Abstract
Recent studies on the immunoglobulin variable heavy chain (IgVH) genes have revealed that B-cell chronic lymphocytic leukemia (B-CLL) consists of at least 2 clinical entities with either somatically mutated or unmutated VH genes. We have analyzed the VH gene mutation status and VH gene usage in 119 B-CLL cases and correlated them to overall survival. A novel finding was the preferential use of the VH3-21 gene in mutated cases, whereas biased VH1-69 gene usage was found in unmutated cases as previously reported. Interestingly, the subset of mutated cases using the VH3-21 gene displayed distinctive genotypic/phenotypic characteristics with shorter average length of the complementarity determining region 3 and clonal expression of λ light chains. In addition, this mutated subset showed significantly shorter survival than other mutated cases and a similar clinical course to unmutated cases. We therefore suggest that B-CLL cases with mutated VH3-21 genes may constitute an additional entity of B-CLL.
Introduction
Biased immunoglobulin variable heavy-chain (IgVH) gene usage has been reported in different entities of B-cell lymphoma and in B-cell chronic lymphocytic leukemia (B-CLL).1-10 This nonrandom usage of individual VH genes has been suggested to reflect that the immunoglobulin structure may play a role in leukemia/lymphoma development, possibly through chronic antigen stimulation.
Prior studies have revealed a preferential usage of the VH1 family member, VH1-69 in B-CLL (12%-21%) compared to normal peripheral blood B cells (1.6%).3,4,7,11,12 Elevated expression of the VH1-69 gene has also been demonstrated as a feature of B-CLL with unmutated VH genes.6,11Interestingly, the VH1-69 rearrangements in B-CLL display distinctive molecular characteristics, such as skewed usage of certain D and JH genes and a significantly longer average length of the complementarity determining region (CDR) 3, which is in contrast to normal VH1-69 expressing peripheral blood B cells.4,7 13 Therefore, it has been proposed that these VH1-69 rearrangements may provide a specific immunoglobulin structure that predisposes the respective B cells to neoplastic transformation, thus suggesting that yet unknown antigens could be involved in leukemogenesis of B-CLL.
We have performed VH gene analysis in 119 B-CLL cases to study the VH gene usage in subsets of mutated and unmutated B-CLL. In addition, we have correlated the VH gene analysis to overall survival.
Study design
Patients
Tumor samples were collected from 119 patients with B-CLL, which had been identified from the archives of the Departments of Pathology and Clinical Immunology at the Uppsala University Hospital and from the Department of Pathology at the Umeå University Hospital between 1981 and 1998. Tumor material was obtained mainly from peripheral blood (64 cases) and bone marrow (44 cases), but in a few cases also from lymph nodes and spleen. According to the Royal Marsden scoring system, the tumor cells typically expressed CD5, CD19, and CD23 and showed a weak expression of immunoglobulin.14In 6 cases, CD23 expression was not analyzed.
There were 82 men and 37 women. The median age at diagnosis was 65 years and the median follow up 90 months. Survival data were obtained in 112 of 119 cases from the Swedish cancer registries in Uppsala and Umeå. Overall survival was calculated from date of diagnosis until death or last follow-up. Kaplan-Meier survival analysis and log-rank test were performed using the Statistica 5.5A software (Stat Soft, Tulsa, OK).
VH gene analysis
VH gene family-specific polymerase chain reaction (PCR) amplification was performed as previously described.15 In the majority of samples clonal PCR products were sequenced directly using the BigDye Terminator Cycle Sequencing Reaction Kit (Perkin-Elmer, ABI, Foster City, CA), but in 13 cases cloning of the PCR products was performed as described elsewhere.15 The sequences were aligned to IgH sequences from the BLAST, V-BASE, and IMGT databases. VH gene sequences deviating more than 2% from the corresponding germline gene were defined as mutated. The distribution ofreplacement mutations and silent mutations within the CDRs and framework regions was assessed according to the multinomial distribution model published by Lossos and colleagues to determine the extent of antigen selection in mutated VH genes.16 The length of the CDR3 was calculated between codon 95 and 102.
Results and discussion
VH gene mutations
A total of 134 IgH gene rearrangements from 119 cases were sequenced, including 15 cases with 2 gene rearrangements. Sixty-nine cases (58%) displayed unmutated VH genes and 50 cases (42%) showed somatically hypermutated VH genes. The median survival was significantly shorter for unmutated cases (71 months, n = 63) compared to mutated cases (124 months, n = 49;P < .001). Indeed, our finding is in line with previous studies and confirms the prognostic significance of VH gene mutational status in B-CLL.11,17,18 Using the multinomial distribution model,16 evidence for antigen selection was indicated in 20 (39.2%) of 51 mutated rearrangements, 8 of which used the VH3-21 gene.
VH gene usage in mutated and unmutated CLL
Overall, the most frequently used VH genes were VH1-69 (n = 21; 15.7%) and VH3-21 (n = 15; 11.2%). In accordance with prior studies, the VH1-69 gene was exclusively used in unmutated B-CLL and demonstrated the highest frequency accounting for 25.3% of unmutated VH genes (Table 1).3,4,7,11 A restricted JH6 gene and D2-2 gene usage was found (75.0% and 23.8%, respectively) and the mean CDR3 length was longer than expected (18.7 codons) in the cases expressing VH1-69.4,13 Our data strongly support the previously reported finding of a VH1-69–using subset with specific molecular features in B-CLL.4 13
The most frequent VH genes used in unmutated and mutated B-CLL
| VH gene usage . | Unmutated, % . | Mutated, % . |
|---|---|---|
| VH1-69 | 25.3 | — |
| VH1-2 | 7.2 | 5.9 |
| VH1-3 | 7.2 | 2.0 |
| VH3-7 | — | 7.8 |
| VH3-9 | 3.6 | 2.0 |
| VH3-21 | 2.4 | 25.5 |
| VH3-23 | — | 5.9 |
| VH3-30 | 3.6 | 7.8 |
| VH3-33 | 4.8 | — |
| VH4-34 | 6.0 | 7.8 |
| Other VHgenes | 39.9 | 35.3 |
| VH gene usage . | Unmutated, % . | Mutated, % . |
|---|---|---|
| VH1-69 | 25.3 | — |
| VH1-2 | 7.2 | 5.9 |
| VH1-3 | 7.2 | 2.0 |
| VH3-7 | — | 7.8 |
| VH3-9 | 3.6 | 2.0 |
| VH3-21 | 2.4 | 25.5 |
| VH3-23 | — | 5.9 |
| VH3-30 | 3.6 | 7.8 |
| VH3-33 | 4.8 | — |
| VH4-34 | 6.0 | 7.8 |
| Other VHgenes | 39.9 | 35.3 |
The VH3-21 gene was used mainly in the mutated (87%, n = 13) versus the unmutated (13%, n = 2) group. The restricted usage of VH3-21 in mutated CLL cases is a novel finding because there are little previous frequency data available on VH3-21 usage in normal B cells or B-CLL. Interestingly, the mutated VH3-21 rearrangements displayed shorter average length of the CDR3 (8.3 codons) compared to the unmutated VH1-69 rearrangements (18.7 codons) and the remaining of the IgH rearrangements (14.8 codons). Four of the mutated VH3-21 cases (nos. 36, 62, 75, and 113) demonstrated identical or almost identical CDR3s (Table2), though having individual VH gene mutation patterns (data not shown). In addition, all mutated VH3-21 cases expressed clonal λ light chains, which is in contrast to the expected frequency in B-CLL. Preliminary sequence data of the Igλ gene rearrangements has revealed a preferential usage of 1 of the Vλ3 family genes (V2-14). The combined usage of the VH3-21 gene and the Vλ3 gene is a new finding not previously described.
Nucleotide sequence analysis in cases using VH3-21
| Case . | VH gene . | Homology, % . | N—D—N* . | D . | JH . |
|---|---|---|---|---|---|
| 1 | VH3-21 | 96.9 | CGAAAGGAG | — | JH6 |
| 4 | VH3-21 | 99.1 | AGCGGTGATCGCA | — | JH6 |
| 11 | VH3-21 | 95.1 | CAGATCGGAACGCTT | — | JH6 |
| 36 | VH3-21 | 97.3 | GATGCAA | — | JH6 |
| 39 | VH3-21 | 97.3 | GATTATAGCAGCAGCT | D6-13 | JH4 |
| 43 | VH3-21 | 96.4 | CAGATCGTAAT | — | JH6 |
| 46 | VH3-21 | 96.4 | GATGCGAACGC | — | JH6 |
| 51 | VH3-21 | 97.3 | TCGTCGGATAGTAGTGGTG | D3-22 | JH4 |
| 62 | VH3-21 | 97.3 | GATGCAA | — | JH6 |
| 75 | VH3-21 | 97.8 | GACGCAA | — | JH6 |
| 78 | VH3-21 | 100 | GATACTCGGGAAGTCGGGCTACTATGGTTCGG | D3-10 | JH6 |
| 87 | VH3-21 | 97.3 | CGGACCGAA | — | JH6 |
| 93 | VH3-21 | 95.1 | GATGGAGCGGGGGCTATGGTTAATGT | D5-5 | JH3 |
| 103 | VH3-21 | 92.8 | GAATCCCGGTTCGTATAACTGGAACTACGGCTAATACAGGTATACT- TATTTATCGTACTATGACAC | D1-7 | JH6 |
| 113 | VH3-21 | 96.9 | GATGCAA | — | JH6 |
| Case . | VH gene . | Homology, % . | N—D—N* . | D . | JH . |
|---|---|---|---|---|---|
| 1 | VH3-21 | 96.9 | CGAAAGGAG | — | JH6 |
| 4 | VH3-21 | 99.1 | AGCGGTGATCGCA | — | JH6 |
| 11 | VH3-21 | 95.1 | CAGATCGGAACGCTT | — | JH6 |
| 36 | VH3-21 | 97.3 | GATGCAA | — | JH6 |
| 39 | VH3-21 | 97.3 | GATTATAGCAGCAGCT | D6-13 | JH4 |
| 43 | VH3-21 | 96.4 | CAGATCGTAAT | — | JH6 |
| 46 | VH3-21 | 96.4 | GATGCGAACGC | — | JH6 |
| 51 | VH3-21 | 97.3 | TCGTCGGATAGTAGTGGTG | D3-22 | JH4 |
| 62 | VH3-21 | 97.3 | GATGCAA | — | JH6 |
| 75 | VH3-21 | 97.8 | GACGCAA | — | JH6 |
| 78 | VH3-21 | 100 | GATACTCGGGAAGTCGGGCTACTATGGTTCGG | D3-10 | JH6 |
| 87 | VH3-21 | 97.3 | CGGACCGAA | — | JH6 |
| 93 | VH3-21 | 95.1 | GATGGAGCGGGGGCTATGGTTAATGT | D5-5 | JH3 |
| 103 | VH3-21 | 92.8 | GAATCCCGGTTCGTATAACTGGAACTACGGCTAATACAGGTATACT- TATTTATCGTACTATGACAC | D1-7 | JH6 |
| 113 | VH3-21 | 96.9 | GATGCAA | — | JH6 |
D sequences are underlined.
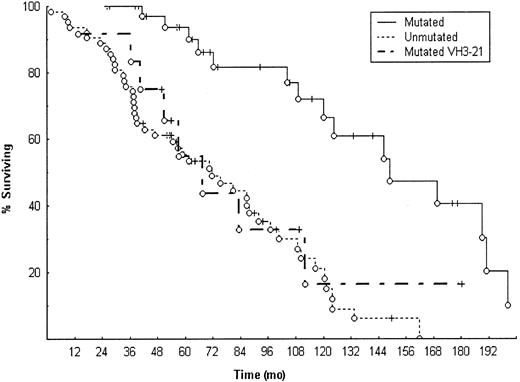
Most interestingly, the survival was significantly shorter for patients with mutated VH3-21 compared to the remaining cases with mutated VH genes (P = .018, Figure1). The median survival for mutated cases with VH3-21 gene usage (n = 12) was 63 months, which was considerably shorter than the remainder of the mutated group excluding VH3-21 (147 months, n = 37). They also showed an overall survival similar to the unmutated group (median survival, 63 versus 71 months). All together, these findings suggest that mutated VH3-21 genes may constitute an additional entity of B-CLL, displaying distinctive genotypic/phenotypic features and a significantly poorer survival than mutated cases in general. However, larger numbers of cases expressing VH3-21 have to be analyzed to further investigate the prognostic impact of VH3-21 usage in mutated B-CLL.
Survival data in unmutated or mutated B-CLL cases and cases using mutated VH3-21 genes.
Median survival for the unmutated cases (n = 63) was 71 months, mutated cases (n = 37, excluding mutated VH3-21 cases) 147 months, and mutated VH3-21 cases (n = 12) 63 months. The difference in median survival was statistically significant between unmutated versus mutated cases, P < .001, and mutated versus mutated VH3-21 cases, P = .018.
Survival data in unmutated or mutated B-CLL cases and cases using mutated VH3-21 genes.
Median survival for the unmutated cases (n = 63) was 71 months, mutated cases (n = 37, excluding mutated VH3-21 cases) 147 months, and mutated VH3-21 cases (n = 12) 63 months. The difference in median survival was statistically significant between unmutated versus mutated cases, P < .001, and mutated versus mutated VH3-21 cases, P = .018.
The VH3-21 gene has been implicated in the production of rheumatoid factors in rheumatoid arthritis.19 More than one third of cases (8 of 20) that showed signs of antigen selection used the VH3-21 gene, which may indicate selective pressure for this particular VH gene. Moreover, the short and in some cases identical CDR3s combined with a preferential Vλ3 gene usage may also suggest a unique binding of an antigen to the VH3-21 encoded Ig heavy chains. It is therefore tempting to speculate on a possible role for antigens in the development of aggressive forms of B-CLL, considering our findings of poor prognostic outcome associated with the usage of the VH1-69 gene as well as the VH3-21 gene. Hypothetically, the biased usage in B-CLL could reflect that these rearrangements encode for specific epitopes to which unknown antigen(s) could bind and stimulate proliferation. However, future studies are necessary to clarify the possible role of specific antigens in the pathogenesis of B-CLL.
The authors are grateful to Anita Lindström and Inger Eriksson for skillful technical assistance and professor Dan Holmberg for scientific advice.
Supported by grants from the Swedish Cancer Society, Lion's Cancer Research Foundation, Umeå University and Uppsala University, the Selanders Research Foundation, Uppsala University and the Research Foundation of the Department of Oncology at Uppsala University.
G.T. and U.T. have contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Richard Rosenquist, Department of Genetics and Pathology, The Rudbeck Laboratory, Uppsala University, SE-751 85 Uppsala, Sweden; e-mail: richard.rosenquist@genpat.uu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal