearrangement signal is seen in the Eμ-A1 SCID samples, but no V(D)J rearrangements were detected. Wild-type bone marrow and kidney are shown for comparison. The V(D)J signal in the kidney sample likely reflects low levels of lymphoid cells in the organ.
To understand potential reasons for the developmental delay of pro–B cells in Eμ-A1 transgenic mice, pro–B cells arrested at this stage by the SCID defect were examined. Cells at this developmental stage express Rag-1 and Rag-2 endonuclease genes, transcribe the unrearranged IgH locus, and express pre–B-cell receptor signaling components. After successful IgH rearrangement and expression of Cμ protein, pre–B-cell receptor signals lead to proliferation and phenotypic changes that characterize the pre–B-cell compartment. We used semiquantitative RT-PCR to characterize the levels of mRNA for Rag-1, Rag-2, Cμ sterile transcripts, and λ5 in bone marrow from Eμ-A1 SCID mice (Figure 7A). Cells at this stage were confirmed to express Eμ-A1 transgene mRNA. We identified no effect of A1 overexpression on the level of these early B-cell transcripts, suggesting that the pro–B-cell phenotype in Eμ-A1 transgenic mice was not caused by defective expression of these components. Bcl-2 expression has been shown to affect cell-cycle entry, and early B cells in Eμ-Bcl-2 mice have reduced proliferation rates.37 38 Because defective cell-cycle progression could affect the pro– to pre–B-cell transition, we analyzed pro–B-cell proliferation in vivo in Eμ-A1 SCID pro–B cells (Figure 7B). We identified no effect of the transgene on the cell cycle profile in this compartment, suggesting that A1 overexpression did not have an overall effect on cell-cycle entry in vivo.
Analysis of bone marrow pro–B cells in SCID and Eμ-A1 SCID mice.
(A) Expression of mRNA for genes required for pre–B-cell development in 6-week-old mice. Bone marrow RNA was amplified using RT-PCR under semiquantitative conditions. Actin mRNA was amplified in parallel as a loading control. Eμ-A1 mRNA transcript is specific for the transgene. (B) Cell-cycle analysis of pro–B cells. Analysis of DNA content by DAPI staining was gated on B220+ cells. Percentages of cells in S and G2+M phases of the cell cycle were determined using FlowJo software.
Analysis of bone marrow pro–B cells in SCID and Eμ-A1 SCID mice.
(A) Expression of mRNA for genes required for pre–B-cell development in 6-week-old mice. Bone marrow RNA was amplified using RT-PCR under semiquantitative conditions. Actin mRNA was amplified in parallel as a loading control. Eμ-A1 mRNA transcript is specific for the transgene. (B) Cell-cycle analysis of pro–B cells. Analysis of DNA content by DAPI staining was gated on B220+ cells. Percentages of cells in S and G2+M phases of the cell cycle were determined using FlowJo software.
Irradiation of newborn SCID mice leads to the partial rescue of thymocyte development, with an increase in thymic cellularity and appearance of CD4+CD8+ immature thymocytes that bear functional T-cell receptor (TCR)-β rearrangements.39 Because A1, like other antiapoptotic Bcl-2 homologs, partially protects against apoptosis induced by DNA damage, we assessed whether the Eμ-A1 transgene might enhance the ability of radiation to rescue T- or B-cell development in the SCID background (Figure 8, Table 5). Progeny of Eμ-A1 SCID × SCID mouse crosses were irradiated within 3 days of birth. After 12 to 22 days, the thymus and bone marrow were examined for markers of T- and B-cell development. We found that radiation-induced rescue of SCID thymocytes was less efficient than previously reported for SCID mice of a distinct genetic background,39 requiring a higher dose and greater observation time to observe robust rescue of double-positive thymocyte development. The degree of rescue varied from individual to individual; however, no overall differences were observed between Eμ-A1 SCID and SCID littermates. We found no evidence for rescue of pre–B- or B-cell development in SCID mice at any dose or time point examined, and the results were identical in Eμ-A1 SCID littermates.
Analysis of B- and T-lymphoid development after irradiation of newborn SCID mice. Mice were analyzed 22 days after irradiation with 150 cGy shortly after birth.
(A) Flow cytometric analysis of bone marrow with markers indicated. No rescue of pre–B-cell development was observed. (B) Rescue of CD4+CD8+ thymocytes, in comparison with unirradiated SCID control. Plots show CD8+ cells (which also expressed CD4) in comparison with CD25+ prothymocytes, which were negative for both CD4 and CD8.
Analysis of B- and T-lymphoid development after irradiation of newborn SCID mice. Mice were analyzed 22 days after irradiation with 150 cGy shortly after birth.
(A) Flow cytometric analysis of bone marrow with markers indicated. No rescue of pre–B-cell development was observed. (B) Rescue of CD4+CD8+ thymocytes, in comparison with unirradiated SCID control. Plots show CD8+ cells (which also expressed CD4) in comparison with CD25+ prothymocytes, which were negative for both CD4 and CD8.
Rescue of CD4+CD8+ thymocytes by radiation in SCID and Eμ-A1 SCID mice
| Group . | Genotype . | Double positive, % . |
|---|---|---|
| Experiment 1 (100 cGy, 16 d) | SCID | 80 |
| SCID | 0.67 | |
| SCID | 0.78 | |
| Eμ-A1 SCID | 1.7 | |
| Eμ-A1 SCID | 9.6 | |
| Eμ-A1 SCID | 2.1 | |
| Experiment 2 (150 cGy, 12 d) | SCID | 1.3 |
| SCID | 19 | |
| SCID | 70 | |
| SCID | 91 | |
| Eμ-A1 SCID | 66 | |
| Experiment 3 (150 cGy, 22 d) | SCID | 84 |
| SCID | 54 | |
| Eμ-A1 SCID | 89 | |
| Eμ-A1 SCID | 12 | |
| Control | SCID | 0.77 ± 0.43 |
| Eμ-A1 SCID | 0.89 ± 0.51 |
| Group . | Genotype . | Double positive, % . |
|---|---|---|
| Experiment 1 (100 cGy, 16 d) | SCID | 80 |
| SCID | 0.67 | |
| SCID | 0.78 | |
| Eμ-A1 SCID | 1.7 | |
| Eμ-A1 SCID | 9.6 | |
| Eμ-A1 SCID | 2.1 | |
| Experiment 2 (150 cGy, 12 d) | SCID | 1.3 |
| SCID | 19 | |
| SCID | 70 | |
| SCID | 91 | |
| Eμ-A1 SCID | 66 | |
| Experiment 3 (150 cGy, 22 d) | SCID | 84 |
| SCID | 54 | |
| Eμ-A1 SCID | 89 | |
| Eμ-A1 SCID | 12 | |
| Control | SCID | 0.77 ± 0.43 |
| Eμ-A1 SCID | 0.89 ± 0.51 |
Offspring of Eμ-A1 SCID × SCID crosses were irradiated at birth with 100 or 150 cGy. At the indicated interval, thymus and bone marrow were analyzed for lymphoid development. Rescue of double-positive thymocytes was calculated as the percentage of cells in lymphoid scatter gates expressing CD4 and CD8 and is given for individual animals. As controls, 5 mice aged 3 to 5 weeks were used, and values reflect the mean ± SD for these groups.
Eμ-A1 mice do not develop lymphoid tumors
Overexpression of Bcl-2 in B cells is associated with the development of tumors as mice age.32,40 Similarly, overexpression of Mcl-1, another cytoprotective member of the Bcl-2 family, also leads to B-cell tumors.41 In multiple lines of Eμ-A1 transgenic mice followed up for as long as 18 months, no B-cell tumors were observed. One Eμ-A1 SCID mouse had thymic lymphoma at age 4 months. These observations suggest that the oncogenic potential for A1 overexpression in lymphoid cells may be less than that for Bcl-2 or Mcl-1.
Discussion
The immune system uses unique forms of targeted genomic instability to randomize parts of the primary sequences of immunoglobulin and TCR genes to create a nearly unlimited repertoire of antigenic specificities. This process generates many out-of-frame or otherwise nonfunctional or deleterious alleles, and selective processes ensure the survival of a primary repertoire of cells with functional and self-tolerant antigen receptors. Further diversification of immunoglobulin genes occurs in B cells during immune responses, requiring additional selection events to ensure survival of the small minority of cells with improved affinity for antigen. These processes illustrate a fundamental aspect of adaptive immunity: survival or death decisions are required on a cell-by-cell basis at several points in immune system development and function. Cell fate decisions in immunity represent a complex integration of signals delivered by antigen receptors, coreceptors, cytokines, and cell death surface receptor proteins such as Fas. Among the intracellular consequences of several of these signals is the modulation of Bcl-2 family proteins, which may promote or antagonize the activation of downstream apoptotic pathways.8 42
Among the antiapoptotic Bcl-2 homologs, little is known regarding the function of A1 in vivo. Analysis of gene-deficient mice is complicated by the apparent redundancy engendered by the presence in mice of 4 nearly identical A1 genes, 3 of which are expressed.43Although these animals can be shown to have increased neutrophil and macrophage apoptosis,21,44 lack of an apparent phenotype in the lymphoid lineage is difficult to interpret because of gene redundancy. An A1-a transgene under the control of the Eμ/H2K expression cassette led to broad overexpression ofA1-a mRNA in the T- and B-cell lineages (Figure 1), consistent with previous results using these control elements.22 We were unable to detect A1 protein in vivo using antibodies available commercially or antibodies generated in our laboratory that interacted specifically with A1 protein in vitro or using a reagent previously shown16 to detect A1 in macrophages with highly inducible A1 expression (P.I.C., D.M.W., unpublished observations, January 2001; May 2001). However, we did find evidence of extended in vitro survival of bone marrow B-lineage cells and thymocytes derived from transgenic mice (Figure 5). For thymocytes, these studies indicated that A1 antagonized apoptosis induced not only by ex vivo culture but also by treatment with death agonist dexamethasone and DNA damage induced by γ-irradiation. In thymocytes, the magnitude of apoptosis inhibition by the Eμ-A1 transgene was considerably less than it was in 2 previous studies of transgenic mice with Bcl-2 overexpression in the thymus.33 34 These data could reflect differences in protein expression or differences in the relative activity of Bcl-2 and A1 in antagonizing cell death. Taken together, these studies indicate that the Eμ-A1 transgene led to the overexpression of functional A1 protein in the developing B- and T-cell compartments, though the degree of overexpression and potential stage-specific differences cannot be accurately quantified. The fact that the phenotype of pro–B-cell expansion was seen in multiple transgenic lines also offers strong genetic evidence for the expression of the transgene, at least within the affected cellular compartment.
Despite evidence for A1 overexpression in the thymus of Eμ-A1 transgenic mice, we saw no discernible effect on thymic cellularity or composition. Maturation of immature CD4+CD8+double-positive T-cell precursors to the CD4+ or CD8+ single-positive stage is largely determined by positive and negative survival signals, which govern the selection of cells bearing major histocompatibility complex–reactive, self-tolerant TCR specificities.30,45 Bcl-xL and A1 are both expressed at high levels at the double-positive stage and are down-regulated with maturation, whereas Bcl-2 expression exhibits the reverse pattern.9,19,46 The limited thymic phenotype of Eμ-A1 mice is similar to that reported for Eμ–Bcl-2 and Eμ–Bcl-xL mice.9,33,34,47-49 Partial inhibition of negative-selection processes and rescue of cells otherwise dying for lack of positive selection has been reported in Eμ–Bcl-2 and Eμ–Bcl-xL mice, suggesting that the modulation of antiapoptotic Bcl-2 homologs may play a role in thymic maturation.33,47,50,51 The antiapoptotic effect of these proteins appears to balance the activities of proapoptotic Bcl-2 homologs.4,8 For example, mice lacking the proapoptotic homolog Bim display an accumulation of thymocytes in early life and a defect in peripheral lymphoid homeostasis.52 In the thymus, CD4+CD8+ pre–T cells undergo massive apoptosis in Bcl-xL–deficient mice, implicating Bcl-xL in thymic selection processes.10,11 This result also indicates that A1 expression at physiologic levels is not functionally redundant with Bcl-xL at this stage. Despite transgene expression in T- and B-cell lineages, including significant increases in A1-amRNA in peripheral lymphoid tissue, we saw no accumulation of T cells in the periphery of Eμ-A1 transgenic mice. Although we cannot rule out poor A1 protein expression in this cellular subset, these results suggest that enforced A1 expression is insufficient to overcome normal homeostatic control of peripheral T-cell populations, similar to findings in Eμ–Bcl-2 mice with T-lineage expression.33,48 In contrast, Eμ–Bcl-xL mice expressing the transgene in the T-cell lineage exhibited 3- to 5-fold increases in lymph node T-cell numbers.9
The major phenotype we observed in Eμ-A1 transgenic mice was the expansion of B-cell progenitors at the pro–B-cell stage. This finding was present in multiple independent mouse lines, indicating that it was a function of the transgene per se, independent of variability conferred by insertional effects (Table 1). The central differentiation event within the pro–B-cell compartment is the V(D)J rearrangement of the IgH locus, which, if successful and in-frame, leads to synthesis of the Igμ protein. Expression of Igμ in the context of the pre–B-cell receptor complex delivers an intracellular signal that leads to cellular proliferation and maturation to the pre–B-cell stage.30 Differentiation events within the pro–B-cell compartment correlate with distinct cellular subfractions defined by the expression of HSA and BP-1.25 29 Within fraction A (HSA− BP-1−), the IgH locus begins to show initial D-to-J rearrangements. In fraction B (HSA+BP-1−), DJ rearrangements are seen and V(D)J rearrangements appear at low levels. Fraction C contains cells with functionally rearranged IgH alleles and includes cells that are rapidly cycling and in transition to the pre–B-cell stage. Expansion of pro–B cells in Eμ-A1 mice specifically affected fraction B (Figure 3) and was accompanied by a relative decrease in fraction D (pre–B cells). These findings might be explained in one of 3 ways: (1) a partial block in the pre–B-cell receptor signal; (2) accumulation of a subset of fraction B cells that might otherwise die without progressing to the pre–B-cell stage; or (3) slowing of the V(D)J rearrangement process.
Mice with defective pre–B-cell receptor signals—for example, through mutation of λ5 or the Syk tyrosine kinase—accumulate pro–B cells that have completed V(D)J rearrangement of the IgH locus.53,54 In contrast, our data indicate that the expanded pro–B-cell pool in Eμ-A1 transgenic mice had normal, low levels of completed V(D)J joins (Figure 4). The pro– to pre–B-cell transition could also be affected if the Eμ-A1 transgene slowed cell-cycle entry that coincides with this event.37 For example, in Eμ–Bcl-2 transgenic mice, the percentage of B-cell precursors in the S+G2+M stages of the cell cycle are substantially diminished. In wild-type mice, cycling pro–B cells were reduced more than 2-fold, whereas in SCID mice, cycling cells were reduced more than 3-fold.38 In contrast, we found no effect of the A1 transgene on cell cycle within the pro–B-cell compartment (Figure 7). Mutational studies suggest that the cell-cycle effects of Bcl-2 can be separated from apoptosis pathways and that they rely on interactions involving the BH4 domain.55 Data from in vitro expression studies suggest that A1 expression does not engender the negative effects on cell cycle seen with Bcl-2.56 Our data suggest that the latter finding is also relevant to properties of A1 in vivo.
Fang et al28 found that pro–B cells were expanded in Eμ–Bcl-xL transgenic mice, similar to the phenotype observed in Eμ-A1 mice. The expanded population of pro–B cells in these mice had increased levels of IgH V(D)J rearrangements and contained an increased frequency of IgH alleles with nonproductive V(D)J rearrangements, leading to the suggestion that pro–B-cell accumulation reflected the survival of cells that had failed to achieve an in-frame IgH rearrangement and that would normally undergo apoptosis. As noted above, the pro–B-cell accumulation in Eμ-A1 mice did not reflect a bias toward cells with completed V(D)J joining of IgH alleles (Figure4), suggesting that this explanation does not apply. A reduction in the efficiency of IgH gene assembly at the pro–B-cell stage could explain the phenotype of Eμ-A1 transgenic mice. We did not observe a significant effect of the transgene on the expression of Rag-1 or Rag-2 mRNA or on germline transcription of the IgH locus. However, these data do not exclude subtle effects or differences at the level of Rag protein expression. A final question is whether the apparent block in pro– to pre–B-cell development could simply reflect differences in expression of the A1 transgene in these compartments. Although this is not the expected expression pattern based on the Eμ control element, the lack of reagents capable of detecting A1 protein in vivo precludes direct testing of this possibility. However, this scenario would not readily explain the absolute reduction in pre–B cells observed in transgenic mice.
To further investigate the properties of A1 overexpression in pro–B cells, we studied the effects of this transgene in the SCID background. The SCID defect impairs rejoining of coding DNA ends in V(D)J recombination, leading to a developmental block at the pro–B stage that is similar to the developmental block in Rag-1−/− or Rag-2−/− mice, except that lymphoid precursor cells carry Rag-mediated DNA breaks within antigen receptor loci.57The SCID defect is leaky, and these DNA breaks may be repaired by alternative mechanisms, permitting the accumulation of mature T and B cells over time. Bcl-2 family members, including A1, antagonize cell death associated with the induction of DNA breaks. In the SCID background, Eμ–Bcl-2 expression leads to a dramatic rescue of pre–B-cell development, a process that involves IgH rearrangement and expression of Igμ or Dμ protein.35,36 58 Despite the significant effect of A1 expression on the pro–B-cell subset in the wild-type background, we did not observe an increase in B220+CD43− pre–B cells when the transgene was expressed on the SCID background (Figure 6). This result corresponded with a failure to rescue V(D)J joining of the IgH locus. One potential explanation for these results is that Bcl-2 and A1 have different spectrums of activity in early B-lineage cells and that cell death pathways activated by unresolved Rag-mediated DNA breaks could be antagonized better by Bcl-2 than by A1. An alternative explanation for these data could involve differences in the relative expression levels of A1 and Bcl-2 in the different transgenic lines.
Exposure of newborn SCID mice to low levels of γ-radiation leads to partial rescue of TCR-β rearrangement and the appearance of CD4+CD8+ thymocytes.39 For unknown reasons, similar effects were not observed in the B-cell compartment. The Eμ-A1 transgene antagonized cell death in thymocytes exposed to γ-irradiation in vitro (Figure 5); however, there appeared to be no potentiation of the radiation-induced rescue of T-cell development in SCID mice (Figure 6). Similarly, we did not observe the rescue of B-cell development in these animals. The mechanism by which SCID thymocyte development is promoted by radiation remains obscure. One explanation invokes increased rejoining of Rag-mediated DNA breaks in early thymocytes by an alternative DNA repair pathway up-regulated by radiation.39 However, the radiation of SCID bone marrow cells can also produce similar results, suggesting that the relevant cellular target for this effect may be one that precedes thymic colonization.59 If this is the case, such a cell might not express the Eμ-A1 transgene.
A1 is normally expressed at low levels in early B cells and is up-regulated in long-lived peripheral B cells, where it may enhance B-cell memory.18 In contrast, Bcl-2 is expressed in pro–B and mature B cells and is down-regulated at the pre–B-cell and immature B-cell stages, whereas Bcl-xL is expressed in pre–B cells and is down-regulated at the mature B stage.12,60,61 A1 and Bcl-xL are up-regulated in response to CD40 signals, by which they appear to contribute to cell survival in the context of B-cell receptor signaling.23,62-68 These data suggest that the physiologic role of A1 is more likely to be important in later stages of B-cell function than at the pro–B-cell stage. It is probable that Eμ-A1 mice express the transgene in peripheral B cells based on the known properties of Eμ-dependent transgenes and on the observed increases in splenic A1-a mRNA. Unexpectedly, we observed a decrease in peripheral B cells in Eμ-A1 transgenic mice, in contrast to B-cell accumulation reported in Eμ–Bcl-2 and Eμ–Bcl-xL transgenic mice.12,28,32 48 This observation corresponded with a modest decrease in serum immunoglobulin levels of switched subclasses; however, serum immunoglobulin levels in mice continued to increase up to 6 months of age, and it remains unknown whether the Eμ-A1 transgene would affect steady state immunoglobulin levels. These observations may reflect the early developmental effects of the transgene in the B-cell lineage rather than indicating a negative effect of A1 overexpression on peripheral B cells.
Bcl-2, Bcl-xL, and A1 share the property of antagonizing cell death. Genetic experiments show that overexpression of Bcl-xL in the T-cell lineage can rescue the Bcl-2 null phenotype in mature T cells,49 illustrating the functional overlap in these proteins in vivo. Because the regulation of these proteins differs, the extent to which they may have distinct individual functions remains unclear. Transgenic mice made under the control of similar expression elements permit comparison of the effects of Bcl-2 homologs on the development and function of the immune system. Our studies indicate that the phenotype of Eμ-A1 mice most resembles that of Eμ–Bcl-xL mice in that the pro–B-cell compartment is strongly affected. However, differences in the IgH rearrangement status of the accumulated pro–B cells suggest that the underlying causes of this phenotype may be different in the 2 strains. In contrast, Bcl-2 appears to have a less prominent effect on the pro–B-cell compartment. Despite these observations, Bcl-2 can rescue the pro– to pre–B-cell transition in SCID mice, whereas A1 does not. Inasmuch as neither the complete spectrum of cellular effects nor the relative activity of antiapoptotic Bcl-2 family members is known, it cannot be ruled out that differences in expression levels among the different transgenic mouse lines reported are responsible for these phenotypic contrasts. However, given the consistency of these phenotypic differences across numerous individual mouse lines, these studies lend further support to the notion that antiapoptotic Bcl-2 homologs interact differentially with intracellular pathways affecting cell fate in lymphoid cells. It is unclear whether all the available data can be accounted for by differences in cell-survival effects of these proteins. Further consideration of the potential roles for Bcl-2 homologs in other cellular pathways are, therefore, warranted in future studies.
Supported by National Institutes of Health grants PHS03174 (J.M.H.), AI41051 (D.M.W.), and CA88075 (D.M.W.) and by an American Lung Association- Washington Affiliate Research Grant (P.I.C.). P.I.C. is the recipient of an American Heart Association Clinician Scientist Award. C.Y.L. is the recipient of a Chang-Gung Memorial Hospital Sabbatical Scholarship Award.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dennis M. Willerford, Division of Hematology, University of Washington School of Medicine, Box 357710, 1959 NE Pacific St, Seattle, WA 98195; e-mail: dwiller@u.washington.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal