Members of the CCAAT/enhancer-binding protein (C/EBP) family are involved in the regulation of cellular differentiation and function of many tissues. Unlike the other members of the family, C/EBPε expression is restricted to granulocytes, macrophages, and lymphocytes. C/EBPε is highly conserved between human and rodents and is essential for terminal granulopoiesis in both species. To study the role that C/EBPε plays in macrophages, wild-type and C/EBPε–deficient (−/−) murine macrophages obtained from thioglycollate-elicited peritoneal lavages and differentiated bone marrow cells were compared. Although macrophage development occurred in both types of mice, the C/EBPε−/− cells had a lower expression of macrophage markers and a morphologic and ultrastructural appearance of immaturity. Phagocytic function, measured by calculating the percentage of internalized opsonized fluorescein isothiocyanate (FITC)–labeled yeast, was significantly impaired in the C/EBPε−/− macrophages compared with their wild-type counterparts. Furthermore, the differential expression of 26 macrophage-specific genes between wild-type and C/EBP−/− mice was analyzed. A subset of genes involved in differentiation, immune, and inflammatory responses was found down-regulated in the C/EBP−/− macrophages. Taken together, this study implicates theC/EBPε gene as an important transcription factor required for normal function and development of macrophages.
Introduction
The CCAAT/enhancer-binding protein ε (C/EBPε) is a member of the basic-leucine zipper transcription factor family. These proteins have a highly homologous C-terminal dimerization domain and a basic DNA-binding domain but differ in the N-terminal transactivation region. The 6 members of the family (C/EBPα, β, γ, δ, ε, ζ) are implicated in the control of cellular proliferation, differentiation, and function of various mammalian cells, including adipocytes, hepatocytes, and myeloid cells.1-3
In the hematopoietic system, C/EBPα expression is prominent in myeloid progenitor cells and during granulocytic differentiation. C/EBPα deletional (−/−) mice are characterized by a lack of mature granulocytes; the differentiation of monocytes and macrophages is not affected.2 4
The C/EBPβ and the C/EBPδ genes are strongly up-regulated by lipopolysaccharide (LPS) and inflammatory cytokines such as interleukin (IL)-1, IL-6, and tumor necrosis factor-α at the transcriptional level.5,6,34 C/EBPβ regulates the transcription of myelomonocytic genes; targeted inactivation of theC/EBPβ gene in the mouse results in macrophage dysfunction, impaired tumor cell killing, and a lymphoproliferative disorder.7-9 Interestingly, though in vitro data suggested that C/EBPβ regulates the expression of a number of cytokines in activated macrophages, cytokine gene expression after LPS stimulation was unchanged in macrophages from wild-type and C/EBPβ knockout mice. In addition, no defect in cytokine production has been detected in macrophages from C/EBPδ knockout mice.10
Unlike the expression of other C/EBP members, C/EBPε expression in humans and mice is restricted to the later stages of granulocyte and macrophage differentiation, and low levels are also detected in the T-lymphoid lineage.11-13,19 C/EBPε knockout mice were initially reported to have a high mortality rate from gram-negative bacterial infections within 5 months of age.14 However, they survive for 1 to 2 years when maintained under strict pathogen-free environment (personal observation). Previous studies revealed functional and maturational defects in their granulocytes and impaired T-cell proliferation.14-17 Ectopic forced expression of C/EBPε in the murine lymphoblastic cell line P388 resulted in the activation of macrophage colony-stimulating factor receptor and LPS-inducible expression of other macrophage-related genes.18 In addition, representational differential analysis using thioglycollate-recruited peritoneal myeloid cells from C/EBPε wild-type and knockout mice revealed a markedly decreased expression of macrophage products, including the chemokines monocyte chemoattractant protein (MCP)–3 and macrophage inhibitory protein 1γ (MIP-1γ), cathepsin L, and the Gal/GalNAC-specific lectin in the C/EBPε−/− mice.19 In this study, we further define the contribution C/EBPε plays in the formation of normal macrophages. As a model system, we compared wild-type and C/EBPε−/− macrophages obtained from either thioglycollate-elicited peritoneal lavage or bone marrow cells cultured in vitro with interleukin 3 (IL-3) and granulocyte macrophage-colony stimulating factor (GM-CSF).
Materials and methods
Mice
C/EBPε −/− mice were generously provided by Drs K. G. Xanthopoulos and Julie Lekstrom Himes (National Institutes of Health). Wild-type 129/SvEv × NIH Black Swiss mice were obtained from Harlan Sprague-Dawley (Indianapolis, IN). The mice were bred under pathogen-free conditions and were killed by cervical neck dislocation at 4 to 8 weeks of age.
Cell culture and cytopreparations
Cells were collected by peritoneal lavage with cold phosphate-buffered saline (PBS) at 1, 3, and 4 days after intraperitoneal injection with 2 mL 4% sterile thioglycollate. After centrifugation, peritoneal cells were resuspended in minimum essential medium α supplemented with 10% fetal calf serum and 50 μM 2-mercaptoethanol. Cytospins were prepared and stained with Wright-Giemsa and nonspecific esterase (NSE), and the percentage of neutrophils and macrophages was determined by light microscopy.
To culture bone marrow–derived macrophages, femurs were flushed using a 26-gauge needle. To remove the stromal cells from the cultured bone marrow, cells were adhered for 3 hours. Nonadherent cells were removed and cultured in the same medium supplemented with 10 ng/mL recombinant murine IL-3 and 10 ng/mL recombinant murine GM-CSF for 11 to 14 days, at which time more than 90% of the bone marrow population was macrophages. As soon as the monocytes started to adhere (6-7 days), all nonadherent cells were washed away from the culture to minimize potential interactions with other cells, such as neutrophils. Macrophages were stimulated with LPS (1 μg/mL), or LPS plus interferon (IFN)-γ (100 U/mL). Cytospins of the cultured bone marrow cells were stained with Wright-Giemsa, NSE (α-naphthyl butyrate esterase), Sudan black, oil-red O, and periodic acid-Schiff with and without diastase and Alcian blue.
Human bone marrow samples were obtained with informed consent from healthy volunteers. Cells were cultured in RPMI media supplemented with 20% fetal calf serum and recombinant human 20 ng/mL GM-CSF for 14 days to induce macrophage differentiation and then were harvested.
Reverse transcription–polymerase chain reaction and Southern blot analysis
Total RNA was extracted with Trizol according to the manufacturer's protocol (Gibco/BRL, Gaithersburg, MD). Three micrograms RNA was treated with RNase-free DNase (1 U; Promega, Madison, WI) and was reverse transcribed with avian myeloblastosis virus RT (Promega) for 60 minutes at 42°C.
Semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed using the following conditions: an initial denaturation step at 94°C for 5 minutes followed by 25 to 35 cycles, 94°C for 30 seconds, 56°C for 40 seconds, and 72°C for 1 minute. Sequences of the primers used in this study are shown in Table1. 18S rRNA was used to ensure the integrity of the cDNA and to normalize the expression of the test genes. For Southern blotting, PCR products were transferred overnight with NaOH (0.4 M) to a nylon membrane (Hybond; Amersham, Arlington Heights, IL), and the filters were probed using internal oligonucleotides for IL-10 (CCAGT- TTTACCTGGTAGAAGTGATG), IL12-p35 (CTGATGCAGTCTCTGAATCATAATG), IL12-p40 (TGAAGTTCAACATCAAGAGCAGTAG), IL6 (AGGCTTAATTACACATGTTCTCTGG), C/EBPε (AAGTGGCACACTGTGGGCAGAC), and 18S (GCAGGCGCGCAAATTACCCA).
Sequence of primers used in RT-PCR assays
| Gene . | Sense primer . | Antisense primer . |
|---|---|---|
| hC/EBPε | CAGACAGGAAGGCGCTGGG | CGGCAGTGGCCAAAGGGGCCTT |
| mC/EBPε | GCTACAATCCCCTGCAGTACC | CACAAGGGCAAGAAGGCA |
| CD14 | CCCGACCCTCCAAGTTTT | ACCTGCTTCAGCCCAGTG |
| Mona | TCTGCAGCATTTCCACCA | GGAATCCCAGCTCGTCCT |
| Fcγ RI | TCGGTGGGGAAGTGGTTA | CTGGCCTCTGGGATGCTA |
| PAI-2 | TTGAGGACGCATCCACTG | GCCTTGCCCTTGTTGAAG |
| IRK1 | CTTGCCTTCGTGCTCTCC | TTGGGCACTCGTCTGTCA |
| VLA5 (α) | GCAGCTGCATTTCCGAGT | CCACAACGGGACACCATT |
| VLA5 (β) | AGCACAACCCCAGCAAAG | GATCCACAAACCGCAACC |
| IL-12p40 | TCATGGCTGGTGCAAAGA | AGGGTCTCCTCGGCAGTT |
| IL-12p35 | AGTTCCAGGCCATCAACG | AAGGCGTGAAGCAGGATG |
| IL-18 | CAGTGAACCCCAGACCAGAC | TCAGGTGGATCCATTTCCTC |
| IL-15 | CTGAGGCTGGCATTCATGT | GCAATTCCAGGAGAAAGCAG |
| IL-6 | GTGACAACCACGGCCTTC | TTCTGCAAGTGCATCATCG |
| IL-10 | TTGGGTTGCCAAGCCTTA | ACCTGCTCCACTGCCTTG |
| MCP-3 | TTCTGTGCCTGCTGCTCA | GCTTCAGCGCAGACTTCC |
| M-CSF | GCCAGACCCTCGAGTCAA | CGTGGAGGGGGAAAACTT |
| c-fms | CCGGGCTCTCAGTCTCAA | CATCTGCATGGACCGTGA |
| MCP-1 | AGCCAATCAGCCATCTGC | CTCTATGGCCTGCGGTGT |
| c-maf | AGGCGGACCCTGAAAAAC | TTTTCTCGGAAGCCGTTG |
| IL-1RA | CCCTGCCACAAACACACA | ATGTGATGCCCTGGTGGT |
| NOS | ACGCTGGCTACCAGATGC | GGGATGCTCCATGGTCAC |
| TNF-α | GTGCTCCTCACCCACACC | GGGCAGATTGACCTCAGC |
| IP-10 | ACGGGCCAGTGAGAATGA | TGTGTGCGTGGCTTCACT |
| ICSBP | ACCAAGAGGAGCCCATCC | TTCAAGGCAGGTGGTGGT |
| MR | CGGCATGGGTTTTACTGC | CACCGAAACGTCCCTTTG |
| SR type 1 | ATTGGCTTCCCTGGAGGT | CACTGGCCTTGGTGGAAG |
| SR type 2 | GGGAGTGTAGGCGGATCA | GCATGGCATGACACAGGA |
| 18S | AAACGGCTACCACATCCAAG | CCTCCAATGGATCCTCGTTA |
| Gene . | Sense primer . | Antisense primer . |
|---|---|---|
| hC/EBPε | CAGACAGGAAGGCGCTGGG | CGGCAGTGGCCAAAGGGGCCTT |
| mC/EBPε | GCTACAATCCCCTGCAGTACC | CACAAGGGCAAGAAGGCA |
| CD14 | CCCGACCCTCCAAGTTTT | ACCTGCTTCAGCCCAGTG |
| Mona | TCTGCAGCATTTCCACCA | GGAATCCCAGCTCGTCCT |
| Fcγ RI | TCGGTGGGGAAGTGGTTA | CTGGCCTCTGGGATGCTA |
| PAI-2 | TTGAGGACGCATCCACTG | GCCTTGCCCTTGTTGAAG |
| IRK1 | CTTGCCTTCGTGCTCTCC | TTGGGCACTCGTCTGTCA |
| VLA5 (α) | GCAGCTGCATTTCCGAGT | CCACAACGGGACACCATT |
| VLA5 (β) | AGCACAACCCCAGCAAAG | GATCCACAAACCGCAACC |
| IL-12p40 | TCATGGCTGGTGCAAAGA | AGGGTCTCCTCGGCAGTT |
| IL-12p35 | AGTTCCAGGCCATCAACG | AAGGCGTGAAGCAGGATG |
| IL-18 | CAGTGAACCCCAGACCAGAC | TCAGGTGGATCCATTTCCTC |
| IL-15 | CTGAGGCTGGCATTCATGT | GCAATTCCAGGAGAAAGCAG |
| IL-6 | GTGACAACCACGGCCTTC | TTCTGCAAGTGCATCATCG |
| IL-10 | TTGGGTTGCCAAGCCTTA | ACCTGCTCCACTGCCTTG |
| MCP-3 | TTCTGTGCCTGCTGCTCA | GCTTCAGCGCAGACTTCC |
| M-CSF | GCCAGACCCTCGAGTCAA | CGTGGAGGGGGAAAACTT |
| c-fms | CCGGGCTCTCAGTCTCAA | CATCTGCATGGACCGTGA |
| MCP-1 | AGCCAATCAGCCATCTGC | CTCTATGGCCTGCGGTGT |
| c-maf | AGGCGGACCCTGAAAAAC | TTTTCTCGGAAGCCGTTG |
| IL-1RA | CCCTGCCACAAACACACA | ATGTGATGCCCTGGTGGT |
| NOS | ACGCTGGCTACCAGATGC | GGGATGCTCCATGGTCAC |
| TNF-α | GTGCTCCTCACCCACACC | GGGCAGATTGACCTCAGC |
| IP-10 | ACGGGCCAGTGAGAATGA | TGTGTGCGTGGCTTCACT |
| ICSBP | ACCAAGAGGAGCCCATCC | TTCAAGGCAGGTGGTGGT |
| MR | CGGCATGGGTTTTACTGC | CACCGAAACGTCCCTTTG |
| SR type 1 | ATTGGCTTCCCTGGAGGT | CACTGGCCTTGGTGGAAG |
| SR type 2 | GGGAGTGTAGGCGGATCA | GCATGGCATGACACAGGA |
| 18S | AAACGGCTACCACATCCAAG | CCTCCAATGGATCCTCGTTA |
Mona indicates monocytic adaptor; FcγRI, Fcγ receptor type I; PAI-2, plasminogen activator inhibitor type-2; IRK1, inwardly rectifying potassium channel; VLA5, fibronectin receptor; c-fms, macrophage colony-stimulating factor receptor; IL-1RA, IL-1 receptor antagonist; NOS, nitric oxide synthase; TNF-α, tumor necrosis factor-α; IP-10, IFN-γ inducible protein 10; ICSBP, IFN consensus sequence-binding protein; MR, mannose receptor; SR, scavenger receptor.
Probes were random primed labeled with α-32P–adenosine triphosphate (ATP) using a Strip EZ DNA kit (Ambion, Austin, TX). Membranes were hybridized overnight at 65°C in Rapid-Hyb buffer (Amersham).
Northern blot analysis
Ten micrograms peritoneal lavage RNA from wild-type and C/EBPε −/− mice was electrophoresed on a denaturing formaldehyde gel and blotted in 20 × SSC overnight to a nylon membrane (Magna Charge; Micron Separations, Westborough, MA). Blots were hybridized for 16 hours at 42°C in Ultra-Hyb buffer (Ambion) and α-32P–dATP-labeled cDNA probes for MCP-3, CD14, M-CSF, plasminogen-activator inhibitor type 2 (PAI-2), and mannose receptor. Washes after hybridization were repeated twice with 2 × SSC and 0.1% sodium dodecyl sulfate (SDS) followed by 2 washes with 0.2 × SSC and 0.1% SDS at 65°C for 20 minutes each. The probes were generated by PCR using the primer pairs described in Table 1.
Western blot analysis
Total cell lysate (30 μg protein) was mixed with an equal volume of 2 × Laemmli sample buffer, boiled for 5 minutes, and electrophoresed on a 10% to 20% SDS polyacrylamide gel (Bio-Rad, Hercules, CA). Proteins were transferred by electroblotting overnight to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Bedford, MA). Membranes were blocked with 5% nonfat dry milk in TBS-T buffer for 1 hour and were incubated with a primary antibody (1 μg/mL; Santa Cruz Biotechnology, Santa Cruz, CA) followed by a secondary horseradish peroxidase–conjugated antibody (Amersham). Detection was performed using an enhanced chemiluminescence detection kit (Pierce, Rockford, IL). To ensure equal loading of cell lysates, the membranes were stripped and reprobed with murine anti–rabbit GAPDH antibody (1:4000 dilution; RDI, Flanders, NJ).
Immunofluorescence analysis
Bone marrow macrophages were washed once with PBS, resuspended in 50 μL staining buffer (minimum essential medium α with 1% fetal bovine serum, 0.1% sodium azide) and incubated with either a phycoerythrin-conjugated rat monoclonal antibody against murine F4/80 (Caltag Laboratories, Burlingame, CA) or an FITC-conjugated antibody against murine CD14 (PharMingen, San Diego, CA) for 30 minutes at 4°C in the dark. After incubation, cells were washed in staining buffer, fixed with 2% paraformaldehyde, and analyzed by flow cytometry.
Enzyme-linked immunosorbent assay
After 10 days of culture, bone marrow–derived macrophages were incubated with culture medium containing varying concentrations of LPS. Cells were counted, and culture supernatants were collected after 12-hour incubation. Secreted IL-18 was quantitated by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (R&D Systems, Minneapolis, MN).
Phagocytosis assay
Phagocytosis of yeast was based on a previously described method.21 Briefly, Candida albicans blastopores (5 × 107 blastospore cells/mL) were ethanol fixed, labeled by incubation with 0.01 mg/mL FITC, and stored at −80°C until use. Opsonization was achieved by suspension of 200 μL wild-type mouse serum with 106 blastospores for 1 hour at 37°C. For the phagocytosis assay, opsonized FITC-labeled blastopores were incubated with either bone marrow– or peritoneal lavage–derived macrophages in a 1:5 ratio. After 1 hour, phagocytosis was stopped by 2 washes with PBS containing 0.02% EDTA. Cells were fixed with 4% paraformaldehyde and were stained with 75% trypan blue to distinguish between internalized C albicans blastopores that remained green versus adherent, noninternalized blastopores that stained blue. The percentage of cells ingesting yeast and the amount of yeast in each cell was determined by counting 500 cells per sample.
Electron microscopy
Thioglycollate-elicited peritoneal lavage and bone marrow–derived macrophages were collected, suspended in cold PBS, and pelleted by centrifugation. The pellet was overlaid with 2.5% glutaraldehyde in cacodylate buffer, washed, and postfixed in 1.0% osmium tetroxide. After dehydration with graded alcohols, the pellets were embedded in Eponate 12 (Ted Pella, Redding, CA). Thick sections were stained with methylene blue–azure 2, and thin sections were stained with uranyl acetate and lead citrate.
Results
C/EBPε expression in human and murine bone marrow–derived macrophages
Although high levels of C/EBPε mRNA were previously detected in murine macrophages, expression in human macrophages has not been clearly shown.20 13 Therefore, using RT-PCR and Southern blot analysis, we examined the expression of C/EBPε in human macrophages and in murine mature bone marrow–derived macrophages with and without IFN-γ and LPS. An easily detected basal level of expression of C/EBPε transcripts was demonstrated in human and murine macrophages, but no change in expression was observed after stimulation of the murine macrophages. As expected, macrophages from the C/EBPε−/− mice had no C/EBPε (Figure 1.)
C/EBPε expression in bone marrow–derived macrophages.
RNA was harvested from murine wild-type and C/EBPε−/− bone marrow–derived macrophages with and without LPS (1 μg/mL) plus IFN-γγg (100 U/mL) stimulation and from human bone marrow macrophages after culture with 20 ng/mL rGM-CSF. RT-PCR–amplified products from human and mouse were blotted and hybridized using a highly conserved internal oligonucleotide probe.
C/EBPε expression in bone marrow–derived macrophages.
RNA was harvested from murine wild-type and C/EBPε−/− bone marrow–derived macrophages with and without LPS (1 μg/mL) plus IFN-γγg (100 U/mL) stimulation and from human bone marrow macrophages after culture with 20 ng/mL rGM-CSF. RT-PCR–amplified products from human and mouse were blotted and hybridized using a highly conserved internal oligonucleotide probe.
C/EBPε is required for GM-CSF– and IL-3–induced macrophage differentiation
Murine bone marrow cells were cultured in the presence of IL-3 and GM-CSF for at least 10 days and then were cultured for 24 hours without or with LPS or IFN-γ plus LPS. NSE staining revealed that more than 90% of cells from the C/EBPε−/− and the wild-type mice were differentiated toward macrophages (Figure2). However, clear differences were noted between the 2 populations. The C/EBPε−/−-derived macrophages stained less prominently with NSE, indicating a reduced enzymic activity, and had more intracytoplasmic vacuoles than wild-type cells (Figure 2B-C). To evaluate the nature of these vacuoles, additional stains were performed. No staining of these cells occurred with periodic acid-Schiff with or without diastase and Alcian blue, suggesting that the vacuoles were negative for glycogen and mucin, respectively. Oil-red O and Sudan black stains indicated an excessive accumulation of lipids in a number of knockout mice macrophages (data not shown). After the marrow cells were cultured in GM-CSF and IL-3 for 14 days, markedly more multinucleated giant cells were observed for the C/EBPε−/− mice than for the wild-type mice (mean percentage multinucleated giant cells ± SD, wild-type 3% ± 0.89, knockout 14% ± 3.69; P < .00054 unpaired, 2-sided t test) (Figure 2A-C). Similar morphologic differences between bone marrow–derived macrophages from wild-type and C/EBP−/− mice were observed before and after stimulation of the cells (Figure 2B-C).
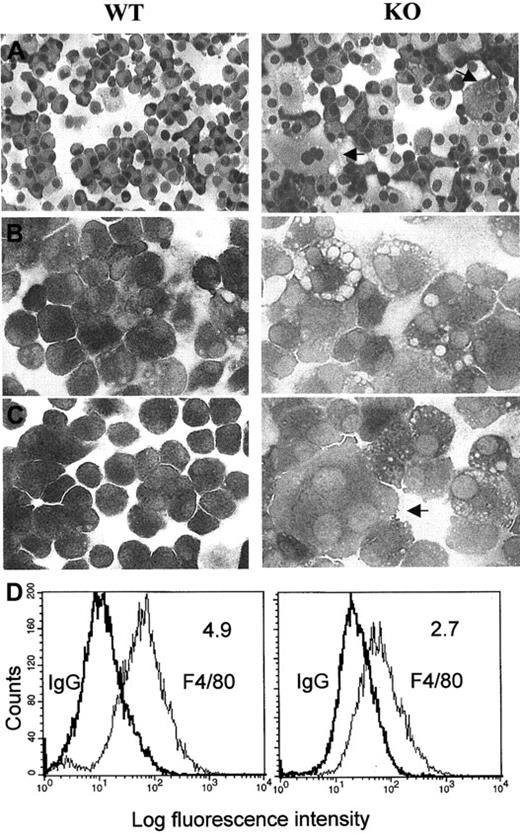
Morphologic characteristics and expression of the macrophage-specific marker F4/80 in macrophages from wild-type (WT) and C/EBPε−/− (KO) mice.
Cytocentrifugation preparations from cultured bone marrow macrophages stained with (A) Wright-Giemsa stain (original magnification, ×200), (B) nonspecific esterase (unstimulated cells, original magnification, ×400), and (C) nonspecific esterase (after stimulation of the cells for 24 hours with LPS). Arrows indicate large multinucleated macrophages (after culturing C/EBPε−/− cells in the presence of rIL-3 and rGM-CSF for 11 to 14 days. These photomicrographs are representative of the results obtained from 3 independent experiments. (D) Histograms of total fluorescence binding of phycoerythrin-conjugated F4/80 (right peak) versus isotype-matched control antibody samples (left peak). Mean fluorescence intensity ratios (total fluorescence divided by control) are shown in the upper right-hand corner of each histogram.
Morphologic characteristics and expression of the macrophage-specific marker F4/80 in macrophages from wild-type (WT) and C/EBPε−/− (KO) mice.
Cytocentrifugation preparations from cultured bone marrow macrophages stained with (A) Wright-Giemsa stain (original magnification, ×200), (B) nonspecific esterase (unstimulated cells, original magnification, ×400), and (C) nonspecific esterase (after stimulation of the cells for 24 hours with LPS). Arrows indicate large multinucleated macrophages (after culturing C/EBPε−/− cells in the presence of rIL-3 and rGM-CSF for 11 to 14 days. These photomicrographs are representative of the results obtained from 3 independent experiments. (D) Histograms of total fluorescence binding of phycoerythrin-conjugated F4/80 (right peak) versus isotype-matched control antibody samples (left peak). Mean fluorescence intensity ratios (total fluorescence divided by control) are shown in the upper right-hand corner of each histogram.
Flow cytometric analysis for expression of the macrophage-specific marker F4/80 on macrophages from the wild-type and knockout bone marrow–derived cells showed that each population had 90% to 99% cells that expressed the antigen. However, the mean fluorescence intensity per cell relative to the immunoglobulin G control was significantly reduced in the C/EBPε−/− macrophages (Figure 2D). In addition, FACS analysis indicated a 50% decrease in the expression of CD14 on the C/EBPε−/− macrophages (data not shown). These data suggest that the C/EBPε-deficient macrophages displayed signs of immaturity.
Ultrastructural analysis by electron microscopy
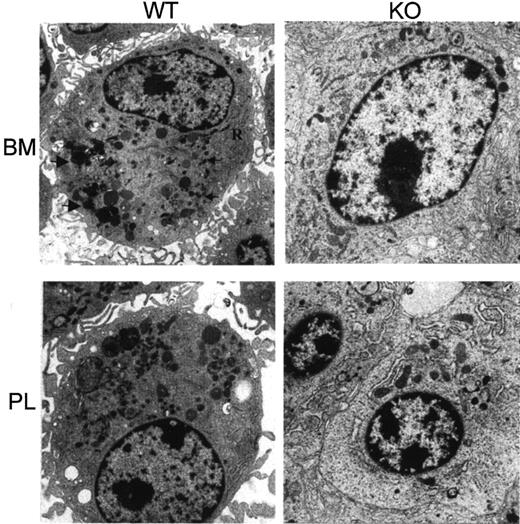
For further study of the morphologic changes, ultrastructural examination was undertaken of bone marrow–derived macrophages and thioglycollate-elicited peritoneal lavage macrophages. Compared with wild type, the impaired development of secondary lysosomes and rough endoplasmic reticulum and a decreased number of primary granules was observed in the C/EBPε−/− mice (Figure3). These changes between wild-type and C/EBPε−/− mice were more prominent in bone marrow–cultured macrophages.
Electron micrograph of macrophages from wild-type and C/EBPε knockout mice.
(Upper panel) Wild-type bone marrow (BM)–derived macrophages that have abundant rough endoplasmic reticulum (R) and numerous primary (small arrowheads) and larger secondary lysosomes (large arrowheads) (×6000). In contrast, the knockout macrophages have fewer cisternae of rough endoplasmic reticulum and a paucity of secondary lysosomes (×8050). (Lower panel) Ultrastructure of wild-type thioglycollate-elicited macrophages (PL) and C/EBPε knockout cells (right)
Electron micrograph of macrophages from wild-type and C/EBPε knockout mice.
(Upper panel) Wild-type bone marrow (BM)–derived macrophages that have abundant rough endoplasmic reticulum (R) and numerous primary (small arrowheads) and larger secondary lysosomes (large arrowheads) (×6000). In contrast, the knockout macrophages have fewer cisternae of rough endoplasmic reticulum and a paucity of secondary lysosomes (×8050). (Lower panel) Ultrastructure of wild-type thioglycollate-elicited macrophages (PL) and C/EBPε knockout cells (right)
Macrophages from C/EBPε−/− mice have impaired phagocytic activity
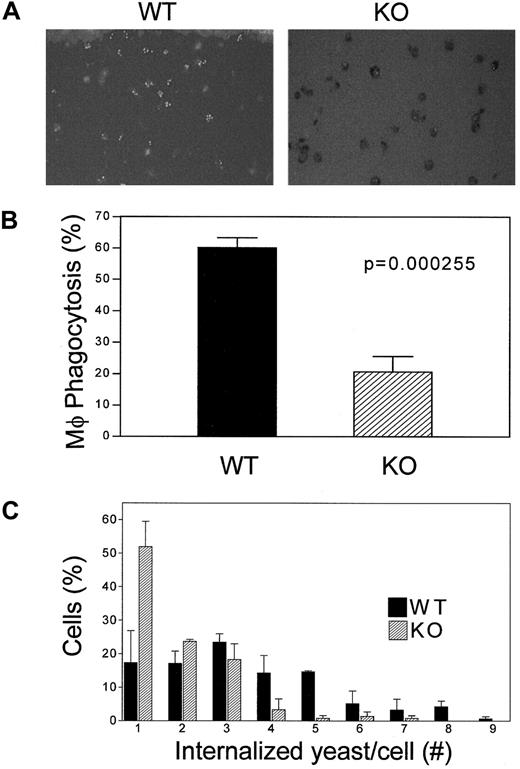
A critical function of macrophages is their ability to phagocytose. To monitor this activity, thioglycollate-elicited peritoneal lavage– and bone marrow–derived macrophages were assessed for their ability to engulf FITC-labeled opsonized C albicans blastopores (Figure 4A). After incubation at 37°C for 1 hour, the mean percentage of phagocytosing cells was markedly lower in C/EBPε−/− peritoneal and bone marrow–derived macrophages than in wild-type macrophages. The mean phagocytic activity of bone marrow–derived macrophages was 58.2% in wild-type animals versus 20.5% in the C/EBPε−/− mice (P < .0002, unpaired, 2-sided t test). A similar reduction was observed in the peritoneal lavage–derived macrophages from C/EBPε−/− mice compared with the wild-type controls (data not shown). Furthermore, the phagocytic capacity, represented by the number of yeast internalized per cell, was decreased in C/EBPε−/− macrophages compared with their wild-type counterparts (Figure 4C).
Analysis of phagocytosis of
C albicans by C/EBP−/− macrophages. Macrophages from thioglycollate-elicited peritoneal lavage and bone marrow were incubated with FITC-labeled opsonized yeast. Cells were stained with trypan blue to distinguish between internalized C albicans blastopores that remained green and nonphagocytosed blastopores that merely adhered to the outer surfaces of the macrophages. (A) Cells were visualized with the use of fluorescence microscopy. (B) Phagocytosis activity of macrophages from bone marrow was calculated as the ratio of cells with fluorescence yeast to the number of total macrophages. (C) Number of internalized C albicans blastopores in each thioglycollate-elicited peritoneal lavage macrophage is shown. (▪) Wild-type macrophages; (▨) knockout macrophages. Both histograms represent the average of 3 separate experiments; each included the analysis of 2 wild-type and 2 knockout mice.
Analysis of phagocytosis of
C albicans by C/EBP−/− macrophages. Macrophages from thioglycollate-elicited peritoneal lavage and bone marrow were incubated with FITC-labeled opsonized yeast. Cells were stained with trypan blue to distinguish between internalized C albicans blastopores that remained green and nonphagocytosed blastopores that merely adhered to the outer surfaces of the macrophages. (A) Cells were visualized with the use of fluorescence microscopy. (B) Phagocytosis activity of macrophages from bone marrow was calculated as the ratio of cells with fluorescence yeast to the number of total macrophages. (C) Number of internalized C albicans blastopores in each thioglycollate-elicited peritoneal lavage macrophage is shown. (▪) Wild-type macrophages; (▨) knockout macrophages. Both histograms represent the average of 3 separate experiments; each included the analysis of 2 wild-type and 2 knockout mice.
Mice with targeted disruption of C/EBPε have a decreased expression of macrophage-specific genes
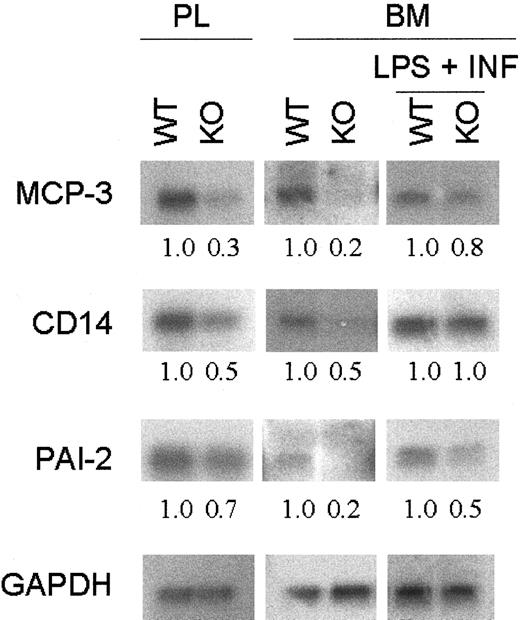
The expression of 26 macrophage-specific genes from bone marrow–derived macrophages and from peritoneal lavage after thioglycollate injection was compared between wild-type and C/EBPε−/− mice. Northern blot analysis revealed differential expression in a subset of these genes. In particular, transcripts for a group of genes that participate in the regulation of the inflammatory response, such as CD14, MCP-3, and PAI-2, had decreased expression in the C/EBPε-deficient macrophages (Figure5). After LPS plus IFN-γ stimulation, the differential expression observed between wild-type and C/EBPε−/− cells for PAI-2 and MCP-3 was reduced but still present. However, differential expression for CD14 was no longer evident.
Expression of macrophage-related genes.
Total RNA was harvested from wild-type and C/EBPε-deficient mice peritoneal lavage (PL) and bone marrow–derived macrophages with and without stimulation with LPS + IFN-γ. Northern blot analysis was performed on 10 μg total RNA. Hybridization with GAPDH confirmed equivalent loading of RNA. Fold change was measured by densitometry and was normalized with the GAPDH ratio.
Expression of macrophage-related genes.
Total RNA was harvested from wild-type and C/EBPε-deficient mice peritoneal lavage (PL) and bone marrow–derived macrophages with and without stimulation with LPS + IFN-γ. Northern blot analysis was performed on 10 μg total RNA. Hybridization with GAPDH confirmed equivalent loading of RNA. Fold change was measured by densitometry and was normalized with the GAPDH ratio.
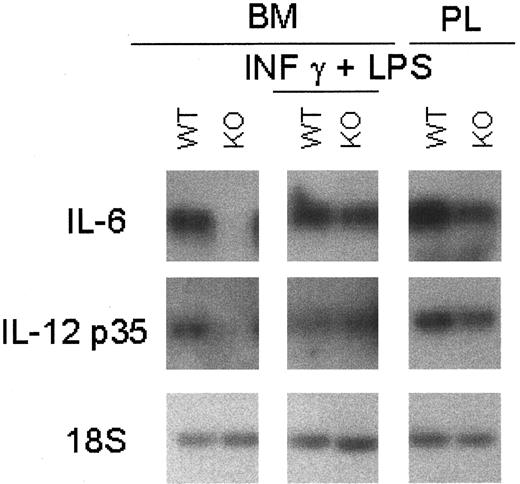
Macrophage-mediated cytokines were also examined with or without stimulation by IFN-γ and LPS. The level of IL-6 mRNA, a cytokine involved in the regulation of granulopoiesis and macrophage differentiation, was less abundant in the unstimulated C/EBPε-deficient macrophages (Figure6). Stimulation with LPS and IFN-γ resulted in macrophages from C/EBPε−/− and wild-type mice having comparable levels of IL-6 mRNA.
Cytokine mRNA expression in C/EBPε−/− macrophages.
Macrophages were harvested from PL 24 hours after thioglycollate challenge and from bone marrow cells cultured for 13 days with and without stimulation with 2-hour LPS (1 μg/mL) plus IFN-γ (100 U/mL) stimulation. Macrophage-mediated cytokines were measured by RT-PCR because of the low basal level of expression. PCR products were blotted and hybridized using internal oligonucleotide 32P-labeled probes specific for the amplified genes.
Cytokine mRNA expression in C/EBPε−/− macrophages.
Macrophages were harvested from PL 24 hours after thioglycollate challenge and from bone marrow cells cultured for 13 days with and without stimulation with 2-hour LPS (1 μg/mL) plus IFN-γ (100 U/mL) stimulation. Macrophage-mediated cytokines were measured by RT-PCR because of the low basal level of expression. PCR products were blotted and hybridized using internal oligonucleotide 32P-labeled probes specific for the amplified genes.
Previous studies suggested that in the presence of C/EBPε−/− macrophages, wild-type murine T lymphocytes activated with anti-CD3 monoclonal antibody, schistosome egg antigen (SEA), or concanavalin A have impaired proliferative responses.17 To explore this observation further, macrophage cytokines important for the regulation and survival of activated T cells were investigated. IL-12 is a pro-inflammatory and an immunoregulatory cytokine critical for Th1 cell response. Bioactive IL-12 (p70) is composed of 2 subunits (p35 and p40) encoded by 2 separated genes. mRNAs encoding these IL-12 subunits were expressed at lower levels in peritoneal and bone marrow–derived −/− macrophages (Figure 6). Stimulation of the C/EBPε−/− and wild-type macrophages with LPS plus IFN-γ resulted in equivalent levels of IL-12 mRNA subunits (Figure 6).
The cytokine IL-18 is important for the function of Th1 cells and, in conjunction with IL-12, synergistically enhances the production of IFN-γ by NK and Th1 cells.25 Analysis of protein expression by ELISA (Figure 7A) and Western blotting (Figure 7B) showed a consistent reduction of IL-18 levels in macrophages from C/EBPε−/− mice compared with those in wild-type animals.
IL-18 production by C/EBPε−/− bone marrow macrophages before and after LPS (100 ng/mL) stimulation.
After 10 days of culturing, the macrophages were extensively washed and incubated for 24 hours in fresh medium containing LPS. Cells were harvested and counted, and IL-18 secretion in the culture medium was quantitated by ELISA. (B) Whole cell protein extracts were prepared from these cells and analyzed by Western blotting using a polyclonal anti–IL-18 antibody. Fold change was measured by densitometry. Blots were probed with anti–GAPDH antibody as a control.
IL-18 production by C/EBPε−/− bone marrow macrophages before and after LPS (100 ng/mL) stimulation.
After 10 days of culturing, the macrophages were extensively washed and incubated for 24 hours in fresh medium containing LPS. Cells were harvested and counted, and IL-18 secretion in the culture medium was quantitated by ELISA. (B) Whole cell protein extracts were prepared from these cells and analyzed by Western blotting using a polyclonal anti–IL-18 antibody. Fold change was measured by densitometry. Blots were probed with anti–GAPDH antibody as a control.
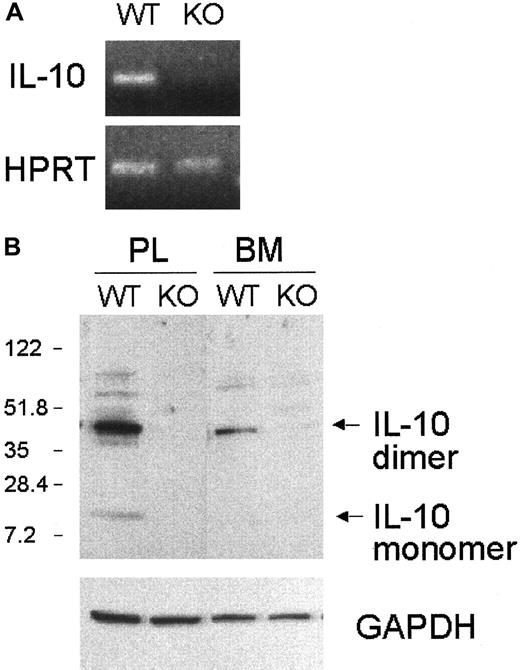
Interestingly, the expression of IL-10, a cytokine that has potent anti-inflammatory and immunosuppressive activity, was absent in the C/EBPε−/− peritoneal macrophages (Figure8A). Additionally, IL-10 protein expression was undetectable in the bone marrow and peritoneal macrophages from the C/EBPε−/− mice (Figure 8B).
IL-10 mRNA and protein expression in C/EBPε−/− macrophages.
(A) RT-PCR was performed on cDNA from peritoneal lavages of wild-type and C/EBPε−/− mice. (B) Total cell lysates were obtained from bone marrow and peritoneal lavage 72 hours after thioglycollate injection. Western blotting was performed using polyclonal anti–IL-10 antibody and anti–GAPDH antibody (control).
IL-10 mRNA and protein expression in C/EBPε−/− macrophages.
(A) RT-PCR was performed on cDNA from peritoneal lavages of wild-type and C/EBPε−/− mice. (B) Total cell lysates were obtained from bone marrow and peritoneal lavage 72 hours after thioglycollate injection. Western blotting was performed using polyclonal anti–IL-10 antibody and anti–GAPDH antibody (control).
Discussion
During the last several decades, a number of studies identified many of the key antimicrobial proteins made by macrophages, which are critical for the prevention of uncontrolled infections. Recently, others and we have begun to define the transcriptional factors that control production of these proteins.7-9,26,27,29,34,35Analyses of animals that have deletion of these transcription factors are a powerful tool to define their relevance in host defense. C/EBPε−/− mice die of overwhelming infections after 3 to 5 months of birth, pointing to the importance of this transcription factor in innate immunity.14 This study begins to define those proteins known to play a critical role in host defense that are regulated by C/EBPε. We found that targeted disruption of C/EBPε leads to macrophage defects in maturation and phagocytosis and in decreased expression of cytokines and other proteins activated during the inflammatory response.
C/EBPε−/− macrophages derived from bone marrow cells cultured with IL-3 and GM-CSF had several morphologic differences compared with the wild-type macrophages, including the formation of multinucleated giant cells. Previously, studies showed that by labeling cell membranes with a fluorochrome, monocytes could fuse with each other after their stimulation with cytokine-containing media but that mature macrophages lost this capacity.30 The higher fusion rates observed in the C/EBPε−/− cells might reflect a defect of macrophage maturation. Other morphologic changes included increased intracytoplasmic vacuoles, some of which contained lipids. Electron microscopy disclosed fewer secondary lysosomes and less prominent rough endoplasmic reticulum and Golgi apparatus in the C/EBPε-deficient macrophages than in wild-type macrophages. These ultrastructural changes suggested a maturation defect in the C/EBPε−/− macrophages that correlated with the attenuated phagocytosis observed in these cells.
Phagocytosis is one of the main effector mechanisms of innate immunity that is activated shortly after infection, and it helps control the replication of the infecting pathogen. The process of phagocytosis is accompanied by the production of a variety of macrophage-related cytokines and costimulatory molecules essential to the adaptive immune response by T and B lymphocytes.26 C/EBPε−/−-derived macrophages displayed impaired internalization of opsonized yeast and generation of phagolysosomes, a process essential for the killing and degradation of phagocytized microorganisms. However, no change was seen in the expression of the receptors known to be involved in the uptake of the microorganisms, such as the mannose receptor, FcγRI, and scavenger receptor types 1 and 2.28 29
Targeted disruption of C/EBPε resulted in macrophages that had diminished expression of genes known to be associated with differentiation of these cells. Normally, CD14 and PAI-2 mRNA levels increase during the differentiation of monocytes to macrophages. PAI-2 serves as a primary regulator of plasminogen activation in the extravascular compartment.23 Responsiveness to LPS (endotoxin) from gram-negative bacteria involves CD14, an integrin receptor expressed on the macrophage surface, which is anchored to the cell surface and binds to the complex of LPS and LPS-binding protein.31,32 Analysis of the CD14 promoter region in vitro demonstrated that C/EBPα binds the promoter in relatively undifferentiated U937 and THP-1 cell lines. However, on differentiation toward monocytic cells with exposure to 1,25 dihydroxy vitamin D3 [1,25(OH)2D3] and transforming growth factor-β treatment, the expression of CD14 was highly up-regulated.33 This paralleled an increase of C/EBP binding to the CD14 promoter. Electrophoretic mobility shift assays indicated that the DNA-C/EBP complex consisted primarily of C/EBPα and C/EBPβ.33 Our studies suggest that in vivo, C/EBPε is also involved in the regulation of basal CD14 levels in macrophages. On the other hand, we did not observe a decrease in the expression of CD14 levels in C/EBPε−/− macrophages after exposure of the cells to LPS and IFN-γ. These data suggest that after stimulation, other members of the C/EBP family can substitute for C/EBPε, possibly as a result of an increase in their expression under these conditions.10,16 34
IL-6 is another gene that contains functional C/EBP-binding sites in its promoter region. Previously, forced expression of C/EBPβ or C/EBPε was shown to be capable of enhancing LPS-inducible expression of IL-6 in the P388 cell line.20,22 Disruption of C/EBPβ in mice, however, leads to an increase in IL-6 production, suggesting that C/EBPβ might even act in vivo as a repressor of IL-6 expression.8 9 Our study shows that C/EBPε is an essential transcription factor for the expression of theIL-6 gene in quiescent macrophages. Therefore, though promoter studies in vitro have shown redundancy in the ability of different members of the C/EBP family to activate target genes, experiments in knockout mice have verified the importance of individual members of the C/EBP family at specific stages of differentiation. The reduction of expression of the CD14 and IL-6genes in the C/EBPε−/− macrophages indicates an overlapping but distinct pattern of cell-, stage-, and stimulus-specific regulation of gene expression by the C/EBP family members.
We have shown that the T-cell receptor–mediated proliferation of T cells was impaired in C/EBPε−/− mice.17 Tissue culture supernatant from concanavalin A–activated C/EBPε−/− splenocytes, but not their wild-type counterparts, partially inhibited or did not support the proliferation of wild-type T cells. In addition, spleen cells from C/EBPε knockout mice expressed lower levels of mRNAs encoding IFN-γ, IL-4, IL-12–p40, and IL-2 than the wild-type counterparts.14 We hypothesized that the C/EBPε knockout mice have a defect in the expression of macrophage-specific genes important in the regulation of T cells. To test this hypothesis, we measured the basal and post–IFN-γ and LPS stimulatory levels of the transcripts coding for several of these key cytokines, including IL-10, IL-12, and IL-18, in C/EBPε+/+ and −/− macrophages.
During the innate immune response, IL-12 is produced primarily by monocytes and macrophages. Together, IL-12 and IL-18 promote the Th1 cell response and induce IFN-γ production from NK and T cells. mRNAs encoding the 2 subunits of IL-12 (IL-12–p35 and IL-12–p40) were expressed at lower levels in the nonstimulated C/EBPε−/− macrophages. In addition, decreased production of IL-18 protein was detected in the macrophages of the C/EBPε knockout mice. Paradoxically, no difference or even higher expression of IL-12 and IL-18 was observed in the knockout macrophages after their treatment by LPS, suggesting that LPS stimulates IL-12 expression by a pathway independent of C/EBPε.34-36
Remarkably, IL-10 mRNA and protein were absent in the peritoneal macrophages from C/EBPε−/− mice. IL-10 promotes the down-regulation of pro-inflammatory cytokine synthesis and the development of the Th2 response. Recent studies demonstrated a role for SP1 and SP3 during activation of the IL-10 promoter in macrophages. Contrary to the regulation of pro-inflammatory cytokine genes, no functionally important binding sites for C/EBP proteins have been identified in the IL-10 promoter.37 38 The observed loss of IL-10 expression may result from the abnormal differentiation of the macrophages in the C/EBPε−/− mice, or perhaps C/EBPε regulates other genes that may directly target IL-10.
Taken together, C/EBPε appears to be required for the function and development of macrophages. Study of the function of individual C/EBP family members is complex because of their overlapping expression patterns and their ability to dimerize with one another and other leucine zipper transcription factors and because each is able to bind to the same canonical C/EBP DNA-binding site. Furthermore, in vitro promoter studies can provide spurious results because the transfected transcription factor is usually markedly overexpressed, which can lead to promiscuous activity not witnessed in vivo. The challenge is to dissect the individual sphere of activity of each C/EBPs. This could occur, at least in part, by the temporal expression of each or of potential dimerizing partner proteins, or in the context of the C/EBP-binding site in the region of the gene (that is, other transcription factors required to bind in the region to mediate effective transcriptional modulation). C/EBP knockout mice provide excellent models for studying the contribution of each member of this family to the differentiation and regulation of cell- and stage-restricted myeloid genes. Nevertheless, this type of analysis is limited if the deleted transcription factor results in a block in differentiation of the lineage. The activity of factors distal to the block cannot be easily studied unless a conditional knockout mouse is constructed. This does not appear to be a major problem for C/EBPε. Although the macrophages are not completely mature, no major lineage block is observed.
Mutations in the C/EBPε gene are the underlying molecular defect in patients with neutrophil-specific granule deficiency. These patients have early and frequent bacterial infections and have abnormalities in neutrophil development and function.39 40Our results demonstrate the importance of C/EBPε in macrophage development in the mouse. Future studies of macrophage function and maturation in patients with neutrophil-specific granule deficiency will further elucidate the role of C/EBPε in humans.
We thank Dr James O'Kelly and Ian Williamson for technical assistance and Dr Susan Spira for assistance with morphology analysis.
Supported in part by a Grant-in-Aid from the Israel Humanitarian Fund Research (S.T.) and by the National Institutes of Health, the Ko-So Fundation, the Parker Hughes Trust, the C. and H. Koeffler Fund, and the Horn Trust. H.P.K. holds the Mark Goodson endowed chair for Cancer Research and is a member of the Jonsson Cancer Center.
S.T. and P.T.V. contributed equally to the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sigal Tavor, Division of Hematology/Oncology, Cedars-Sinai Medical Center, UCLA School of Medicine, 8700 Beverly Blvd, D-5065, Los Angeles, CA 90048; e-mail: koeffler@cshs.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal