Abstract
The detection of leukemia cells on newborn genetic screening cards (“Guthrie cards”) of a small group of patients and several sets of identical twins developing acute lymphoblastic leukemia (ALL) with identical phenotypic and chromosomal markers has provided evidence that childhood ALL cases may arise in utero. We conducted a retrospective study of a randomly selected group of childhood B-precursor ALL patients to determine the frequency of the presence of “leukemic” clones prenatally in ALL cases by testing newborn screening cards. The 17 ALL patients analyzed had a median age of 46 months (range, 18 months to 13 years) and had median presenting white blood cell (WBC) counts of 10 950/μL (range, 2900-70 300/μL) at diagnosis. A clonal rearrangement of the immunoglobulin heavy chain (IgH) gene was identified in diagnostic lymphoblasts and sequenced and patient-specific primers were used to amplify DNA from blood samples on the patient's newborn screening cards. Twelve of the 17 (71%) analyzed newborn cards had detectable IgH rearrangements amplified by seminested polymerase chain reaction. DNA sequencing confirmed that the IgH rearrangements detected matched the IgH sequences identified from diagnostic leukemia cells, indicating the presence of a “leukemic” clone at birth. There were no differences in age or presenting WBC counts between the cases with or without positive newborn screening cards. All 6 patients with hyperdiploid ALL had detectable “leukemic” clones on their cards. The results of our study support the notion that a high proportion of childhood B-precursor ALL cases arise in utero, although postnatal events are also important factors in leukemogenesis.
Introduction
Acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer and thus identifying factors linked to leukemogenesis have major implications for health care for children. Possible exogenous factors that have been variably linked to the development of childhood leukemia include ionizing radiation, pesticides, nitrites, electromagnetic fields, and abnormal immunologic responses to common infectious agents.1-6
Epidemiologic studies have also suggested that leukemogenesis may occur as a multistep process with the initiating event occurring in utero and subsequent genetic “hits” occurring postnatally.2,4,7Several sets of identical twins developing the same type of ALL, with identical genomic fusion sequences of rearranged oncogenes, have confirmed the concept of an in utero origin for childhood leukemia.8-16 Three groups have recently reported the retrospective identification of “leukemic” clones in blood spots of archival newborn genetic screening cards from a limited number of patients, particularly ALL cases with t(4;11) and t(12;21).17-20
In this report we describe the results of a retrospective study of a heterogeneous, randomly selected group of 17 B-precursor ALL patients, representative of our clinical population of ALL cases (excluding infant ALL), to determine the frequency of ALL cases originating prenatally. To detect “leukemic” clones, we used patient/leukemia-specific IgH gene rearrangements, rather than translocation breakpoint genomic sequences as a clonal marker for leukemia. Our study analyzed the largest sample of newborn screening cards of ALL patients to date with diverse biologic features and revealed a remarkably high frequency of B-precursor ALL patients with detectable “leukemic” clones on their cards, indicative of a prenatal origin of leukemia. Long latency periods until clinical diagnosis suggest possible roles for postnatal factors in leukemogenesis.
Patients, materials, and methods
Patients
Patients eligible for this study included (1) individuals diagnosed with B-precursor ALL at Children's Hospital of Michigan and born in the State of Michigan from 1982 to 1999, (2) age 18 months or older at diagnosis, and (3) available cryopreserved diagnostic leukemia cells or bone marrow slides and newborn screening cards. Informed consent was obtained from all parents. The patient characteristics are described in Table1. This project was approved by the Wayne State University Human Investigation Committee and the State of Michigan Department of Community Health Human Subjects Committee.
Patient characteristics and results
| Patient . | Sex . | Age at diagnosis . | Race . | WBC (/μL) . | Karyotype . | Leukemic clone detected on newborn card . |
|---|---|---|---|---|---|---|
| 1 | M | 18 mo | W | 4 000 | DNA index 1.13 | + |
| 2 | M | 25 mo | AA | 34 000 | 46,XY[8] | + |
| 3 | F | 27 mo | W | 32 100 | 51-52,XX,+ X,+ 6,+ 14,+ 18,+ 21,+ 21[13]/46,XX[8] | + |
| 4 | M | 30 mo | W | 36 900 | 47,XY,+ 21[10]/46,XY[22] | − |
| 5 | F | 33 mo | W | 9 400 | 48,XX,+ X,+ 21[17]/46,XX[17] | + |
| 6 | F | 34 mo | W | 4 400 | 56,XX,+ X,+ 4,+ 10,+ 14,+ 14,+ 18,+ 21,+ 22,+ mar,+ mar[1]/46,XX[19]; DNA index 1.20 | + |
| 7 | M | 35 mo | AA | 8 200 | 46,XY[29] (t(12;21) detected by FISH) | + |
| 8 | M | 42 mo | Asian | 8 500 | 55,XY,+ X,+ 4,+ 6,+ 9,+ 10,+ 14,+ 15,+ 21,+ 21[11]/46,XY[9] | + |
| 9 | F | 46 mo | W | 2 900 | 46,XX[22] | − |
| 10 | F | 4 y | W | 50 500 | 46,XX[21] | − |
| 11 | F | 4 y | W | 3 200 | 47,XX,t(10;12)(q22;p12),+ der(10)t(10;12)(q22;p12)[16]/46,XX[9] | − |
| 12 | M | 5 y | W | 5 700 | DNA index > 1.16 | + |
| 13 | F | 8 y | AA | 12 500 | 46,XX,del(6)(q15q23)[17]/46,XX[10] | + |
| 14 | M | 9 y | W | 2 300 | 55-57[4]/46,XY[23] | + |
| 15 | M | 9 y | W | 33 000 | 47,XY,+ 21[32] (Down syndrome) | − |
| 16 | M | 10 y | AA | 70 300 | 47,XY,+ X,t(4;11)(q21;q23),der(9;14)(q10;q10)[19]/46,XY[1] | + |
| 17 | F | 13 y | W | 46 400 | 46,XX,t(1;19)(q23;p13)[4]/46,idem,add(9)(p22)[3]/46,XX[3] | + |
| Patient . | Sex . | Age at diagnosis . | Race . | WBC (/μL) . | Karyotype . | Leukemic clone detected on newborn card . |
|---|---|---|---|---|---|---|
| 1 | M | 18 mo | W | 4 000 | DNA index 1.13 | + |
| 2 | M | 25 mo | AA | 34 000 | 46,XY[8] | + |
| 3 | F | 27 mo | W | 32 100 | 51-52,XX,+ X,+ 6,+ 14,+ 18,+ 21,+ 21[13]/46,XX[8] | + |
| 4 | M | 30 mo | W | 36 900 | 47,XY,+ 21[10]/46,XY[22] | − |
| 5 | F | 33 mo | W | 9 400 | 48,XX,+ X,+ 21[17]/46,XX[17] | + |
| 6 | F | 34 mo | W | 4 400 | 56,XX,+ X,+ 4,+ 10,+ 14,+ 14,+ 18,+ 21,+ 22,+ mar,+ mar[1]/46,XX[19]; DNA index 1.20 | + |
| 7 | M | 35 mo | AA | 8 200 | 46,XY[29] (t(12;21) detected by FISH) | + |
| 8 | M | 42 mo | Asian | 8 500 | 55,XY,+ X,+ 4,+ 6,+ 9,+ 10,+ 14,+ 15,+ 21,+ 21[11]/46,XY[9] | + |
| 9 | F | 46 mo | W | 2 900 | 46,XX[22] | − |
| 10 | F | 4 y | W | 50 500 | 46,XX[21] | − |
| 11 | F | 4 y | W | 3 200 | 47,XX,t(10;12)(q22;p12),+ der(10)t(10;12)(q22;p12)[16]/46,XX[9] | − |
| 12 | M | 5 y | W | 5 700 | DNA index > 1.16 | + |
| 13 | F | 8 y | AA | 12 500 | 46,XX,del(6)(q15q23)[17]/46,XX[10] | + |
| 14 | M | 9 y | W | 2 300 | 55-57[4]/46,XY[23] | + |
| 15 | M | 9 y | W | 33 000 | 47,XY,+ 21[32] (Down syndrome) | − |
| 16 | M | 10 y | AA | 70 300 | 47,XY,+ X,t(4;11)(q21;q23),der(9;14)(q10;q10)[19]/46,XY[1] | + |
| 17 | F | 13 y | W | 46 400 | 46,XX,t(1;19)(q23;p13)[4]/46,idem,add(9)(p22)[3]/46,XX[3] | + |
W indicates white; AA, African American; FISH, fluorescence in situ hybridization.
Diagnostic specimens
Genomic DNAs were extracted from lymphoblasts obtained from diagnostic bone marrow aspirations that were cryopreserved in liquid nitrogen or Wright-Giesma–stained diagnostic bone marrow slides maintained at room temperature. DNAs from cryopreserved cells were extracted with the Puregene DNA Isolation Kit (Gentra, Minneapolis, MN) according to the manufacturer's instructions; DNA was extracted from bone marrow slides when cryopreserved cells were unavailable. For the latter, bone marrow coverslips were removed by immersion inp-xylene for 24 to 48 hours, the cellular material was scraped off the slides, and DNA was extracted with a modified protocol using the Puregene kit. DNA extracted from bone marrow slides was quantitated with Nucleic Acid QuickSticks (Clontech, Palo Alto, CA).
IgH identification
Rearrangements of the IgH gene were used as a patient/leukemia-specific molecular marker, to identify “leukemic” clones in blood samples on newborn screening cards. This method could identify leukemia cells with and without acquired structural chromosomal rearrangements, as used in minimal residual disease detection of ALL.21,22 Amplification of rearranged IgH genes was performed using a panel of 6 VH-segment family (VH 1-6) forward primers with a JH-segment consensus reverse primer.23 24 The polymerase chain reaction (PCR) consisted of 100 to 500 ng genomic DNA extracted from the diagnostic lymphoblast samples, in a total volume of 50 μL using the GC-RICH PCR System (Roche, Mannheim, Germany), 0.2 mM of each deoxyribonucleoside triphosphate and 0.2 μM of each primer. The following PCR parameters were used for all combinations of the JH- and VH-segment primers. After an initial 5-minute denaturation at 95°C, PCR was performed for 35 to 40 cycles of 30 seconds at 95°C, 30 seconds at 60°C, 1 minute at 72°C, followed by a final extension at 72°C for 7 minutes in a Perkin-Elmer GeneAmp 9600 (PE Applied Biosystems, Foster City, CA). Aliquots of the PCR (40 μL) were analyzed on agarose gels (2%) with ethidium bromide staining.
The most intense band representative of the clonal IgH rearrangement was excised, subcloned into the pGEM-T Easy vector system (Promega, Madison, WI), and sequenced by automated sequencing using the Bigdye DNA Sequencing Kit (PE Applied Biosystems). Sequencing was performed by the DNA sequencing facility at the Center of Molecular Biology and Genetics (Wayne State University). For each diagnostic patient sample, a minimum of 3 identical clones was identified to represent the patient/leukemia-specific IgH clonal rearrangement. Patient clonal IgH sequences were analyzed by BLAST (National Center for Biotechnology Information, Bethesda, MD) and compared to known VH, DH, JH sequences to determine the location of the VH, DH, JH segments and the position of N insertion nucleotides.
Based on patient-specific clonal IgH rearrangements identified from diagnostic leukemia samples, 2 patient-specific reverse primers were designed and used in a seminested PCR reaction to amplify genomic DNA from the newborn screening cards. The first primer was designed from the patient's DH-N-JH sequence across the junction of the DH and JH segments, whereas a second primer was designed from the N-DH-N segment where N denotes the random insertion nucleotides at the junction of the VH, DH, and JH sequences. Both patient-specific primers were used as the reverse primer together with the forward VH primer used previously to obtain the IgH clonal gene rearrangement.
The sensitivity and specificity of each set of patient-specific PCR primers was tested with serial dilutions of genomic DNA extracted from the patient's diagnostic leukemia cells mixed with genomic DNA extracted from the peripheral blood from a healthy control.
Newborn screening card analysis
The newborn screening (“Guthrie”) cards, prepared from heel sticks of newborns, of the leukemia patients enrolled in this study were obtained from the State of Michigan Newborn Screening Program in Lansing, where they are stored at room temperature for 21.5 years.
To prevent any potential contamination of PCRs between diagnostic leukemia samples and the newborn screening cards (which could lead to false-positive results), all reagents and equipment used for PCR analysis of the patient's diagnostic leukemia IgH rearrangement were kept in separate rooms from the reagents used for the newborn screening card PCR analysis.25
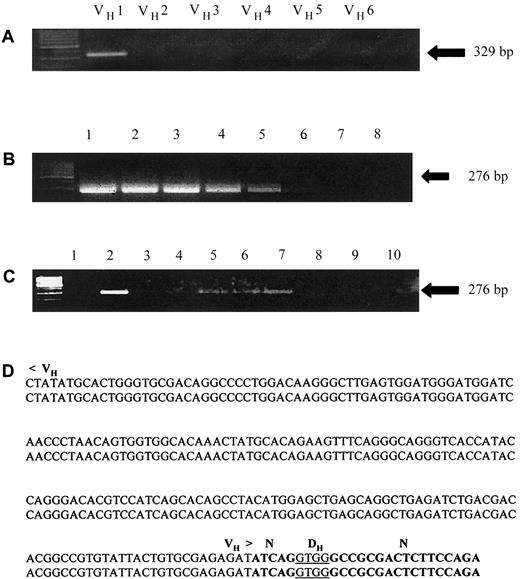
For each PCR, genomic DNA was amplified directly from 2-mm diameter punches obtained from the newborn screening card blood samples (consisting of an approximate 1.3-cm diameter blood spot). Each punch was washed with the Instagene Dry Blood Kit (Biorad, Hercules, CA) to remove PCR inhibitors including red blood cells prior to the amplification.26 Seminested PCR was used with 25 cycles for the first round, followed by 40 cycles for the second round, with 1 μL of a 1:100 dilution of the primary PCR product as template. PCR products of appropriate size representing rearranged IgH genes, were excised, sequenced and compared to the previously identified IgH sequences of the patient's diagnostic leukemia sample. For all PCRs, control amplifications included genomic DNAs from healthy controls (or newborn screening cards from siblings, if available), and blanks lacking genomic DNA. In addition, to verify the ability to directly amplify genomic DNA from newborn card punches, the ubiquitously expressed gene, reduced folate carrier, was amplified as a control from each patient's newborn screening card blood sample. A representative analysis of a patient leukemia sample and newborn screening card is shown in Figure 1.
Identification of clonal IgH rearrangement in diagnostic lymphoblasts and detection of leukemia/patient-specific IgH rearrangement in blood samples of archival newborn screening cards.
(A) Genomic DNA extracted from diagnostic lymphoblasts was amplified using a panel of 6 VH-segment family primers (VH 1-6) and a JH-segment consensus reverse primer (see “Patients, materials, and methods”). Lane 1 represents a patient/leukemia-specific clonal IgH gene rearrangement that was excised and sequenced. (B) Seminested PCR testing the specificity and sensitivity of the patient-specific primers designed across the DH-N-JH and N-DH-N segments based on the clonal IgH rearrangement identified in panel A. Lane 1 represents amplification of 100 ng of the patient's diagnostic genomic DNA; lanes 2 through 5 represent serial 1:10 dilutions of the patient's genomic DNA with genomic DNA extracted from peripheral blood lymphocytes of a healthy control. Lane 6 represents a PCR using control genomic DNA, and lanes 7 and 8 represent blank controls from the first and second rounds of the seminested PCRs, respectively. (C) PCR amplification of 2-mm punches obtained from the patient's newborn screening card blood sample using a seminested PCR with primers based on the IgH sequence identified in panel A. Lanes 2, 5, 6, and 7 represent punches in which the patient/leukemia-specific IgH rearrangement was amplified by seminested PCRs using a set of patient/leukemia-specific primers as described in “Patients, materials and methods.” Lane 8 represents a PCR using a punch from an artificial control newborn card and lanes 9 and 10 represent blank controls. (D) DNA sequence identifying identical IgH genomic sequences from the patient's newborn screening card blood samples amplified in panel C (lower sequence) and from lymphoblasts obtained at diagnosis (upper sequence).
Identification of clonal IgH rearrangement in diagnostic lymphoblasts and detection of leukemia/patient-specific IgH rearrangement in blood samples of archival newborn screening cards.
(A) Genomic DNA extracted from diagnostic lymphoblasts was amplified using a panel of 6 VH-segment family primers (VH 1-6) and a JH-segment consensus reverse primer (see “Patients, materials, and methods”). Lane 1 represents a patient/leukemia-specific clonal IgH gene rearrangement that was excised and sequenced. (B) Seminested PCR testing the specificity and sensitivity of the patient-specific primers designed across the DH-N-JH and N-DH-N segments based on the clonal IgH rearrangement identified in panel A. Lane 1 represents amplification of 100 ng of the patient's diagnostic genomic DNA; lanes 2 through 5 represent serial 1:10 dilutions of the patient's genomic DNA with genomic DNA extracted from peripheral blood lymphocytes of a healthy control. Lane 6 represents a PCR using control genomic DNA, and lanes 7 and 8 represent blank controls from the first and second rounds of the seminested PCRs, respectively. (C) PCR amplification of 2-mm punches obtained from the patient's newborn screening card blood sample using a seminested PCR with primers based on the IgH sequence identified in panel A. Lanes 2, 5, 6, and 7 represent punches in which the patient/leukemia-specific IgH rearrangement was amplified by seminested PCRs using a set of patient/leukemia-specific primers as described in “Patients, materials and methods.” Lane 8 represents a PCR using a punch from an artificial control newborn card and lanes 9 and 10 represent blank controls. (D) DNA sequence identifying identical IgH genomic sequences from the patient's newborn screening card blood samples amplified in panel C (lower sequence) and from lymphoblasts obtained at diagnosis (upper sequence).
Results
Eighty current B-precursor ALL patients at Children's Hospital of Michigan were enrolled on the study of whom 25 patients were further analyzed based on the availability of newborn screening cards and diagnostic leukemia cells. Seventeen of the 25 patients had a clonal IgH gene rearrangement identified in genomic DNA extracted from diagnostic lymphoblast samples from either cryopreserved cells (n = 8) or bone marrow slides (n = 9). Following sequencing of the clonal IgH rearrangement, patient-specific primers were designed to amplify unique IgH sequences by seminested PCR from blood samples on the newborn screening cards (Figure 1). The 17 patients ranged in age from 18 months to 13 years (median age, 46 months) at the time of leukemia diagnosis and included 12 whites, 4 African Americans, and 1 Asian American. The presenting white blood cell (WBC) counts ranged from 2900 to 70 300/μL (median, 10 950 μL) and all diagnostic leukemia samples comprised at least 85% lymphoblasts (Table1).
Clonal rearrangements of the IgH gene were detected in 12 of the 17 (71%) newborn cards analyzed using patient-specific primers. DNA sequencing confirmed that the IgH rearrangements detected from the blood samples of the newborn screening cards matched the IgH sequences identified from diagnostic leukemia cells, suggesting the presence of a “leukemic” clone at birth (Table 1 and Figure 1). Additionally, there was no amplification of control DNAs, confirming the specificity of the PCR primers in only detecting the patient/leukemia-specific IgH rearrangement. These results support the notion that a “leukemic” clone was present at birth in 12 of the 17 ALL patients and that an initiating leukemic event occurred in utero for these patients.
There were no differences in age at diagnosis or presenting WBC count between the 12 patients in whom “leukemic” clones were detected at birth and the 5 patients whose newborn cards tested negative (median age 39 months versus 4 years, P = .67 and median WBC count 8950/μL versus 33 000/μL, P = .83; Table 1). The 12 patients with detectable clones included 6 with hyperdiploidy (patients 1, 3, 6, 8, 12, and 14), 2 patients with normal karyotypes (1 with the cryptic t(12;21) detected by fluorescence in situ hybridization), 1 patient each with t(4;11) and t(1;19), and 1 patient with acquired trisomy 21 (Table 1). The cases without detectable clones on newborn cards included 2 patients with normal karyotypes, 1 patient with acquired trisomy 21, 1 patient with a t(10;12) translocation, and 1 patient with Down syndrome. There were no differences in birth weight nor maternal age between the 2 groups (data not shown).
An estimation of the number of “leukemic” clones present on the newborn screening cards can be made. From the estimated number of mononuclear cells in the dried blood of one circle (540 000, based on an average WBC count of a healthy newborn infant being 18 000/μL27 and approximately 30 μL whole blood filling the area of one 1.3-cm circle on a screening card) and the DNA content of a single cell (6 pg),22 we estimate that the “leukemic” clone was present in a range from 1:363 cells (patient 1), 1:788 cells (patient 7) to 1:1166 cells (patient 16). This is based on the level of sensitivity of each set of patient-specific PCR primers tested with serial dilutions of the patients' leukemic DNAs and the intensity of bands on gel electrophoresis analysis from the PCR amplification of individual 2-mm punches from the newborn cards. These results are in keeping with our level of detection using “artificial” newborn screening cards spotted with serial dilutions of the REH B-precursor ALL cell line mixed with control blood samples (data not shown).
Discussion
This study was designed to determine the frequency of a possible in utero origin of childhood B-precursor ALL in a group of patients representative of the clinical ALL population seen at our center and not biased toward a particular ALL cytogenetic subtype (eg, infant ALL cases with t(4;11) were excluded). We used clonal rearrangements of the IgH gene representative of the lymphocyte clone from which the leukemia arose, as a molecular marker to be amplified from newborn screening cards. Clonal IgH rearrangements have been widely used as a leukemia/patient-specific marker for the detection of minimal residual disease in ALL21 22 and allow for the detection of “leukemic” clones that lack structural chromosomal alterations (ie, translocations). At the current time, newborn screening cards (which can be used to amplify DNA though not RNA) can only be used for retrospective studies and cannot prospectively identify patients at risk for developing leukemia.
Seventeen patients with B-precursor ALL were analyzed for whom diagnostic leukemia cells and newborn screening cards were available. In each case, a patient/leukemia-specific clonal IgH gene rearrangement from diagnostic leukemia cells was identified, which could detect low levels of leukemia cells by PCR amplification. Twelve of the 17 (71%) patients analyzed had detectable “leukemic” clones amplified from their newborn cards, which matched their diagnostic leukemia IgH sequences, suggesting an in utero origin of the “leukemic” clone in these cases. It is conceivable that the 5 patients in our study who did not have leukemia IgH rearrangements detected on their newborn cards may have had “leukemic” clones present, albeit below the detection level of our PCR assay, or clonal evolution in the IgH gene rearrangement may have occurred postnatally.
The 12 patients with detectable “leukemic” clones on their newborn screening cards presented at a median age of 39 months (range, 18 months to 13 years), indicating the potential for a long latency period from the initiation of leukemogenesis (ie, in utero) to the development of clinically detectable leukemia. A long latency period from leukemogenesis in utero to the clinical diagnosis of ALL has also been reported in 2 sets of identical twins; those in one set with T-ALL were diagnosed at the ages of 9 and 11 years and those in the other set with t(12;21) ALL were diagnosed at the ages of 5 and 14 years.12 13 All 6 patients with hyperdiploid ALL karyotypes in this cohort (> 50 chromosomes or DNA index > 1.0), had detectable “leukemic” clones on their newborn screening cards, though it is unknown whether the clone detected contained the hyperdiploid chromosome complement. Technically, it is impossible to determine whether the leukemic clones present on the cards contained quantitative chromosomal abnormalities, due to their low cell numbers.
A semiquantitative estimation can be made as to the number of “leukemic” clones that were present on the newborn screening cards ranging from 1:363 cells (patient 1 diagnosed at the age of 18 months), 1:788 cells (patient 7 diagnosed at the age of 35 months) to 1:1166 cells for (patient 16 diagnosed at the age of 10 years). These estimated cell numbers are in keeping with the previous published studies17 19 and suggest that the “leukemic load” at birth was much greater in patients diagnosed at younger ages than the load in patients diagnosed at older ages. This would argue that the results of our study are indeed detecting a “leukemic” or preleukemic clone rather than a nonmalignant B lymphocyte that had undergone rearrangement of its IgH gene.
Our study represents the largest and most biologically diverse sample of ALL patients analyzed to date for the presence of “leukemic” clones at birth on newborn screening cards. The 4 previously published studies analyzing newborn screening cards from leukemia patients included a total of 22 patients with B-precursor ALL (including 7 patients with t(4;11) of which 6 were under the age of 1 year and 9 cases of t(12;21) ALL) and 2 patients with T-ALL17-20(Table 2). Eighteen of the 24 patients had “leukemic” clones detected retrospectively on newborn cards by PCR amplification of patient-specific genomic fusion sequences of either the t(4;11) or t(12;21) rearranged oncogenes17,18 or the detection of clonal leukemia-specific rearrangements of the T-cell receptor gene or IgH gene similar to our approach.19 20
Previously published results detecting leukemic clones at birth from newborn screening cards
| . | Age at diagnosis . | Detected at birth . | ALL subtype . |
|---|---|---|---|
| Translocation breakpoint fusion sequence used as leukemic clone marker | |||
| Gale et al17; Marker: t(4;11) | 5 mo | + | BP-ALL |
| 6 mo | + | BP-ALL | |
| 24 mo | + | BP-ALL | |
| Wiemels et al18; Marker: t(12;21) | 25 mo | + | BP-ALL |
| 34 mo | + | BP-ALL | |
| 39 mo | + | BP-ALL | |
| 40 mo | − | BP-ALL | |
| 41 mo | − | BP-ALL | |
| 42 mo | + | BP-ALL | |
| 47 mo | − | BP-ALL | |
| 51 mo | + | BP-ALL | |
| 61 mo | + | BP-ALL | |
| Clonal IgH/TcR gene rearrangement used as leukemic clone marker | |||
| Fasching et al19; Marker: IgH/TcR | 6 mo | + | t(4;11) |
| 12 mo | + | t(4;11) | |
| 56 mo | + | BP-ALL | |
| 25 mo | + | T-ALL | |
| 26 mo | + | T-ALL | |
| Yagi et al20; Marker: IgH/TcR | 14 d | + | t(4;11) |
| 2 mo | + | t(4;11) | |
| 2 y | + | BP-ALL | |
| 2.2 y | + | BP-ALL | |
| 1.5 y | − | BP-ALL | |
| 9 y | − | BP-ALL | |
| 2 y | − | BP-ALL |
| . | Age at diagnosis . | Detected at birth . | ALL subtype . |
|---|---|---|---|
| Translocation breakpoint fusion sequence used as leukemic clone marker | |||
| Gale et al17; Marker: t(4;11) | 5 mo | + | BP-ALL |
| 6 mo | + | BP-ALL | |
| 24 mo | + | BP-ALL | |
| Wiemels et al18; Marker: t(12;21) | 25 mo | + | BP-ALL |
| 34 mo | + | BP-ALL | |
| 39 mo | + | BP-ALL | |
| 40 mo | − | BP-ALL | |
| 41 mo | − | BP-ALL | |
| 42 mo | + | BP-ALL | |
| 47 mo | − | BP-ALL | |
| 51 mo | + | BP-ALL | |
| 61 mo | + | BP-ALL | |
| Clonal IgH/TcR gene rearrangement used as leukemic clone marker | |||
| Fasching et al19; Marker: IgH/TcR | 6 mo | + | t(4;11) |
| 12 mo | + | t(4;11) | |
| 56 mo | + | BP-ALL | |
| 25 mo | + | T-ALL | |
| 26 mo | + | T-ALL | |
| Yagi et al20; Marker: IgH/TcR | 14 d | + | t(4;11) |
| 2 mo | + | t(4;11) | |
| 2 y | + | BP-ALL | |
| 2.2 y | + | BP-ALL | |
| 1.5 y | − | BP-ALL | |
| 9 y | − | BP-ALL | |
| 2 y | − | BP-ALL |
BP-ALL indicates B-precursor acute lymphoblastic leukemia; TcR, T-cell receptor.
The particular strength of our study of 17 patients derives from (1) the larger sample size; (2) the heterogeneous patient population based on age of diagnosis, presenting WBC count, and variety of leukemia cytogenetics; (3) the exclusion of infant ALL cases; and (4) the reporting of an actual frequency of positive and negative cases. The significance of one report in which 2 infant ALL cases diagnosed at the age of 2 weeks and 2 months of life (with WBC counts > 100 000/μL), who had “leukemic” clones detected on screening cards,20 is unclear because it is not unlikely that leukemia cells could be detected by PCR amplification of blood samples obtained 14 days and 2 months previously in these 2 cases.
Combining the results of the above-cited studies with our study, 30 of 41 (73%) of pediatric ALL patients had detectable “leukemic” clones present in blood samples from newborn screening cards. Two possible hypotheses are likely. The first would suggest that in a high proportion of childhood B-precursor ALL cases, the initiating leukemogenic event occurs in utero with a variable latency period for the full expression of the disease. Alternatively, the presence of multiple lymphocyte clones may be a normal feature of newborns and the physiologic postnatal switch from TH-2 to TH-1 of immunologic transition eliminates these clones in the vast majority of cases. In the remainder, an aberrant immune response to an infection, as proposed by Greaves, may allow for the persistence of a “preleukemic” clone with later emergence of leukemia.4
The results of our retrospective study of a heterogeneous group of childhood B-precursor ALL patients have revealed a high frequency of patients having evidence of “leukemic” clones present at birth and detectable in blood samples on newborn screening cards. This appears to indicate that a high proportion of B-precursor ALL cases arises in utero, though the wide variation in latency period to the time of clinical diagnosis of leukemia suggests that postnatal factors, including the acquisition of additional genetic “hits,” play an important role in leukemogenesis as well. Prospective studies such as those initiated by Mori et al28 of screening for TEL-AML1, t(12;21), fusion transcripts by reverse transcription–PCR in random umbilical cord blood samples and subsequent follow-up may provide further insights into the frequency of “leukemic” clones present in newborns and the mechanisms of their subsequent disappearance or emergence as leukemia.
We are indebted to Carolyn Scheer, RN, for assistance in contacting and helping to enroll patients for the study.
Supported in part by the Children's Research Center of Michigan, Children's Leukemia Foundation of Michigan, Art Gagnon Memorial Fund, BPCT Golf Charity, and Leukemia Research, Life, Inc. (Detroit, MI). Y.R. is supported by the George Ginopolis Memorial Fund.
J.W.T. and M.A.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jeffrey W. Taub, Children's Hospital of Michigan, 3901 Beaubien Blvd, Detroit, Michigan 48201; e-mail:jtaub@med.wayne.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal