The ETV6 gene (first identified as TEL) is a frequent target of chromosomal translocations in both myeloid and lymphoid leukemias. At present, more than 40 distinct translocations have been cytogenetically described, of which 13 have now also been characterized at the molecular level. These studies revealed the generation of in-frame fusion genes between different domains of ETV6 and partner genes encoding either kinases or transcription factors. However, in a number of cases—including a t(6;12)(q23;p13), the recurrent t(5;12)(q31;p13), and some cases of the t(4;12)(q11-q12;p13) described in this work—functionally significant fusions could not be identified, raising the question as to what leukemogenic mechanism is implicated in these cases. To investigate this, we have evaluated the genomic regions at 4q11-q12 and 5q31, telomeric to the breakpoints of the t(4;12)(q11-q12;p13) and t(5;12)(q31;p13). The homeobox geneGSH2 at 4q11-q12 and the IL-3/CSF2locus at 5q31 were found to be located close to the respective breakpoints. In addition, GSH2 and IL-3 were found to be ectopically expressed in the leukemic cells, suggesting that expression of GSH2 and IL-3 was deregulated by the translocation. Our results indicate that, besides the generation of fusion transcripts, deregulation of the expression of oncogenes could be a variant leukemogenic mechanism for translocations involving the 5′ end of ETV6, especially for those translocations lacking functionally significant fusion transcripts.
Introduction
Cytogenetic and molecular characterization has demonstrated that nonrandom chromosomal rearrangements in hematologic malignancies are associated with specific oncogenic mechanisms.1,2 Translocations and inversions can result in the generation of oncogenic fusion genes or deregulated expression of protooncogenes.1,2 The latter mechanism has been mainly described for translocations and inversions involving the immunoglobulin heavy and light chain (IgH/L) loci or T-cell receptor (TCR) loci in lymphoid leukemias and lymphomas. In these cases, expression of protooncogenes is deregulated by their juxtaposition to, respectively, B-cell– or T-cell–specific regulatory sequences of the IgH/L or TCR loci.2 This mechanism is less common in myeloid leukemias, although some examples, such as the inv (3)(q21q26) with RPN1-mediated ectopic expression of EVI1,have been described.3,4 The other mechanism, the generation of oncogenic fusion proteins, is observed in both lymphoid and myeloid malignancies and frequently involves the CBFA2(21q22), MLL (11q23), and ETV6 (12p13) loci, which are known to be fused to a wide variety of partner genes.5
Translocations involving the ETV6 gene are among the most intriguing aberrations found in hematologic malignancies, not only because so many different translocation partners—more than 405,6—have been cytogenetically described, but also because this gene contributes to leukemogenesis through at least 3 different mechanisms. One mechanism is the modulation of the activity of the transcription factors such as CBFA2 and ARNT or the activation of the kinases PDGFRB, ABL1, ABL2, JAK2, NTRK3, and SYK by the generation of a fusion protein including the homodimerization (pointed) domain of ETV6.7-16 A second mechanism is found in the MN1-ETV6 fusion protein that contains the DNA binding domain of ETV617 and was recently demonstrated to function as a chimeric transcription factor with oncogenic properties.18Finally, the ETV6-MDS1/EVI1 and ETV6-CDX2 fusions only contain the transcription/translation start ofETV6.19,20 These fusions are believed to represent ectopic expression of the transcription factorsEVI1 and CDX2, which are not normally expressed in the hematopoietic system. These data clearly illustrate that most cloned translocations involving ETV6 fit to the paradigm of the generation of fusion proteins, either chimeric kinases or chimeric transcription factors. However, the molecular analysis of a t(6;12)(q23;p13)21 and the recurrent t(5;12)(q31;p13),22 with the 5q31 breakpoints ranging from the 5′ end to the 3′ end of the long fatty acyl-CoA synthetase 2 (ACS2) gene (official gene symbolKIAA0837), has indicated a lack of functionally significant fusion proteins, suggesting that another mechanism is involved in these translocations.
We previously described the in-frame fusion of a novel gene,CHIC2 (BTL), to ETV6 in 1 case of myeloid/natural killer (NK) cell leukemia and 3 cases of acute myeloid leukemia (AML)-M0 with a t(4;12)(q11-q12;p13).23 Although we have characterized CHIC2 as a palmitoylated membrane-associated protein,24 its function remains unknown and the oncogenic features of CHIC2-ETV6 are not yet understood. We describe here a further molecular analysis of the recurrent t(4;12)(q11-q12;p13) based on the characterization of 2 additional cases. Surprisingly, although the breakpoints in these cases were located within the region spanned by the P1-based artificial chromosome (PAC) contig at theCHIC2 locus,23 theCHIC2-ETV6 fusion was absent and no other meaningful fusions could be detected. Based on this observation and taking into account that the recurrent t(5;12)(q31;p13) is also lacking a functionally significant fusion transcript,22 we hypothesized that these recurrent translocations may involve the ectopic expression of protooncogenes at the partner chromosomes. To evaluate this possibility, the genomic regions telomeric to the breakpoints at 4q11-q12 and 5q31 were investigated for the presence of candidate protooncogenes, and the expression of the identified candidate genes was investigated. Our results suggest that ectopic expression of the homeobox gene GSH2 and the growth factor gene interleukin-3 (IL-3), located close to the breakpoints at 4q11-q12 and 5q31, respectively, may be the underlying leukemogenic factors explaining the pathogenesis of the t(4;12)(q11-q12;p13) and t(5;12)(q31;p13).
Patients, materials, and methods
Case reports
Case 1 is a myeloid/NK cell leukemia, and cases 2, 3 and 4 are AML-M0 cases with a t(4;12)(q11-q12;p13), which were previously described.23 Two additional cases with a t(4;12)(q11-q12;p13) were collected at the University Hospital of Navarra, Spain, (case 5) and the University Hospitals Leuven, Belgium (case 6). Case 5 (male, age 47) was diagnosed as AML-M2 and was previously described.6 The material was obtained at relapse. Case 6 (male, age 25) presented with a de novo acute lymphocytic leukemia (ALL) at 11 years of age. He received chemotherapy over a period of 3 years. Fourteen years after the first diagnosis of ALL, he presented with therapy-related AML. Complete remission was obtained, but the patient relapsed and died of leukemia 1 year after the diagnosis of AML. Case 7 (male, age 49) was diagnosed as atypical CML (aCML) with karyotype 46,XY, t(5;12)(q31;p13) and was previously described.25 All studies were performed on bone marrow samples, except for case 3, for which we obtained a peripheral blood sample containing less than 10% blasts.
RNA isolation and cDNA synthesis
Total RNA was isolated using Trizol (Life Technologies, Paisley, United Kingdom). Poly(A)+ RNA from control bone marrow, peripheral blood, and fluorescence-activated cell sorted (FACS) cells was obtained using the mRNA isolation kit for blood/bone marrow (Roche, Mannheim, Germany). Poly(A)+ RNA from fetal brain was purchased from Clontech, Palo Alto, CA. First-strand complementary DNA (cDNA) was prepared from total RNA or poly(A)+ RNA using Moloney murine leukemia virus reverse transcriptase (RT) (Life Technologies) and random hexamer primers.
Rapid amplification of cDNA ends
Rapid amplification of cDNA ends (RACE) was performed as previously described.23 For 5′-RACE on case 5, nested polymerase chain reaction (PCR) was performed using the primers ETV6R3a (5′-GAATGAGGAGATCGATAGCG-3′) and ETV6R3b (5′-TCCCTGCTCCAGTAAATTGGCTGCAAG-3′). For 3′-RACE on case 5, nested PCR was performed using the primers ETV6F1a (5′-TGAGACATGTCTGAGACTCCTGCT-3′) and ETV6F2 (5′-CCTCCAGAGAGCCCAGTGCCGAGT-3′). For 3′-RACE on case 7, nested PCR was performed using the primers ETV6F1a and ETV6F1b (5′-ACTCCTGCTCAGTGTAGCATTAAG-3′).
Reverse transcriptase–polymerase chain reaction
Standard RT-PCR was performed to assess the expression of theCHIC2-ETV6 fusion gene and the expression ofIL-3, CSF2, IL-5, and IL-9.The primers used were IL-3F (5′-AACCTGGAGGCATTCAACAG-3′) and IL-3R (5′-ACGTCAGTTTCCTCCGGAAT-3′), CSF2F (5′-AGGATGTGGCTGCAGAGC-3′) and CSF2R (5′-AGGGCAGTGCTGCTTGTAGT-3′), IL-5F (5′-CAAACGCAGAACGTTTCAGA-3′) and IL-5R (5′-CAGTACCCCCTTGCACAGTT-3′), and IL-9F (5′-CCTCTGACAACTGCACCAGA-3′) and IL-9R (5′-CTTGCCTCTCATCCCTCTCA-3′). TaqMan PCR was performed to analyze the expression of GSH2using the TaqMan PCR mix (Perkin Elmer, Wellesley, MA) and 10 pM of the primers GSH2TMF (5′-AGATTCCACTGCCTCACCATG-3′), GSH2TMR (5′-TCTCTCTCCAGCTCCAGGAGTT-3′), GAPDTMF (5′-AGCCTCAAGATCATCAGCAATG-3′), and GAPDTMR (5′-ATGGACTGTGGTCATGAGTCCTT-3′) together with 10 pM of the probes GSH2TMP (5′-FAM-CTCTGACGCCAGCCAGGTACCCA-TAMRA-3′) and GAPDTMP (5′-JOE-CCAACTGCTTAGCACCCCTGGCC-TAMRA-3′). The detection of GAPD served as internal control. The CT values given in the text represent the number of the PCR cycle in which the fluorescent signal significantly exceeds background levels.
Generation of sequence tagged site markers
Sequence tagged sites (STSs) were designed based on available genomic sequences. The primers used were as follows: for STS-G, 5′-GCACCGCCACCACCTACAAC-3′ and 5′-GGGGATGTGAGGGAGGAGGC-3′; for STS-K, 5′-AAGCAGGAACTGTGGAAGGTG-3′ and 5′-GTGGCCAAGATAAGGGGCTA-3′; for STS-5A, 5′-TGGGGTGCACTCTGTACATC-3′ and 5′-GAACCAGTAGGCAAGGATGG-3′; for STS-3A, 5′-AAATGTTCCCCTGTGTGTGC-3′ and 5′-CCCATCATTTGTAGGCTGTTG-3′; for STS-I, IL-3F and IL-3R; and for STS-C, CSF2F and 5′-ACAGGCCCACATTCTCTCAC-3′. PCR was performed with an annealing temperature of 60°C and 30 cycles.
Constructs
The construct murine stem cell virus (MSCV)-GSH2, containing the full open reading frame of GSH2, was obtained by ligating a genomic fragment containing exon 1 to a PCR product containing exon 2 using the NcoI restriction site at the border of the 2 exons into the retroviral vector MSCV-puromycin (kindly provided by Dr D.G. Gilliland, Harvard, Boston, MA). The MSCV-GSH2delN construct, containing only the homeobox domain, and the MSCV-GSH2delH construct, containing the open reading frame of GSH2 with a deletion in the homeobox domain, were generated by PCR. The empty MSCV-puromycin vector (MSCV) served as control.
Cell culture, viral vector production, and transduction
The HEK293T and NIH3T3 cell lines were cultured in Dulbecco modified Eagle medium containing 10% fetal calf serum. NIH3T3 cells were obtained from the American Tissue Culture Collection (ATCC, Manassas, VA). Viral vectors were produced by cotransfection of HEK293T cells with a mixture of the respective MSCV plasmids and an ecotropic packaging plasmid (Cell Genesys, Foster City, CA) using Fugene-6 (Roche). Transduction of NIH3T3 cells was performed in 6-well plates in the presence of polybrene (8 μg/mL) (Sigma, St Louis, MO). After 8 hours, the cells were washed with phosphate-buffered saline and fresh medium was added. After 24 hours, selection was started using puromycin (3 μg/mL). All experiments were started within 2 weeks after the transduction and were performed at least in double using independent transductions.
FACS of bone marrow cells
Bone marrow cells were obtained from healthy volunteers. Mononuclear cells, obtained by using a Ficoll density gradient, were separated in a CD34+ and CD34− fraction by use of the magnetic-activated cell separation procedure (Miltenyi Biotec, Auburn, CA). Further fractionation was performed by incubation with the appropriate monoclonal antibodies and subsequent separation by FACS. The antibodies used were fluorescein isothiocyanate– or phycoerythrin-conjugated anti-CD3, anti-CD7, anti-CD13, anti-CD33, anti-CD34, and anti-CD38 (Becton Dickinson, Franklin Lakes, NJ).
Genomic clones
PAC clones for the CHIC2 locus were previously described.23 The bacterial artificial chromosome (BAC) clone 434C1, containing more than 100 kilobases (kb) upstream and 40 kb downstream of exon 1 of ETV6, was identified by database searches and obtained from the RPCI11 library (Roswell Park Cancer Institute, Buffalo, NY). Other clones from the ETV6 locus were previously described.26,27 The PAC clones P3701 and P3704 were identified by screening of the RPCI5 library (Roswell Park Cancer Institute) using a probe derived from the 5′ end of theACS2 gene. The localization of cosmid c33g8 (kindly provided by Dr M. Lovett) at the IL-3 locus was previously described.28
Results
Molecular cloning of additional t(4;12)(q11;p13) cases
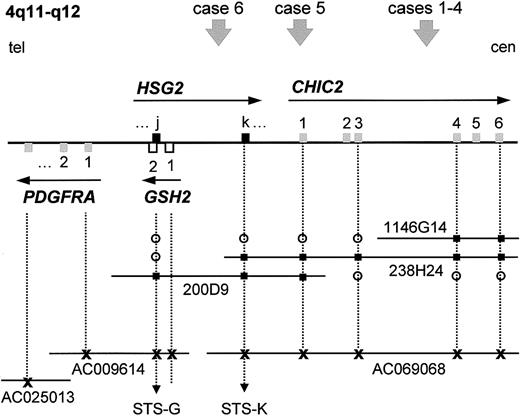
We previously described the molecular analysis of a myeloid/NK cell leukemia and 3 AML-M0 cases with a t(4;12)(q11-q12;p13).23 All cases were characterized by the presence of a fusion of the first 3 exons of CHIC2(BTL) to exons 2 to 8 of ETV6 (TEL). For 3 of these cases, the 4q breakpoint was determined by fluorescence in situ hybridization (FISH) and mapped to the region spanned by PAC 1146G14 (Figure 1). For this study, 2 additional AML cases with a t(4;12)(q11-q12;p13) were collected. Investigation of these cases by FISH revealed the breakpoints at 12p13 to be located in intron 2 of ETV6 for case 5 and close to exon 1 (upstream or downstream of exon 1) of ETV6 for case 6 (Table 1). Unlike the previously reported cases, for which FISH showed split signals for PAC 1146G14, the 4q11-q12 breakpoint of case 5 was located telomeric to PAC 1146G14 and in the region covered by PAC 238H24, while the breakpoint in case 6 was located in the region spanned by PAC 200D9 and telomeric to PAC 238H24 and thus upstream of CHIC2 (Table 1, Figure 1). This directly excluded the presence of a CHIC2-ETV6 fusion in case 6. Attempts to amplify a CHIC2-ETV6 fusion transcript in case 5 using primers in exon 1 of CHIC2 and in exon 3 ofETV6 yielded no product, whereas the control (case 1, previously described23) was positive, indicating that also in case 5 no CHIC2-ETV6 fusion transcript was present (results not shown).
PAC/BAC contig covering the
CHIC2 locus at 4q11-q12 and localization of the different breakpoints of the t(4;12)(q11;p13). We previously reported the generation of a PAC contig (PACs 200D9, 238H24, and 1146G14) spanning the CHIC2 locus at 4q11-q12.23 Database searches identified several partially sequenced BAC clones containing the CHIC2 andGSH2 genes, which were aligned to the PAC contig (drawing not to scale). Using PCR and database searches, GSH2 and the 2 exons of HSG2 were located on this contig. The localization of the breakpoints of 6 t(4;12) cases is indicated by an arrowhead and numbered according to the cases as described in the text. Exons are represented by open or filled squares, and the transcriptional orientation is indicated by an arrow. The sequenced BAC clones are represented by their accession numbers. Tel indicates telomeric side; cen, centromeric side; open circle, negative by hybridization or PCR; filled square, positive by hybridization or PCR; x, positive by electronic PCR/database searches.
PAC/BAC contig covering the
CHIC2 locus at 4q11-q12 and localization of the different breakpoints of the t(4;12)(q11;p13). We previously reported the generation of a PAC contig (PACs 200D9, 238H24, and 1146G14) spanning the CHIC2 locus at 4q11-q12.23 Database searches identified several partially sequenced BAC clones containing the CHIC2 andGSH2 genes, which were aligned to the PAC contig (drawing not to scale). Using PCR and database searches, GSH2 and the 2 exons of HSG2 were located on this contig. The localization of the breakpoints of 6 t(4;12) cases is indicated by an arrowhead and numbered according to the cases as described in the text. Exons are represented by open or filled squares, and the transcriptional orientation is indicated by an arrow. The sequenced BAC clones are represented by their accession numbers. Tel indicates telomeric side; cen, centromeric side; open circle, negative by hybridization or PCR; filled square, positive by hybridization or PCR; x, positive by electronic PCR/database searches.
Summary of the FISH results obtained for the 2 novel t(4;12)(q11-q12;p13) cases
| Probes . | (Localization) . | Case 5 . | Case 6 . |
|---|---|---|---|
| 4q11 | CHIC2 | ||
| 200D9 | Exon 1 | der(12) | split or der(12)* |
| 238H24 | Exons 1-6 | split | der(4) |
| 1146G14 | Exons 4-6 | der(4) | der(4) |
| 12p13 | ETV6 | ||
| 434C1 | Telomeric to exon 1/exon 1 | der(4) | |
| 209J1 | Exon 1/intron 1 | der(12) | |
| 179A6 | Exon 1/intron 1 | der(4) | |
| 50F4 | Intron 1/exon 2 | der(4) | |
| TEL11 | Intron 2/exon 3 | split | |
| 54D5 | Exons 5-8 | der(12) | |
| 148B6 | Exon 8 | der(12) | der(12) |
| Probes . | (Localization) . | Case 5 . | Case 6 . |
|---|---|---|---|
| 4q11 | CHIC2 | ||
| 200D9 | Exon 1 | der(12) | split or der(12)* |
| 238H24 | Exons 1-6 | split | der(4) |
| 1146G14 | Exons 4-6 | der(4) | der(4) |
| 12p13 | ETV6 | ||
| 434C1 | Telomeric to exon 1/exon 1 | der(4) | |
| 209J1 | Exon 1/intron 1 | der(12) | |
| 179A6 | Exon 1/intron 1 | der(4) | |
| 50F4 | Intron 1/exon 2 | der(4) | |
| TEL11 | Intron 2/exon 3 | split | |
| 54D5 | Exons 5-8 | der(12) | |
| 148B6 | Exon 8 | der(12) | der(12) |
Only part of the metaphases showed split signals. This is indicative for a breakpoint close to the centromeric side of the region covered by PAC 200D9.
We next investigated the presence of other fusion transcripts in case 5. Using 5′-RACE starting from exon 3 of ETV6, transcripts were detected fusing a novel sequence to exon 2 or directly to exon 3 of ETV6 (Figure 2). This translocation seemed to be complex, because 3′-RACE experiments showed the presence of a transcript fusing exon 2 of ETV6 to the same sequence as identified by 5′-RACE (Figure 2), indicative of a duplication of both 12p and 4q sequences associated with the translocation. No DNA from this case was available to confirm this by Southern blotting. Unfortunately, due to lack of sufficient material, a molecular analysis could not be performed for case 6.
RACE results for case 5 indicating the different
HSG2-ETV6 andETV6-HSG2 fusions. The 5′- and 3′-RACE, performed on cDNA obtained from case 5, revealed the existence of 3 differentHSG2-ETV6 fusion transcripts and an ETV6-HSG2fusion transcript. Exon 2 of ETV6 and exon k ofHSG2 are present in both types of fusions, which can only be explained by a duplication event associated with the translocation. Exon k of HSG2 is underlined; sequences from ETV6are in italics; stop codons are in bold.
RACE results for case 5 indicating the different
HSG2-ETV6 andETV6-HSG2 fusions. The 5′- and 3′-RACE, performed on cDNA obtained from case 5, revealed the existence of 3 differentHSG2-ETV6 fusion transcripts and an ETV6-HSG2fusion transcript. Exon 2 of ETV6 and exon k ofHSG2 are present in both types of fusions, which can only be explained by a duplication event associated with the translocation. Exon k of HSG2 is underlined; sequences from ETV6are in italics; stop codons are in bold.
Cloning of human GSH2/HSG2
Database searches revealed 100% identity of part of the sequence obtained by 5′-RACE from case 5 to several human expressed sequence tags. These represent the GSH2 gene; they are highly similar to the murine Gsh2 cDNA sequence. However, the sequence fused to ETV6 was derived from the complementary noncoding strand of GSH2. Part of this novel sequence overlapped with exon 2 of GSH2 and was found to be colinear with the sequence of BAC AC009614, while the other part of the novel sequence was found to be colinear with BAC AC069068. Consensus splice donor and acceptor sequences were present at the genomic sequences, indicating that these 2 parts are 2 exons of a novel gene that is located at theGSH2 locus but is transcribed in the opposite direction asGSH2 and hence was named HSG2 (Figure 1). Stop codons were found to be present in the 3 different reading frames, indicating that these exons are derived from the 5′- or 3′-untranslated region of HSG2 or that HSG2 is a noncoding transcript. The exact identity and complete cDNA sequence ofHSG2 was not further analyzed. Because the completeHSG2 transcript is currently unknown, the 2 exons ofHSG2 identified in this work could not be numbered and were named exon j and exon k.
Next, the 2 exons of GSH2 were cloned from aHindIII fragment of PAC 200D9. Similar to murineGsh2, the complete 915 bp open reading frame ofGSH2 was found to be present in 2 exons (sequence submitted to GenBank), separated by an intron of 2 kb. Several expressed sequence tags were found to match 100% with the exon sequences determined by us and confirmed the splice sites. In addition, RT-PCR on cDNA from human fetal brain using primers in the different exons confirmed the expression and splicing of GSH2 (results not shown).
An STS derived for exon 2 of GSH2 (STS-G) and for exon k ofHSG2 (STS-K), respectively, was used to assess their localization in our PAC contig spanning the CHIC2 locus.GSH2, and thus also exon j of HSG2, was found to be present only in PAC 200D9, whereas exon k of HSG2 was located in the region shared between the PACs 200D9 and 238H24 (Figure1).
Taken together, these data indicated that GSH2 was located in front of CHIC2 and that not GSH2, but its antisense gene, HSG2, was fused to ETV6 in case 5. Because the HSG2 transcript contained stop codons in all reading frames, no HSG2-ETV6 fusion protein can be generated. Thus, in case 5, no functionally significant fusion transcript was generated by the translocation. However, knowing that the GSH2 gene was in close proximity to the breakpoint in the t(4;12)(q11-q12;p13) cases and taking into account that ectopic expression of homeobox genes is a known mechanism involved in leukemia, we hypothesized that ectopic expression of GSH2 could represent an alternative leukemogenic event. To investigate this, we analyzed the expression ofGSH2 in these cases and investigated its transforming properties using a focus formation assay in the NIH3T3 cell line.
Expression of GSH2 in the t(4;12)
We analyzed the expression of GSH2 by use of TaqMan real-time quantitative RT-PCR using GAPD as an internal control. As expected, expression of GSH2 was detected in control cDNA derived from fetal brain. No expression was detected in any of the hematopoietic tissues analyzed or in the myeloid cell line HL60. However, expression of GSH2 was detected in the 4 cases that we previously described to contain theCHIC2-ETV6 fusion (cases 1-4) and in the AML-M2 case (case 5), which contains the HSG2-ETV6 fusion but lacks the CHIC2-ETV6 fusion (Table2). Although the CT value for GSH2 in case 5 was rather high, this was still different from the negative control samples. RNA from this case was extracted from fixed cells and was of poor quality, as is also clear from the higher CT value forGAPD. To exclude that the expression of GSH2detected in these cases was the result of a clonal expansion of a CD34+ cell type expressing GSH2, we also analyzed the expression of GSH2 in CD34+fractions of bone marrow that was highly enriched for the respective markers (Table 1). Expression of GSH2 could not be detected in any of these subpopulations. Both GAPD (Table 1) and the expression of the homeobox gene NKX2-3 (data not shown) were easily detected, indicating that the negative result forGSH2 was not due to a technical problem and that the RNA extraction from these FACS-sorted cells was adequate.
TaqMan real-time quantitative RT-PCR analysis of the expression of GSH2
| Sample . | CTvalue* . | ΔCT . | |
|---|---|---|---|
| GAPD . | GSH2 . | ||
| Fetal brain | 15.5 | 26.0 | 10.5 |
| Bone marrow | 19.6 ± 0.2 | Greater than 40 | No expression |
| Peripheral blood | 23.4 ± 0.2 | Greater than 40 | No expression |
| Spleen | 22.6 ± 0.3 | Greater than 40 | No expression |
| HL60 cell line | 16.0 ± 0.4 | Greater than 40 | No expression |
| CD34+ | 21.1 ± 0.9 | Greater than 40 | No expression |
| CD34+CD38− | 33.3 ± 0.6 | Greater than 40 | No expression |
| CD34+CD38+ | 24.5 ± 1.3 | Greater than 40 | No expression |
| CD34+CD7+ | 26.4 ± 0.2 | Greater than 40 | No expression |
| CD34+CD13+CD33+ | 23.2 ± 0.5 | Greater than 40 | No expression |
| CD34− | 18.0 ± 1.0 | Greater than 40 | No expression |
| CD34−CD3+ | 27.3 ± 0.6 | Greater than 40 | No expression |
| CD34−CD13+ | 30.3 ± 0.6 | Greater than 40 | No expression |
| CD34−CD33+ | 27.7 ± 1.2 | Greater than 40 | No expression |
| BM from case 1 | 22.7 | 25.4 | 3.3 ± 0.8 |
| BM from case 2 | 24.9 | 30.3 | 5.7 ± 0.3 |
| PB from case 3 | 25.0 | 33.8 | 9.6 ± 0.7 |
| BM from case 4 | 22.2 | 26.9 | 4.2 ± 0.7 |
| BM from case 5† | 28.3 | 37.1 | 9.0 ± 0.5 |
| BM from case 6 | Nd | nd | nd |
| Water‡ | Greater than 40 | Greater than 40 | No expression |
| Sample . | CTvalue* . | ΔCT . | |
|---|---|---|---|
| GAPD . | GSH2 . | ||
| Fetal brain | 15.5 | 26.0 | 10.5 |
| Bone marrow | 19.6 ± 0.2 | Greater than 40 | No expression |
| Peripheral blood | 23.4 ± 0.2 | Greater than 40 | No expression |
| Spleen | 22.6 ± 0.3 | Greater than 40 | No expression |
| HL60 cell line | 16.0 ± 0.4 | Greater than 40 | No expression |
| CD34+ | 21.1 ± 0.9 | Greater than 40 | No expression |
| CD34+CD38− | 33.3 ± 0.6 | Greater than 40 | No expression |
| CD34+CD38+ | 24.5 ± 1.3 | Greater than 40 | No expression |
| CD34+CD7+ | 26.4 ± 0.2 | Greater than 40 | No expression |
| CD34+CD13+CD33+ | 23.2 ± 0.5 | Greater than 40 | No expression |
| CD34− | 18.0 ± 1.0 | Greater than 40 | No expression |
| CD34−CD3+ | 27.3 ± 0.6 | Greater than 40 | No expression |
| CD34−CD13+ | 30.3 ± 0.6 | Greater than 40 | No expression |
| CD34−CD33+ | 27.7 ± 1.2 | Greater than 40 | No expression |
| BM from case 1 | 22.7 | 25.4 | 3.3 ± 0.8 |
| BM from case 2 | 24.9 | 30.3 | 5.7 ± 0.3 |
| PB from case 3 | 25.0 | 33.8 | 9.6 ± 0.7 |
| BM from case 4 | 22.2 | 26.9 | 4.2 ± 0.7 |
| BM from case 5† | 28.3 | 37.1 | 9.0 ± 0.5 |
| BM from case 6 | Nd | nd | nd |
| Water‡ | Greater than 40 | Greater than 40 | No expression |
The CT value was determined in at least 3 independent experiments. It is indicated as mean ± SD when no expression of GSH2 was detected. When expression ofGSH2 was detected, individual CT values are given from 1 representative experiment and the mean ± SD is determined from the ΔCT values obtained from the independent experiments.
RNA was extracted from fixed cells.
Water was used as “no template control” in each experiment.
Overexpression of GSH2 transforms NIH3T3 cells
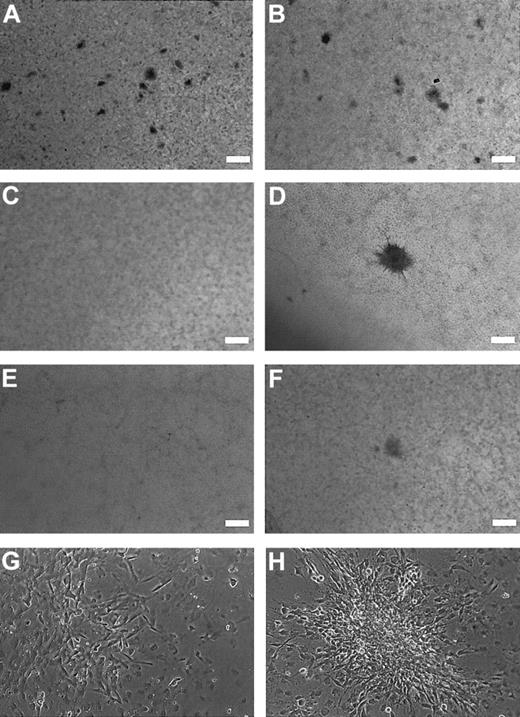
To investigate the transforming properties of GSH2 in vitro, we retrovirally transduced the NIH3T3 cell line with the empty MSCV vector or with the MSCV vector containing wild-typeGSH2 (MSCV-GSH2), GSH2 with a deletion of the N-terminus (MSCV-GSH2delN), or GSH2 with a deletion of the homeobox domain (MSCV-GSH2delH), respectively. Focus formation was determined as the ability to induce a significant larger number of foci than observed after transduction of the empty vector. Experiments with the different constructs were performed at the same time, with the same batch of cells. The number of spontaneous foci observed for the empty vector was 10 at maximum. In contrast, overexpression of wild-typeGSH2 resulted in the presence of more than 200 small foci per 10 cm dish (Figure 3A,B, Table 2). The deletion mutants of GSH2 induced only a small number of foci, comparable to the empty vector (Table 3). Typically, these foci were larger in size than the foci induced by overexpression ofGSH2 (Figure 3D), although microscopically the foci induced by GSH2 were clearly composed of transformed cells but apparently grew more slowly (Figure 3GH). We also tested whether expression of the CHIC2-ETV6 fusion (MSCV-CHIC2ETV6) had transforming properties, but expression ofCHIC2-ETV6 did not induce focus formation in the NIH3T3 cell line (results not shown).
Focus formation assay illustrating the oncogenic potential of
GSH2. Monolayer of cells overexpressing wild-typeGSH2 (A,B: 2 different frames of the same dish), indicating a high number (> 200 foci/10 cm dish) of relatively small foci, compared with the monolayer of cells transduced with the empty MSCV vector (C). In the latter case, spontanous foci (0-10 foci/10 cm dish) were observed. Typically, these foci were relatively larger than the foci observed with GSH2 overexpression (compare panels A and B with D). The N-terminal deletion mutant and homeobox deletion mutant of GSH2 induced only a low number of foci, similar to the empty vector. The monolayers observed with these construct are shown in panels E and F. (A-F: scale bar = 2 mm). The morphology and size of the foci observed for these constructs were similar to the foci observed for the empty vector. Although the cells overexpressingGSH2 generated relatively small foci, higher-magnification pictures (G,H) clearly show the presence of transformed cells growing on top of the monolayer.
Focus formation assay illustrating the oncogenic potential of
GSH2. Monolayer of cells overexpressing wild-typeGSH2 (A,B: 2 different frames of the same dish), indicating a high number (> 200 foci/10 cm dish) of relatively small foci, compared with the monolayer of cells transduced with the empty MSCV vector (C). In the latter case, spontanous foci (0-10 foci/10 cm dish) were observed. Typically, these foci were relatively larger than the foci observed with GSH2 overexpression (compare panels A and B with D). The N-terminal deletion mutant and homeobox deletion mutant of GSH2 induced only a low number of foci, similar to the empty vector. The monolayers observed with these construct are shown in panels E and F. (A-F: scale bar = 2 mm). The morphology and size of the foci observed for these constructs were similar to the foci observed for the empty vector. Although the cells overexpressingGSH2 generated relatively small foci, higher-magnification pictures (G,H) clearly show the presence of transformed cells growing on top of the monolayer.
Molecular analysis of an atypical CML case with a t(5;12)(q31;p13)
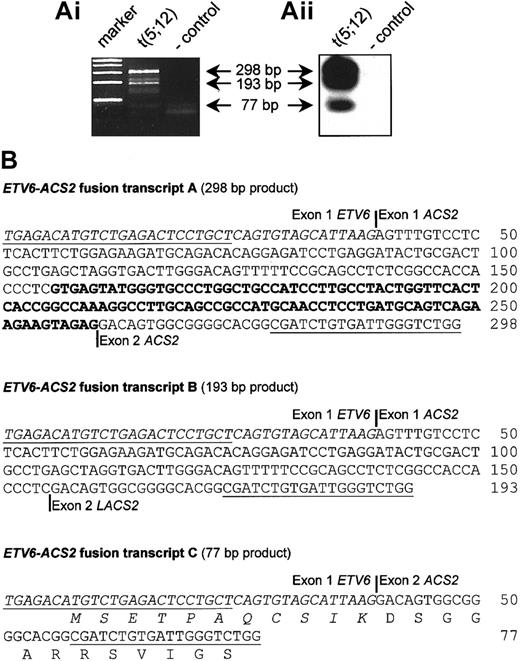
The t(5;12)(q31;p13) has been described as a recurrent translocation occurring in various myeloid malignancies (at least 18 cases were previously described), often associated with eosinophilia.22,25,29-32 We collected an atypical CML (aCML) case with a t(5;12)(q31;p13) as the sole chromosomal abnormality and investigated the presence of a possible fusion transcript. This case was described previously, and the breakpoint in ETV6was mapped to intron 1.25 RACE experiments revealed the presence of an ETV6-ACS2 fusion, confirming that this case was similar to the cases reported by Yagasaki et al.22 Further analysis by RT-PCR revealed the presence of 3 different ETV6-ACS2 fusion transcripts, among which was an in-frame fusion fusing the first 11 amino acids of ETV6 to almost the complete ACS2 protein (Figure4). According to the RT-PCR results, the breakpoint in ACS2 was at the 5′ end of this gene, similar to case 3 described by Yagasaki and coworkers.22
Detection of
ETV6-ACS2 fusion transcripts in the aCML case with a t(5;12)(q31;p13) (case 7). (Ai) RT-PCR confirmed the fusion of ETV6 to ACS2, as identified by 3′-RACE in the t(5;12) case (case 7). (Aii) Hybridization with an internal oligonucleotide revealed the existence of 3 different fusion transcripts, which was confirmed by cloning and sequencing of these RT-PCR products, shown in the lower panel. (B) Representation of the identified fusion transcripts. Open reading frames were determined on the basis of the start codon of ETV6. Only transcript C is an in-frame fusion. Exon numbering of ACS2 is according to Yagasaki et al.22
Detection of
ETV6-ACS2 fusion transcripts in the aCML case with a t(5;12)(q31;p13) (case 7). (Ai) RT-PCR confirmed the fusion of ETV6 to ACS2, as identified by 3′-RACE in the t(5;12) case (case 7). (Aii) Hybridization with an internal oligonucleotide revealed the existence of 3 different fusion transcripts, which was confirmed by cloning and sequencing of these RT-PCR products, shown in the lower panel. (B) Representation of the identified fusion transcripts. Open reading frames were determined on the basis of the start codon of ETV6. Only transcript C is an in-frame fusion. Exon numbering of ACS2 is according to Yagasaki et al.22
However, in the 3 cases studied by Yagasaki and colleagues, only out-of-frame ETV6-ACS2 fusion transcripts were identified, showing that the t(5;12)(q31;p13) does not generate a common in-frame fusion gene. The exact leukemogenic factor responsible for the pathogenesis of the t(5;12)(q31;p13) thus remained unresolved. According to these observations, the only invariant result of this translocation is the juxtaposition of the region directly telomeric toACS2 with the ETV6 locus. We therefore investigated whether this could lead to the juxtaposition of a protooncogene on 5q31 to ETV6 and if this could better explain the leukemogenic mechanism of this translocation.
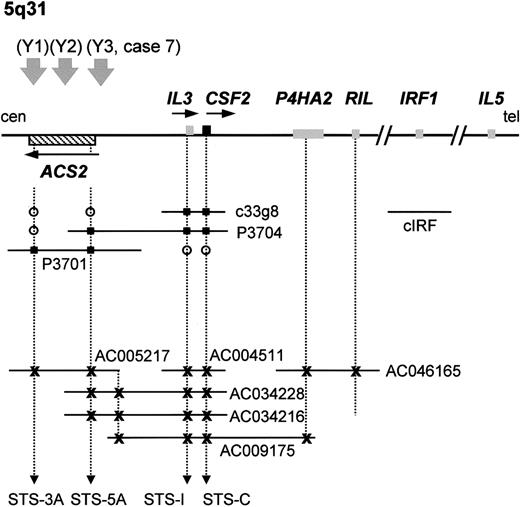
Analysis of the region telomeric to the breakpoint at 5q31 revealed the presence of the IL-3/CSF2 locus in the genomic sequence of BAC CTD-2198K16 (accession no. AC034228) that also contained the 5′ end of ACS2. This was confirmed by further analysis of this region using the available genomic sequences (accession numbers are listed in Figure5) and by mapping of 2 PAC clones (P3701 and P3704) and a cosmid (c33g8) by PCR using STS markers specific for the 5′ end of ACS2 (STS-5A) and the 3′ end of ACS2(STS-3A), IL-3 (STS-I), and CSF2 (STS-C) (Figure 5). A complete 1 Mb contig encompassing the ACS2, IL-3, CSF2, IRF1, IL-5, andIL-4 loci was constructed previously,33although the exact orientation of this contig remained unclear. This was due mainly to the small physical distance between theIL-3 and IL-5 loci, which made FISH-based mapping difficult and resulted in contradictions in the literature.28,34 The cloning of 4 t(5;12) cases with anETV6-ACS2 fusion and the known telomere to centromere orientation of ETV626 are strongly suggestive for a telomere to centromere orientation of ACS2,enabling orientation of the contig described by Frazer et al33 as centromere–ACS2–IL3–CSF2–IRF1–IL5–IL4–telomere. In agreement with this, Yagasaki et al22 mapped theIRF1 gene telomeric to the 5q31 breakpoint by FISH, and Grimaldi et al35 cloned the t(5;14)(q31;q32) breakpoint, also confirming that CSF2 is located telomeric toIL-3. Taken together, all these data are consistent with a localization of IL-3 immediately telomeric to ACS2,as shown in Figure 5. Because ACS2 and IL-3were both present in PAC P3704 and according to the contig reported by Frazer et al,33 the distance between these genes is most probably 100 kb or less.
PAC/BAC/cosmid contig covering the
ACS2 andIL-3/CSF2 loci at 5q31 and localization of the different breakpoints of the t(5;12)(q31;p13).Schematic illustration of the small PAC/cosmid contig linkingACS2 with IL-3/CSF2 (not to scale). Database searches identified several partially or completely sequenced genomic clones containing the ACS2, IL-3, CSF2, and P4HA2 genes, confirming the PAC contig. These clones are represented by their accession numbers. Yagasaki et al22 mapped IRF1 telomeric to the breakpoints of the t(5;12). A 1 Mb contig was previously described33that links ACS2 to IL-5 and is in agreement with the map shown here. The 3 t(5;12)(q31;p13) breakpoints described by Yagasaki et al22 are numbered as Y1, Y2, and Y3. The breakpoint described in this work is indicated with case 7, as described in the text. Genes are represented by boxes, and their transcriptional orientation is indicated by an arrow. The sequenced BAC clones are represented by their accession numbers. tel indicates telomeric side; cen, centromeric side; open circle, negative by hybridization or PCR; filled square, positive by hybridization or PCR; x, positive by electronic PCR/database searches.
PAC/BAC/cosmid contig covering the
ACS2 andIL-3/CSF2 loci at 5q31 and localization of the different breakpoints of the t(5;12)(q31;p13).Schematic illustration of the small PAC/cosmid contig linkingACS2 with IL-3/CSF2 (not to scale). Database searches identified several partially or completely sequenced genomic clones containing the ACS2, IL-3, CSF2, and P4HA2 genes, confirming the PAC contig. These clones are represented by their accession numbers. Yagasaki et al22 mapped IRF1 telomeric to the breakpoints of the t(5;12). A 1 Mb contig was previously described33that links ACS2 to IL-5 and is in agreement with the map shown here. The 3 t(5;12)(q31;p13) breakpoints described by Yagasaki et al22 are numbered as Y1, Y2, and Y3. The breakpoint described in this work is indicated with case 7, as described in the text. Genes are represented by boxes, and their transcriptional orientation is indicated by an arrow. The sequenced BAC clones are represented by their accession numbers. tel indicates telomeric side; cen, centromeric side; open circle, negative by hybridization or PCR; filled square, positive by hybridization or PCR; x, positive by electronic PCR/database searches.
Expression of IL-3 in the t(5;12)(q31;p13)
The expression of IL-3, CSF2, IL-5, and IL-9 was analyzed in a bone marrow sample obtained from the aCML case (case 7). Only expression ofIL-3 could be clearly detected (Figure6), indicating a good correlation between the position of the breakpoint and the expression of IL-3 in this myeloid sample. Although CSF2 is only 10 kb distal toIL-3, no expression of this gene was observed. The expression of IL-5 and IL-9, which are not located in the immediate surroundings of the t(5;12) breakpoint, was investigated as a control. In this way a “contamination” of the sample with normal T cells expressing IL-3 was excluded because expression of IL-5 and IL-9 would then also be detected.
Expression of
IL-3 in the aCML case with a t(5;12)(q31;p13) (case 7). Expression of IL-3 but not of CSF2, IL-5, or IL-9 was detected in a bone marrow sample from the patient with aCML (case 7). By contrast, high-level expression of IL-3 could not be detected in control bone marrow cells (BM), indicating ectopic expression of IL-3 in the t(5;12) case. The faint band observed at 394 bp in control bone marrow is due to a genomic contamination of the RNA sample.
Expression of
IL-3 in the aCML case with a t(5;12)(q31;p13) (case 7). Expression of IL-3 but not of CSF2, IL-5, or IL-9 was detected in a bone marrow sample from the patient with aCML (case 7). By contrast, high-level expression of IL-3 could not be detected in control bone marrow cells (BM), indicating ectopic expression of IL-3 in the t(5;12) case. The faint band observed at 394 bp in control bone marrow is due to a genomic contamination of the RNA sample.
Discussion
We report here a further molecular study of the recurrent translocations t(4;12)(q11-q12;p13) and t(5;12)(q31;p13). Our results described in this work together with our previous study23and the study of Yagasaki and colleagues22 clearly indicate heterogeneity in the localization of the breakpoints at both 4q11-q12 (upstream, at the 5′ end or in intron 3 of CHIC2) and 5q31 (at the 5′ end, in the middle or at the 3′ end ofACS2). In correspondence with this, no common fusion transcript that could explain the pathogenic character of these translocations is generated by either of these translocations. In addition, the HSG2-ETV6 transcript detected in case 5 with the t(4;12) and the ETV6-ACS2transcripts described by Yagasaki et al22 in cases with a t(5;12) do not represent in-frame fusion transcripts, raising the question as to how these transcripts contribute to the pathogenesis of the respective translocations. Two other AML cases with a t(4;12)(q11-q12;p13) and an HSG2-ETV6 fusion have been reported,36 further confirming the heterogeneity of the 4q11-q12 breakpoints.
In fact, the only invariant result of these translocations is the juxtaposition of the chromosomal regions telomeric, respectively, toCHIC2 on 4q11-q12 and ACS2 on 5q31 to theETV6 locus on 12p13. The juxtaposition of protooncogenes to the RPN1 locus, the IgH/L loci, or the TCR loci has been well illustrated and results in the deregulated expression of these protooncogenes.2,3 37 We identified the homeobox geneGSH2 and the growth factor genes IL-3 andCSF2 in the regions adjacent to the breakpoints at, respectively, 4q11-q12 and 5q31 of the t(4;12)(q11-q12;p13) and the t(5;12)(q31;p13). Ectopic expression of GSH2 was detected in all t(4;12)(q11;p13) cases, with or without theCHIC2-ETV6 fusion, and expression ofIL-3 was observed in the aCML case with a t(5;12)(q31;p13), also characterized by an ETV6-ACS2 fusion. These results suggest that ectopic expression of GSH2 andIL-3 could be the underlying leukemogenic mechanisms of the t(4;12)(q11-q12;p13) and t(5;12)(q31;p13) cases, independent of the presence of fusion transcripts.
The Gsh2 gene was previously characterized as a brain-specific homeobox gene important in forebrain and hindbrain formation of the mouse.38,39 We did not find evidence thatGSH2 is also expressed during hematopoiesis. Interestingly,GSH2, belonging to the ParaHox genes,40contains a homeobox domain that is very similar to the homeobox of the clustered HOX genes, which are involved in both normal and abnormal hematopoiesis.41 In addition, overexpression ofGSH2 was found to induce focus formation in the NIH3T3 cell line (Figure 3), illustrating its oncogenic potential.
Evidence for the involvement of IL-3 in leukemia was described previously by the analysis of the t(5;14)(q31;q32) in B-ALL, involving ectopic expression of IL-3 as a consequence of its translocation to the IgH locus,35,42 and by analysis of mutations found in the murine WEHI-3B cell line, which illustrated that ectopic expression of HoxB8 andIL-3 together results in AML in a mouse model.43 Although expression of IL-3 alone produced only a nonneoplastic myeloproliferative disease in this mouse model, it is clear from these studies that ectopic expression ofIL-3/IL-3 leads to a proliferative defect and may be involved in the multistep process of leukemogenesis.43,44 In addition, because IL-3is a growth factor regulating eosinophil proliferation and differentation,45 ectopic expression of IL-3could also explain the eosinophilia associated with the t(5;12) cases,22,25,29-32 as was also observed for the t(5;14)(q31;q32).35,42 This is supported by the observation that continuous administration of IL-3 leads to a dose- and time-dependent eosinophilia.46
Although we clearly detected expression of GSH2 andIL-3 in the t(4;12) and t(5;12) cases, several remarks must be taken under consideration when describing this as an abnormal expression. Bone marrow is a heterogeneous mixture of different cell types of which in leukemia only one particular cell was originally mutated and expanded. It is thus possible that the expression of a gene, which is expressed only in a rare bone marrow cell type, would become detectable by RT-PCR in leukemic bone marrow as a consequence of the clonal expansion of this cell type. Second, translocations are known to occur in open chromatin structures, suggesting that the genes found at the breakpoints are transcriptionally active, thus arguing that the detected expression of GSH2 and IL-3 in the leukemias might not be abnormal. However, the breakpoints of the t(4;12) and t(5;12) are not very close to GSH2 orIL-3 but occur in or at least close to, respectively, theCHIC2 and ACS2 genes, which are expressed in the hematopoietic system22,23 and thus most probably are located in an open chromatine region, providing space for the translocations to occur. The fact that GSH2 expression could not be detected in any of the analyzed subpopulations highly enriched for CD34+ cells further indicates that GSH2 is not expressed in the hematopoietic system, although we cannot exclude its expression in a particularly rare bone marrow cell type. However, even if GSH2 would be expressed in a rare bone marrow cell type that escaped our detection, this would not directly excludeGSH2 from its involvement in the t(4;12). Transformation might then be the result of the inability to down-regulateGSH2, rather than its induced expression. In this regard, the ectopic expression of Hoxa9 by retroviral insertion in BXH2 mice is a well-characterized example illustrating that the ectopic expression of a gene that is normally expressed in bone marrow progenitor cells can nevertheless be leukemogenic.47 48
It has been well established that IL-3 is expressed by activated T cells, NK cells, and mast cells.49 Detection of its expression in a CML sample, containing mainly myeloid cells, is suggestive for its abnormal expression in this cell type. If expression of IL-3 would be the result of the presence of other IL-3–producing cells, then expression of other growth factors such asCSF2, IL-5, and IL-9 also would be expected, which was not the case (Figure 6). Although IL-3and CSF2 are located very close to each other, no expression of CSF2 was detected in the aCML case (Figure 6). Interestingly, this was also observed in B-cell ALL cases with a t(5;14)(q31;q32) involving the IgH locus.42 AlthoughIL-3 and CSF2 have an overlapping expression pattern, CSF2 expression can also be regulated separately.49 50 Elucidation of the mechanism by which expression of IL-3 becomes activated in these leukemias could provide further explanation for this.
The exact mechanism by which expression of GSH2 andIL-3 is activated is currently not known. It has become clear that the final transcription level of a gene is determined by the combined action of regulatory sequences that might be either positive or negative and the accessibility of these sequences determined by the chromatin structure and the nuclear architecture.51,52Therefore, it might be expected that regulatory sequences as enhancers in ETV6 and silencers at the partner chromosome can be major contributors, with additional effects from the change in chromatin structure around the breakpoint. Well-known examples involving enhancer-mediated ectopic expression are the translocations involving the RPN1 locus, the IgH/L loci, or the TCR loci.2,3,37,53 These studies have also clearly illustrated that the breakpoints can be located up to 200 kb away from the deregulated protooncogene.3 Other examples of how translocations can influence the expression of genes have been described for the translocations associated with Beckwith-Wiedemann syndrome, which involve aberrant expression of the IGF2 gene by loss of imprinting,54,55 and for chromosome rearrangements at the Kit/Pdgfra loci in mutant mice, affecting the expression ofKit.55 56 In addition to the effects of enhancers and silencers, all translocations are likely to change local chromatin structure and nuclear architecture in its whole, which may be associated with changes in gene expression.
Taken together, our results indicate that the common mechanism of the t(4;12)(q11-q12;p13) cases, with or without theCHIC2-ETV6 fusion, may be the ectopic expression of GSH2. Further analysis of additional cases with breakpoints at the 5′ end of CHIC2 and analysis of the in vivo leukemogenic properties of GSH2 will be necessary to fully understand the importance of its expression in the t(4;12)(q11-q12;p13) cases. In addition, our results suggest that the AML-M0 cases represent a subgroup in the t(4;12)(q11-q12;p13) characterized by both the expression of GSH2 and theCHIC2-ETV6 fusion. Although expression ofCHIC2-ETV6 does not transform the NIH3T3 cell line, it will be of interest to investigate whether the presence of theCHIC2-ETV6 fusion plays an additional role in determining the immature phenotype of the AML-M0 cases. With respect to the reported t(5;12)(q31;p13) cases, both our data and the known leukemia-related properties of IL-3 are supportive for the ectopic expression of IL-3 as an important transforming mechanism in these cases. Although we have only been able to investigate a single case, the 3 cases described by Yagasaki et al22 also involve a translocation of IL-3 to the der,12 supporting the possibility ofIL-3–induced expression by ETV6 regulatory sequences.
Although many translocations involving ETV6 have been shown to result in the generation of an in-frame fusion gene, encoding a fusion protein with oncogenic properties, our results indicated the importance of a variant mechanism: the ectopic expression of a protooncogene. Other translocations, such as the reported t(6;12)(q23;p13)21 and t(7;12)(p15;p13)57 and the t(10;12)(q24;p13) and t(12;13)(p13;q12) (P. M., unpublished results, 2001), seem not to result in the generation of functionally significant fusion genes. It will be of further interest to investigate whether these cases involve ectopic expression of a protooncogene. Interestingly, several cases of the t(3;12)(q26;p13) have been described, with the 3q26 breakpoints either 5′ or 3′ toEVI1, a gene that is ectopically expressed in myeloid malignancies with the inv (3)(q21q26) or the t(3;3)(q21;q26). One of the t(3;12)(q26;p13) cases was cloned and revealed the fusion ofETV6 to MDS1/EVI1.19However, at least one of the other cases described by Raynaud et al58 had breakpoints centromeric to EVI1 and thus could involve ectopic expression of EVI1 by a similar mechanism as proposed in this work. Identification of candidate protooncogenes for other translocations involving breakpoints at the 5′ end of ETV6 will be greatly facilitated by the availability of the complete human genome sequence.
We thank Vic Van Duppen for the FACS analysis and Betty Emanuel for performing FISH experiments. We are very grateful to Drs G. Verhoef, C. Cabrol, P. Talmant, and C. Bilhou-Nabera for providing valuable patient material.
Supported by grant G.0121.00 of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. J.C. is an aspirant of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter Marynen, Center for Human Genetics, Campus Gasthuisberg O&N 06, Herestraat 49, B-3000 Leuven, Belgium; e-mail:peter.marynen@med.kuleuven.ac.be.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal