RP105 is a B-cell surface molecule that has been recently assigned as CD180. RP105 ligation with an antibody induces B-cell activation in humans and mice, leading to proliferation and up-regulation of a costimulatory molecule, B7.2/CD86. RP105 is associated with an extracellular molecule, MD-1. RP105/MD-1 has structural similarity to Toll-like receptor 4 (TLR4)/MD-2. TLR4 signals a membrane constituent of Gram-negative bacteria, lipopolysaccharide (LPS). MD-2 is indispensable for TLR4-dependent LPS responses because cells expressing TLR4/MD-2, but not TLR4 alone, respond to LPS. RP105 also has a role in LPS responses because B cells lacking RP105 show hyporesponsiveness to LPS. Little is known, however, regarding whether MD-1 is important for RP105-dependent LPS responses, as MD-2 is for TLR4. To address the issue, we developed mice lacking MD-1 and generated monoclonal antibodies (mAbs) to the protein. MD-1–null mice showed impairment in LPS-induced B-cell proliferation, antibody production, and B7.2/CD86 up-regulation. These phenotypes are similar to those of RP105-null mice. The similarity was attributed to the absence of cell surface RP105 on MD-1–null B cells. MD-1 is indispensable for cell surface expression of RP105. A role for MD-1 in LPS responses was further studied with anti–mouse MD-1 mAbs. In contrast to highly mitogenic anti-RP105 mAbs, the mAbs to MD-1 were not mitogenic but antagonistic on LPS-induced B-cell proliferation and on B7.2 up-regulation. Collectively, MD-1 is important for RP105 with respect to B-cell surface expression and LPS recognition and signaling.
Introduction
The innate immune response is the first line of defense against microbial pathogens.1-3 The principal challenge for the immune system is to recognize pathogens and to mount an immediate defense response. A wide variety of bacterial components are capable of stimulating the innate immune system. These include lipopolysaccharide (LPS), peptidoglycan, lipoteichoic acid, lipoarabinomannan, lipopeptides, and bacterial DNA. LPS is a principal component of Gram-negative bacteria that potently activates the innate immune system, and it is one of the best-studied molecules among microbial products.4
Toll-like receptors (TLRs), mammalian homologues ofDrosophila Toll, have been implicated in innate recognition and signaling of these microbial products in humans and mice.5,6 TLR4, for example, signals LPS. The mutations of the TLR4 gene lead to LPS hyporesponsiveness in humans and mice.7-10 TLR4 expression, however, does not confer LPS responsiveness on cell lines, suggesting a requirement for an additional molecule.11 We recently cloned a molecule, MD-2, that is associated with the extracellular domain of TLR4. Coexpression of MD-2 imparts LPS responsiveness to TLR4.11
MD-2 was originally identified as a molecule similar to MD-1. MD-1 is associated with the extracellular domain of RP105.12 RP105 is a type 1 transmembrane protein with extracellular leucine-rich repeats (LRRs) and a short cytoplasmic tail.13 RP105 is similar to Drosophila Toll in the extracellular LRRs. In this regard, RP105 is the first mammalian molecule to be described as similar to Toll. In contrast to ubiquitous expression of TLR4, RP105 expression is largely restricted to immune cells including mature B cells and macrophages.13 Antibody-mediated cross-linking of RP105 induces resistance against irradiation-induced apoptosis, robust B-cell proliferation, and up-regulation of a costimulatory molecule, B7.2, revealing RP105 as a potent regulator of B-cell activation.14-17 RP105 has an important role in B-cell activation by LPS because B cells lacking RP105 are impaired in LPS-induced proliferation and antibody production.18Little is known, however, about a role for MD-1 in RP105-dependent LPS responses. We addressed the issue by establishing monoclonal antibodies (mAbs) to mouse MD-1 and mice lacking MD-1.
Materials and methods
Animals
BALB/c and C57BL/6 mice were obtained from Japan SLC (Hamamatsu, Japan). The mice lacking RP105 or MD-1 were maintained in the animal facility at Saga Medical School.
Cells and derivation of the monoclonal antibodies to mouse MD-1
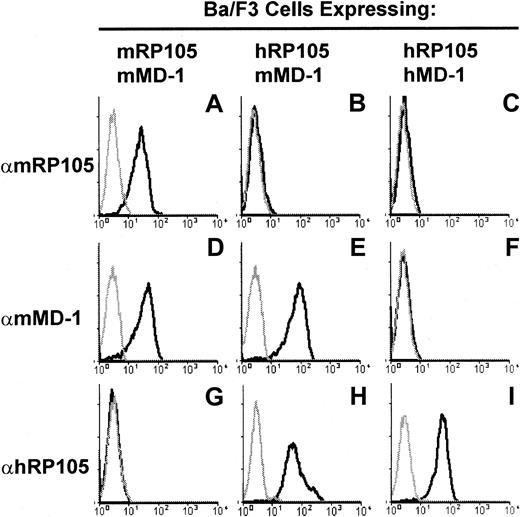
Normal rat kidney (NRK) cells stably expressing mouse RP105 and MD-1 were established by transfecting the pEFBOS expression vectors encoding these molecules.19 Ba/F3 cells20expressing mouse RP105/MD-1 and human RP105/MD-1 were described previously.12 15 We further established Ba/F3 cells expressing human RP105 and mouse MD-1 by transfecting pEFBOS vectors encoding these molecules. These 2 molecules were able to associate with each other and were expressed on the cell surface (Figure 1).
Establishment of anti–MD-1 mAbs.
The specificity of anti–MD-1 mAbs is shown with flow cytometry staining of Ba/F3 cells expressing either mouse RP105/mouse MD-1 (A, D, G), human RP105/mouse MD-1 (B, E, H), or human RP105/human MD-1 (C, F, I). These cells were stained with the mAbs to either mouse RP105, RP/14 (A-C), mouse MD-1, MD14 (D-F), or human RP105, MHR73 (G-I). FITC-labeled goat anti–rat IgG or anti–mouse IgG was used for the second reagent. Gray lines depict staining with the second antibody alone.
Establishment of anti–MD-1 mAbs.
The specificity of anti–MD-1 mAbs is shown with flow cytometry staining of Ba/F3 cells expressing either mouse RP105/mouse MD-1 (A, D, G), human RP105/mouse MD-1 (B, E, H), or human RP105/human MD-1 (C, F, I). These cells were stained with the mAbs to either mouse RP105, RP/14 (A-C), mouse MD-1, MD14 (D-F), or human RP105, MHR73 (G-I). FITC-labeled goat anti–rat IgG or anti–mouse IgG was used for the second reagent. Gray lines depict staining with the second antibody alone.
Rats were immunized with NRK cells expressing mouse RP105/MD-1, and mAbs were screened that stained RP105/MD-1–expressing Ba/F3 cells specifically. We next excluded the mAbs reactive with RP105 by selecting the mAbs that stained Ba/F3 cells expressing human RP105/mouse MD-1, but not those expressing human RP105/human MD-1 (Figure 1).9 Monoclonal antibodies were cloned and purified for further studies. We used 2 mAbs, MD14 (rat IgG2a/κ) and MD113 (rat IgG2b/κ) in the current study.
Generation of the MD-1–deficient mice
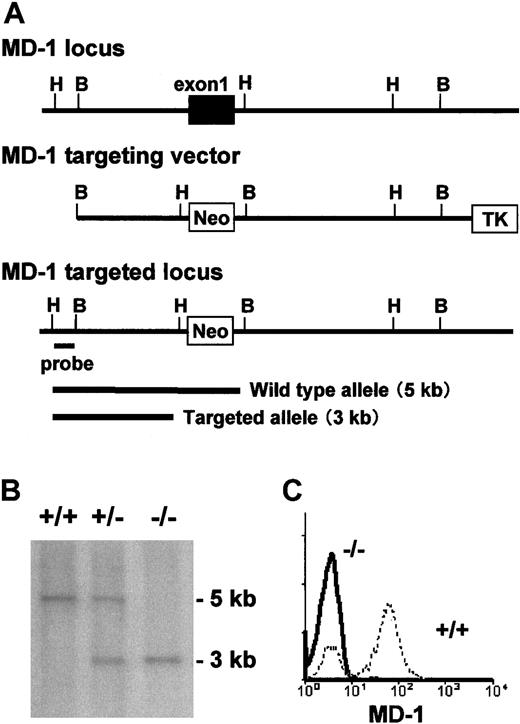
The MD-1 genomic clones were purchased from Genome Systems (St Louis, MO), and the MD-1 gene was subcloned into the pBluescript II vector (Stratagene, La Jolla, CA). The first exon of theMD-1 gene was replaced by the neomycin resistance(neor) gene, which is under the control of the mouse phosphoglycerate kinase-1 (PGK-1) gene promoter (Figure2A). The herpes simplex virus thymidine kinase (HSV-tk) gene, driven by the PGK-1 promoter, was introduced at the 3′ end of the targeting vector. Linearized DNA of the targeting vector was transfected into the E14.1 embryonic stem cells; this was followed by selection of the G418 and ganciclovir-resistant embryonic stem cell clones, as described previously.18Clones of double-resistant cells were collected and genotyped by Southern blot analysis using the probe located outside the targeting construct (Figure 2A). A homologous recombinant embryonic stem clone was used to produce chimeric mice by an aggregation method, as described previously.21 Male chimera mice were then bred with C57BL/6 female mice for the generation of heterozygous MD-1± mice, which were further bred with each other to generate homozygous MD-1−/− mice (Figure 2B). Wild-type or heterozygous littermates were used as controls. Mice were kept under conventional conditions in the animal facility at Saga Medical School. Experiments were carried out according to institutional ethical guidelines for animal experiments and according to safety guidelines for gene manipulation experiments.
Targeted disruption of the murine
MD-1 gene by homologous recombination. (A) Schematic representation of the MD-1 gene, the targeting construct, and the targeted MD-1 locus. The first exon of theMD-1 gene is shown as a closed box. The indicated external probe detects 5.0 and 3.0 kb of the HindIII-digested genomic DNA of the wild-type and targeted MD-1 genes, respectively. (B) Southern blot analysis of the MD-1 mutation in mice. Tail DNA isolated from the wild-type, MD-1+/−, and MD-1–−/− mice was digested with HindIII, electrophoresed on an agarose gel, blotted onto nylon membrane, and hybridized with the probe, which is shown in panel A. (C) Spleen cells from wild-type (broken line) or MD-1−/− mice (thick line) were stained with an anti–MD-1 mAb MD14, followed by FITC-labeled goat anti–rat IgG.
Targeted disruption of the murine
MD-1 gene by homologous recombination. (A) Schematic representation of the MD-1 gene, the targeting construct, and the targeted MD-1 locus. The first exon of theMD-1 gene is shown as a closed box. The indicated external probe detects 5.0 and 3.0 kb of the HindIII-digested genomic DNA of the wild-type and targeted MD-1 genes, respectively. (B) Southern blot analysis of the MD-1 mutation in mice. Tail DNA isolated from the wild-type, MD-1+/−, and MD-1–−/− mice was digested with HindIII, electrophoresed on an agarose gel, blotted onto nylon membrane, and hybridized with the probe, which is shown in panel A. (C) Spleen cells from wild-type (broken line) or MD-1−/− mice (thick line) were stained with an anti–MD-1 mAb MD14, followed by FITC-labeled goat anti–rat IgG.
Cell staining and flow cytometry
Single-cell suspensions were incubated at 2 × 105cells/100 μL on ice in staining buffer (phosphate-buffered saline containing 0.5% bovine serum albumin and 0.01% NaN3) with optimal amounts of antibodies for 15 minutes. For unlabeled antibodies, cells were washed in staining buffer, and were incubated with fluorescein isothiocyanate (FITC)–conjugated goat anti–rat immunoglobulin G (IgG) or anti–mouse IgG antibody (American Qualex, San Clemente, CA) for 10 minutes. The mAbs used were phycoerythrin-conjugated rat anti–mouse B7.2 (Cedarlane Laboratories, Ontario, Canada); FITC-conjugated rat anti–mouse CD23 (PharMingen, San Diego, CA), rat anti–mouse RP105 (RP/14),14 and mouse anti–human RP105 (MHR73).15 We also stained cells that had been treated with membrane permeabilization.22 Flow cytometric analysis was performed on a FACScan cytometer (Becton Dickinson, Mountain View, CA).
Analysis of in vitro B-cell activation
Splenic B cells were purified by negative selection with the anti-CD43 mAb S7 conjugated to Dynabeads M-450 sheep anti–rat IgG (Dynal, Lake Success, NY) as described.18 B-cell purity has been always higher than 95%, as judged by flow cytometry analyses (data not shown). Wild-type and mutant B cells were incubated at 2 × 105/well in 96-well, flat-bottomed plates with: goat anti–mouse IgM F(ab')2 (Organon Teknika, Tokyo, Japan); rat anti–mouse CD40 mAb LB429S23; rat anti–mouse RP105 mAb RP/14; or LPS. For the analysis of LPS responses, B cells were treated with LPS derived from Salmonella minnesota Re595 or Escherichia coli 055:B5. Lipid A was of bacterial origin (S minnesota). For proliferation, B cells were stimulated for 3 days, pulsed with 1 μCi/well (37 000 Bq) [3H]-TdR, and harvested onto a glass filter. The uptake of [3H]-TdR was counted as described previously.18
Analysis of humoral immune responses
Littermate control and MD-1–deficient mice were immunized intraperitoneally with 20 μg trinitrophenyl (TNP)–LPS in phosphate-buffered saline (5 mice per group). Serum concentrations of TNP-specific antibodies at different time points were measured by enzyme-linked immunosorbent assay (ELISA). ELISA was performed by coating plastic plates with TNP-bovine serum albumin (10 μg/mL), and serial serum dilutions were applied onto the plate. Bound antibodies were revealed by goat antibodies specific for IgM or IgG3 isotype (Caltag Laboratories, Burlingame, CA).
Statistical analysis
Significance was evaluated using the Student t test for unpaired data.
Immunoprecipitation and immunoprobing
The interleukin-3 (IL-3)–dependent line Ba/F3 was transfected with the pEFBOS expression vector encoding a variety of mouse TLR4, mouse MD-2, mouse RP105, and mouse MD-1. Ba/F3 cells expressing RP105/MD-1; RP105 and MD-2; and RP105/MD-1 + TLR4/MD-2 were established as described previously.11,18 Cells were washed and lysed in lysis buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 50 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 10 μg/mL soybean trypsin inhibitor, 2 mM MgCl2, and 2 mM CaCl2. After 30 minutes of incubation on ice, the lysate was centrifuged and nuclei were removed. Beads coupled with anti-RP105 mAb RP/14 or with the anti-flag mAb M2 were added to the cell lysate and were rotated for 4 hours at 4°C. Beads were washed in the lysis buffer, and bound proteins were subjected to SDS-PAGE (10% acrylamide under nonreduced conditions) and Western blot analysis. Precipitated proteins were detected with the anti-flag mAb M2 (Sigma, St Louis, MO) and Supersignal chemiluminescent substrate (Pierce, Rockford IL). Cell surface biotinylation was also used to detect RP105, as described previously.13
Results
Derivation of monoclonal antibodies to mouse MD-1
To detect cell surface MD-1, we made mAbs to mouse MD-1 and showed the specificity of mAbs by fluorescence staining (Figure 1). Anti–mouse MD-1 mAbs reacted with mouse RP105/MD-1 but did not cross-react with human RP105/MD-1. Whereas the anti-RP105 mAb did not react with the complex consisting of human RP105 and mouse MD-1, the anti–MD-1 mAbs did (Figure 1B,E), confirming their specificity to MD-1. Anti–MD-1 mAbs were used for fluorescence staining of spleen cells. Positive cells were mainly restricted to mature B cells (Figure2C and data not shown), which were similar to the results with the anti-RP105 mAb.14
B cells lacking MD-1 are hyporesponsive to lipopolysaccharide
To address a role of MD-1 in the immune system, we made mice lacking MD-1. The first exon encodes a signal sequence and the following 27 amino acids. The exon was replaced with the neomycin resistance gene in the targeted allele (Figure 2A). Truncated products, even if aberrantly produced, should stay inside the cells. Mice homozygous with the targeted allele, which was confirmed with Southern hybridization (Figure 2B), did not express MD-1 as judged by flow cytometry staining of spleen cells with the anti–MD-1 mAb (Figure2C).
Flow cytometry analyses of lymphoid organs, including the bone marrow, spleen, lymph node, and thymus, did not reveal any differences between homozygous and wild-type mice, demonstrating normal lymphocyte and myeloid cell development in the absence of MD-1 (data not shown). Because MD-1 was expressed on mature spleen B cells, we stained a variety of B cell markers, including CD45, CD21, CD22, CD23, surface IgM, and surface IgD. Again, we could not see any difference between wild-type and MD-1–null B cells. Splenic B cell maturation was not altered in the absence of MD-1.
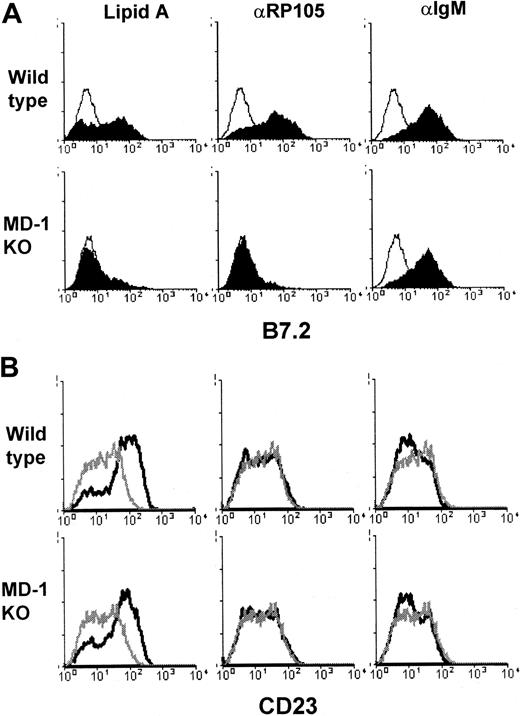
B-cell responsiveness to a variety of stimuli was next studied. MD-1–deficient B cells were not impaired in proliferation induced by anti-CD40 or anti-IgM antibody, whereas they revealed low responses in proliferation driven by anti-RP105 mAb, lipid A, or LPS prepared fromS minnesota or E coli (Figure3). These results prompted us to further study B-cell responses to LPS or the anti-RP105 mAb. In addition to proliferation, LPS-induced B-cell activation led to up-regulation of a variety of cell surface molecules, such as B7.2/CD86. B7.2 is a costimulatory molecule that facilitates an interaction with T cells (reviewed in Lenschow et al24). LPS, therefore, acts as an adjuvant in this context. Lipid A or anti-RP105 mAb induced B7.2 up-regulation on wild-type B cells, but not on those lacking MD-1 (Figure 4). The lack of MD-1, however, did not affect anti-IgM–induced B7.2 up-regulation, revealing the selective impairment in the responses to LPS or the anti-RP105 mAb (Figure 4). Not all LPS responses were impaired in B cells lacking MD-1. CD23 is a B-cell activation antigen involved in the regulation of B-cell proliferation and the production of IgE. Interestingly, CD23 was up-regulated with lipid A but not with the anti-RP105 mAb or anti-IgM Ab. Lipid A-induced up-regulation of CD23 was not impaired in MD-1–null mice (Figure 4, and “Discussion”).
MD-1–deficient B cells are impaired in LPS-induced B-cell proliferation.
Purified spleen B cells prepared from wild-type (■) or MD-1–deficient mice (▪) were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). [3H]-TdR uptake was counted as described in “Materials and methods.” Results were shown as mean values of count per minute (cpm) from triplicate wells with standard errors. When compared with wild-type B cells, MD-1–deficient B cells showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .001; lipid A 0.1 μg/mL, P < .002; lipid A 1.0 μg/mL, P < .003; Re595 0.01 μg/mL,P < .011; Re595 0.1 μg/mL, P < .001; Re595 1.0 μg/mL, P < .0001; 055:B5 0.01 μg/mL,P < .11; 055:B5, 0.1 μg/mL, P < .002; 055:B5 1.0 μg/mL, P < .001.
MD-1–deficient B cells are impaired in LPS-induced B-cell proliferation.
Purified spleen B cells prepared from wild-type (■) or MD-1–deficient mice (▪) were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). [3H]-TdR uptake was counted as described in “Materials and methods.” Results were shown as mean values of count per minute (cpm) from triplicate wells with standard errors. When compared with wild-type B cells, MD-1–deficient B cells showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .001; lipid A 0.1 μg/mL, P < .002; lipid A 1.0 μg/mL, P < .003; Re595 0.01 μg/mL,P < .011; Re595 0.1 μg/mL, P < .001; Re595 1.0 μg/mL, P < .0001; 055:B5 0.01 μg/mL,P < .11; 055:B5, 0.1 μg/mL, P < .002; 055:B5 1.0 μg/mL, P < .001.
LPS-induced up-regulation of B7.2/CD86, but not CD23, is impaired in MD-1–deficient B cells.
Purified B cells from wild-type (upper panels) or MD-1–deficient (lower panels) mice were stimulated with lipid A (1.0 μg/mL), anti-RP105 (10 μg/mL), or anti-IgM (10 μg/mL), as indicated. After 1 day, cells were harvested and stained with anti-B7.2 (A) or anti-CD23 (B). Open histograms (A) and gray lines (B) show cells stimulated with medium alone. Similar results were obtained from 3 independent experiments.
LPS-induced up-regulation of B7.2/CD86, but not CD23, is impaired in MD-1–deficient B cells.
Purified B cells from wild-type (upper panels) or MD-1–deficient (lower panels) mice were stimulated with lipid A (1.0 μg/mL), anti-RP105 (10 μg/mL), or anti-IgM (10 μg/mL), as indicated. After 1 day, cells were harvested and stained with anti-B7.2 (A) or anti-CD23 (B). Open histograms (A) and gray lines (B) show cells stimulated with medium alone. Similar results were obtained from 3 independent experiments.
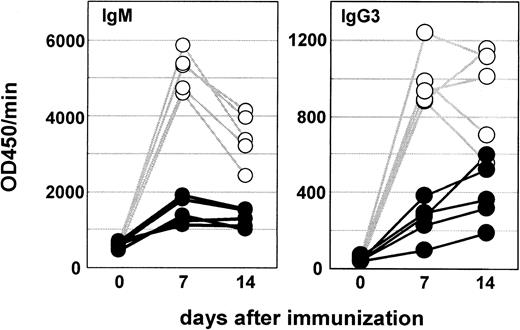
We also studied an in vivo B-cell response to LPS. Mice were immunized with TNP-LPS, and the production of TNP-specific antibodies was measured with ELISA. We show the results of IgM or IgG3 isotypes, which were dominant in response to TNP-LPS. Consistent with in vitro studies, MD-1–deficient mice revealed hyporesponsiveness in antibody production to TNP-LPS (Figure 5). We also studied antibody responses to a thymus-independent type 2 antigen TNP-Ficoll. MD-1–deficient mice were not affected in the antibody response (data not shown).
MD-1–deficient mice were impaired in LPS-induced antibody production.
Littermate control (○) and MD-1–deficient mice (●) were immunized intraperitoneally with TNP-LPS (20 μg per mouse) and bled 1 and 2 weeks later. Five mice were used for each group. Titers of TNP-specific IgM or IgG3 were measured with ELISA as described in “Materials and methods.”
MD-1–deficient mice were impaired in LPS-induced antibody production.
Littermate control (○) and MD-1–deficient mice (●) were immunized intraperitoneally with TNP-LPS (20 μg per mouse) and bled 1 and 2 weeks later. Five mice were used for each group. Titers of TNP-specific IgM or IgG3 were measured with ELISA as described in “Materials and methods.”
MD-1 is indispensable for RP105 expression on the B-cell surface
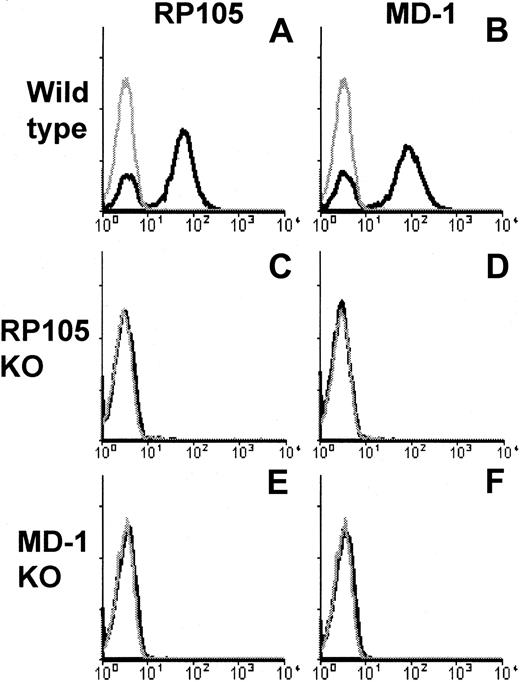
Our results clearly showed that B cells lacking MD-1 were hyporesponsive to LPS or anti-RP105 mAb. In this regard, B cells lacking MD-1 are similar to RP105-null B cells.18 RP105 and MD-1 are associated with each other, and we previously showed that transfected MD-1 up-regulated cell surface RP105 that had been transfected beforehand,12 15 suggesting an important role for MD-1 in cell surface expression of RP105. LPS hyporesponsiveness in B cells lacking MD-1 may be attributed to impaired expression of RP105. We stained RP105 on B cells lacking MD-1 and found the complete absence of RP105 (Figure 6). We further sought the RP105 protein, which might have been produced but stayed inside the cells because of the lack of MD-1. We stained MD-1–deficient B cells that had been subjected to membrane-permeabilizing treatment with the aim to detect intracellular RP105, but we could not detect it (data not shown). We next treated B cells lacking MD-1 with recombinant soluble MD-1, which, however, did not complement cell surface RP105 expression (data not shown). Collectively, the coexpression of MD-1 is indispensable for cell surface RP105 expression in vivo. In the meantime, MD-1 was not detected on B cells lacking RP105 (Figure 6). MD-1 may be membrane-anchored only by RP105 on the B-cell surface.
MD-1 is required for cell surface expression of RP105.
Spleen cells from wild-type (A, B), RP105-deficient (C, D), or MD-1–deficient (E, F) mice were stained with anti-RP105 mAb RP/14 (left column) or anti–MD-1 mAb MD14 (right column), followed by goat anti–rat IgG-FITC. Stained cells were analyzed on FACScan. Gray lines show the histograms obtained from cells stained with the second reagent alone.
MD-1 is required for cell surface expression of RP105.
Spleen cells from wild-type (A, B), RP105-deficient (C, D), or MD-1–deficient (E, F) mice were stained with anti-RP105 mAb RP/14 (left column) or anti–MD-1 mAb MD14 (right column), followed by goat anti–rat IgG-FITC. Stained cells were analyzed on FACScan. Gray lines show the histograms obtained from cells stained with the second reagent alone.
Antagonistic effect of the anti–MD-1 monoclonal antibody on B-cell responses to lipopolysaccharide
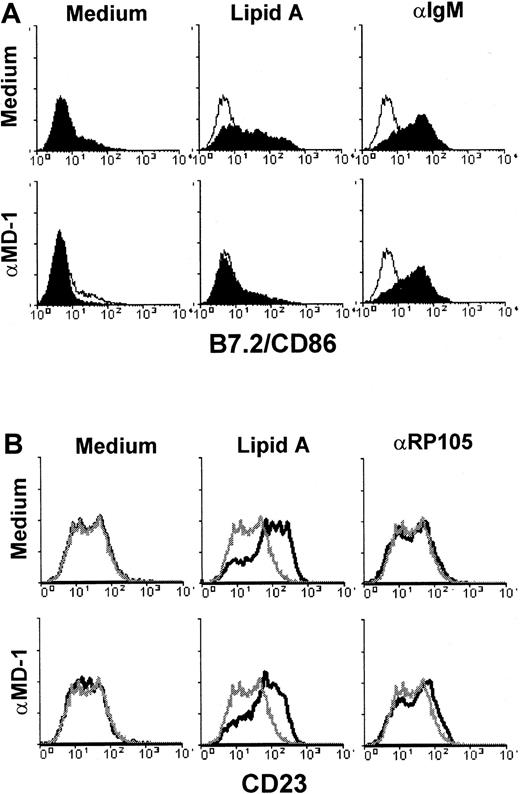
We further asked whether MD-1 contributed functionally to B-cell activation by antibody-mediated RP105 ligation or LPS. Anti–MD-1 mAbs were included in in vitro B-cell activation studies. We have established as many as 20 independent mAbs to RP105, all of which induced robust B-cell proliferation (Miyake et al14 and data not shown). In sharp contrast, none of the 9 mAbs to MD-1 was mitogenic (Figure 7 and data not shown). In this regard, all the anti–MD-1 mAbs we obtained were functionally distinct from the highly mitogenic anti-RP105 mAbs. Anti–MD-1 mAbs were included in B-cell proliferation driven by a variety of stimuli (Figure 7). They also were inhibitory of B-cell proliferation driven by LPS and by an anti-RP105 mAb, and they also suppressed LPS-induced up-regulation of B7.2 (Figure 8). Inhibition was specific because the anti–MD-1 mAb had no effect on the anti–IgM-induced B7.2 up-regulation. LPS-induced up-regulation of CD23 was barely influenced by the anti–MD-1 mAb, showing consistency with the results using B cells lacking MD-1.
Effect of an anti–MD-1 mAb on LPS-induced B-cell proliferation.
Purified spleen B cells were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-IgM (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). Anti–MD-1 mAb MD88 (10 μg/mL) was included in the groups shown with closed bars. [3H]-TdR uptake was counted as described in “Materials and methods.” Results were represented by mean cpm values from triplicate wells with standard errors. When compared with B cells cultured without antibody, B cells cultured in the presence of the anti–MD-1 mAb showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .003; Re595 1.0 μg/mL, P < .012; Re595 0.1 μg/mL, P < .010; Re595 0.01 μg/mL,P < .003; 055:B5 0.01 μg/mL, P < .005; 055:B5 0.1 μg/mL, P < .001.; 055:B5 1.0 μg/mL,P < .001.
Effect of an anti–MD-1 mAb on LPS-induced B-cell proliferation.
Purified spleen B cells were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-IgM (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). Anti–MD-1 mAb MD88 (10 μg/mL) was included in the groups shown with closed bars. [3H]-TdR uptake was counted as described in “Materials and methods.” Results were represented by mean cpm values from triplicate wells with standard errors. When compared with B cells cultured without antibody, B cells cultured in the presence of the anti–MD-1 mAb showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .003; Re595 1.0 μg/mL, P < .012; Re595 0.1 μg/mL, P < .010; Re595 0.01 μg/mL,P < .003; 055:B5 0.01 μg/mL, P < .005; 055:B5 0.1 μg/mL, P < .001.; 055:B5 1.0 μg/mL,P < .001.
Effect of an anti–MD-1 mAb on up-regulation of B7.2/CD86 and CD23.
Purified B cells were stimulated with lipid A (1.0 μg/mL), anti-RP105 (10 μg/mL), or anti-IgM (10 μg/mL) in the presence (lower panels) or absence (upper panels) of the anti–MD-1 mAb MD113 (10 μg/mL), as indicated. After 1 day, cells were harvested and stained with anti-B7.2 (A) or anti-CD23 (B). Open histograms (upper panels) and gray lines (lower panels) show cells stimulated with medium alone. Similar results were obtained from 3 independent experiments.
Effect of an anti–MD-1 mAb on up-regulation of B7.2/CD86 and CD23.
Purified B cells were stimulated with lipid A (1.0 μg/mL), anti-RP105 (10 μg/mL), or anti-IgM (10 μg/mL) in the presence (lower panels) or absence (upper panels) of the anti–MD-1 mAb MD113 (10 μg/mL), as indicated. After 1 day, cells were harvested and stained with anti-B7.2 (A) or anti-CD23 (B). Open histograms (upper panels) and gray lines (lower panels) show cells stimulated with medium alone. Similar results were obtained from 3 independent experiments.
RP105 does not interact with MD-2
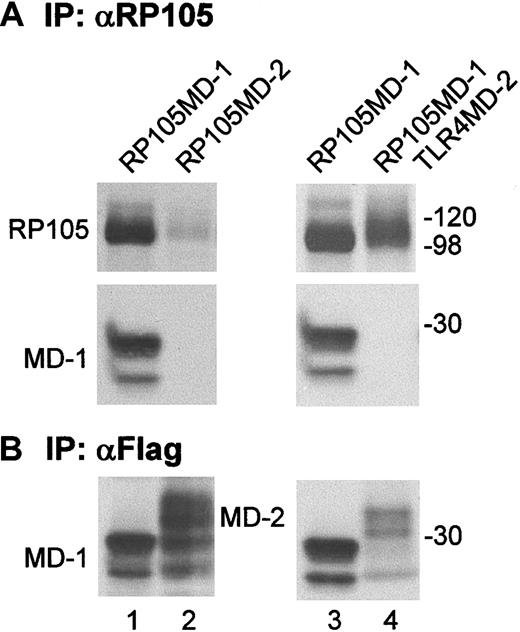
Finally, we sought to determine whether RP105 interacts with MD-2. Ba/F3 cells expressing RP105 and MD-2 were established and used for immunoprecipitation with anti-RP105 mAb (Figure9A). We could not, however, detect coprecipitation of MD-2, which was abundantly expressed as shown by immunoprecipitation and immunoprobing with the anti-flag mAb (Figure 9B, lane 2). The RP105 signal from Ba/F3 cells expressing RP105 and MD-2 was low compared with the RP105 signal from Ba/F3 cells expressing RP105/MD-1 (Figure 9A, lanes 1 and 2). Because cell surface RP105 is dependent on the coexpression of MD-1,12 we could not obtain Ba/F3 cells expressing MD-2 and RP105 whose expression is as high as on those expressing RP105/MD-1. This result is consistent with the results that cell surface RP105 is absent on MD-1–deficient B cells (Figure 6). To obtain higher expression of RP105 on the cell surface, we used Ba/F3 cells expressing RP105/MD-1 and TLR4/MD-2 (lane 4). RP105 was precipitated with anti-RP105 mAb, and coprecipitated MD-2, if there was any, was probed with anti-flag mAb. We still could not see MD-2 coprecipitation. Taken together with the absence of RP105 on B cells lacking MD-1 but expressing endogenous MD-2,11 RP105 was not likely to substitute MD-2 for MD-1.
RP105 does not interact with MD-2.
Ba/F3 cells expressing RP105 + MD-1 (lane 1), RP105 + MD-2 (lane 2), RP105/MD-1 (lane 3), or RP105/MD-1 + TLR4/MD-2 (lane 4) were used for immunoprecipitation with anti-RP105 mAb RP/14 (A) or with the anti-flag mAb M2 (B). RP105 was detected with cell surface biotinylation as described in “Materials and methods.” MD-1 (lanes 1, 3) and MD-2 (lanes 2, 4) were tagged with the Flag epitope and detected by immunoprobing with the anti-flag mAb.
RP105 does not interact with MD-2.
Ba/F3 cells expressing RP105 + MD-1 (lane 1), RP105 + MD-2 (lane 2), RP105/MD-1 (lane 3), or RP105/MD-1 + TLR4/MD-2 (lane 4) were used for immunoprecipitation with anti-RP105 mAb RP/14 (A) or with the anti-flag mAb M2 (B). RP105 was detected with cell surface biotinylation as described in “Materials and methods.” MD-1 (lanes 1, 3) and MD-2 (lanes 2, 4) were tagged with the Flag epitope and detected by immunoprobing with the anti-flag mAb.
Discussion
The current study underscored a role for MD-1 in B-cell responsiveness to LPS by producing mice that lacked MD-1. MD-1–deficient mice are similar to RP105-deficient mice in B-cell phenotype. Both types showed hyporesponsiveness in LPS-induced B-cell proliferation and antibody production. This similarity between the 2 mutant mice is attributed to the lack of cell surface RP105/MD-1 (Figure 6). MD-1 is indispensable for cell surface expression of RP105. We previously established Ba/F3 cells expressing RP105 alone by transfecting the expression vector encoding RP105.12Expression was enhanced by further transfection of MD-1. A role for MD-1 in vivo was found to be more critical than those in vitro results. Considering that RP105 is not expressed on B cells lacking MD-1 but expressing endogenous MD-2 (Figure 6 and 11), RP105 does not seem to substitute MD-2 for MD-1. In keeping with this, RP105 expression was low in Ba/F3 cells overexpressing RP105 and MD-2, and we could not see an association between RP105 and MD-2 (Figure 9). To achieve higher expression of RP105 and MD-2 on the cell surface, we also used Ba/F3 cells expressing RP105/MD-1 and TLR4/MD-2. We could not see the association between MD-2 and RP105 or RP105/MD-1 (Figure 9, lanes 3 and 4). These results revealed a specific interaction between RP105 and MD-1 but not with MD-2.
The heavy chain of major histocompatibility complex (MHC) class I is associated with β2-microglobulin (β2m). MD-1 and β2m are similar in that β2m does not have a transmembrane portion; each is attached to another molecule on the cell surface. The absence of β2m leads to misfolding of the MHC class I heavy chain. The misfolded heavy chain is not allowed to proceed beyond the endoplasmic reticulum (ER) and is subject to degradation (reviewed in Pamer and Cresswell25). RP105 alone may be similarly unstable in ER. Ectopically expressed human RP105 alone, which stayed inside the Ba/F3 cells, appeared in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses to be less glycosylated than the RP105 associated with MD-1 on the Ba/F3 cell surface (Miura et al15 and data not shown). RP105 alone may not be able to proceed beyond ER and may be subject to ER-associated degradation.26 If this is the case, it is reasonable that exogenously added recombinant soluble MD-1, which would not reach ER, had no effect on recovering RP105 expression (data not shown). RP105 expression was not detected with mixed cell culture of spleen cells lacking either RP105 or MD-1, which was expected to provide B cells lacking MD-1 with soluble MD-1 from B cells lacking RP105 (data not shown). It seems important for MD-1 to be coexpressed with RP105 in the same cells.
Flow cytometry staining with cell membrane permeabilization did not detect intracellular RP105 in B cells lacking MD-1. Misfolded RP105 would not accumulate but is likely to be degraded. Even after the treatment of spleen cells lacking MD-1 with proteasome inhibitors such as lactacystin and N-carbobenzoxyl-leucinyl-leucinal,27 we could not detect intracellular RP105 by flow cytometry staining with cell membrane permeabilization. Additional biochemical studies using metabolic labeling and immunoprecipitation of RP105 would be required for revealing an intracellular fate of RP105 in the absence of MD-1.
We established as many as 20 independent mAbs to RP105, all of which induced robust B-cell proliferation (Miyake et al14 and data not shown). In sharp contrast, none of the 9 anti–MD-1 mAbs was mitogenic (Figure 7 and data not shown). The anti–MD-1 mAbs should be similar to the anti-RP105 mAbs in cross-linking RP105/MD-1; nonetheless, only anti-RP105 mAbs are mitogenic. Cross-linking of RP105/MD-1 is insufficient for triggering B-cell activation. It must be noted that the cytoplasmic portion of RP105 consists of only 11 amino acids, which would be too short to deliver an activation signal. Another molecule may deliver an activation signal for RP105/MD-1. The anti–MD-1 mAbs might disrupt a link between RP105/MD-1 and such a putative signal transducer, whereas the anti-RP105 mAbs could strengthen the association and trigger an activation signal through the putative signal transducer. A search for the signal transducer is underway.
Using anti–MD-1 mAbs, we showed a functional role for MD-1 in RP105-dependent and LPS-triggered B-cell activation. The anti–MD-1 mAbs were found to be antagonistic on LPS-induced proliferation and B7.2 up-regulation. These results are consistent with LPS hyporesponsiveness in mice lacking either RP105 or MD-1, and they suggest a functional role for MD-1 in B-cell responses to LPS. We believe that MD-1 is not just a molecule that helps RP105 to come out on the cell surface but that MD-1 helps RP105 to signal LPS, as MD-2 does TLR4.
LPS-induced up-regulation of CD23 was not impaired in B cells lacking MD-1 (Figure 4). Anti–MD-1 mAb was unable to inhibit LPS-induced CD23 up-regulation (Figure 8). TLR4 is, on the other hand, indispensable for this response because B cells derived from TLR4-mutated C3H/HeJ mice do not show CD23 up-regulation in response to LPS (data not shown). CD23 up-regulation seems to require the LPS signaling through TLR4 but not through RP105/MD-1. In keeping with this, anti-RP105, antibody-dependent ligation of RP105/MD-1 did not lead to CD23 up-regulation (Figure 8B). These results reveal that LPS responses in B cells are not simple but are heterogeneous in their requirement for the signal through RP105/MD-1. B cells require the signal through RP105/MD-1 and also through TLR4/MD-2 for B7.2 up-regulation, proliferation, and antibody production, but not for CD23 up-regulation.
It is important to note that tissue distribution of TLR4/MD-2 and RP105/MD-1 is not similar. TLR4/MD-2 is broadly expressed in organs including heart, kidney, brain, and liver,5,6,8 whereas RP105/MD-1 is restricted to immune cells such as B cells, macrophages, and dendritic cells.12,28 Nonimmune cells are likely to use only TLR4/MD-2 for LPS recognition and signaling. B cells, on the other hand, have an additional recognition or signaling pathway through RP105/MD-1. Gorczynski et al29 showed that MD-1 down-regulation with the antisense oligodeoxynucleotides led to impairment in LPS-induced CD80/CD86 up-regulation on bone marrow-derived dendritic cells. The antisense treatment is likely to result in the down-regulation of RP105/MD-1. Dendritic cells may be similar to B cells in their requirement for RP105/MD-1 in LPS recognition and signaling. Further studies about mice lacking MD-1 will focus on cell lineages other than B cells, including macrophages and dendritic cells.
Supported by Special Coordination Funds of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government (Monbukagakusho); Uehara Memorial Foundation; Yamanouchi Foundation for Research on Metabolic Disorders; Mitsubishi-Tokyo Pharmaceutical; and Sankyo.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kensuke Miyake, Department of Microbiology and Immunology, Division of Infectious Genetics, The Institute of Medical Science, The University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639; e-mail: kmiyake@ims.u-tokyo.ac.jp.

![Fig. 3. MD-1–deficient B cells are impaired in LPS-induced B-cell proliferation. / Purified spleen B cells prepared from wild-type (■) or MD-1–deficient mice (▪) were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). [3H]-TdR uptake was counted as described in “Materials and methods.” Results were shown as mean values of count per minute (cpm) from triplicate wells with standard errors. When compared with wild-type B cells, MD-1–deficient B cells showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .001; lipid A 0.1 μg/mL, P < .002; lipid A 1.0 μg/mL, P < .003; Re595 0.01 μg/mL,P < .011; Re595 0.1 μg/mL, P < .001; Re595 1.0 μg/mL, P < .0001; 055:B5 0.01 μg/mL,P < .11; 055:B5, 0.1 μg/mL, P < .002; 055:B5 1.0 μg/mL, P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/5/10.1182_blood.v99.5.1699/5/m_h80522217003.jpeg?Expires=1765893535&Signature=qMTQ3wx0XR~-1s4ahQQ2ibrwpV6mNqwcJkImEdxXB2kH0dHZqOwC8a1fhtsTtfkm1WJw8MVV5v4~R3Rpsx-7GKglL6MBk2hJey32SI8wO7PFrCQinVviUqsTzaAyyPHtRsYraNG0w0OgdcFt2T~EzYxYll3dwCQotckNN4eltxn263Fmnqpyaaoh3Y9dO8pFiH9YMFWVYtb~Pk1af2I89Utsb7Q1Pf6C-qlV9Sc~6y1ZlB1L5bIGubCoahf9ankgOE-9ZdND1YmBZAachIjAQO01AYJleSNAAPn99wt3pz-N0gUXL1UT1jxrngAm3XqSacoUYdHDSV5rWOcCy1c2eg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Effect of an anti–MD-1 mAb on LPS-induced B-cell proliferation. / Purified spleen B cells were stimulated for 3 days with anti-CD40 (10 μg/mL); anti-IgM (10 μg/mL); anti-RP105 (2 μg/mL); lipid A (0.01, 0.1, and 1.0 μg/mL); LPS from S minnesota Re595 or E coli 055: B5 (0.01, 0.1, and 1.0 μg/mL). Anti–MD-1 mAb MD88 (10 μg/mL) was included in the groups shown with closed bars. [3H]-TdR uptake was counted as described in “Materials and methods.” Results were represented by mean cpm values from triplicate wells with standard errors. When compared with B cells cultured without antibody, B cells cultured in the presence of the anti–MD-1 mAb showed significant hyporesponsiveness to LPS as revealed by the following P values: lipid A 0.01 μg/mL,P < .003; Re595 1.0 μg/mL, P < .012; Re595 0.1 μg/mL, P < .010; Re595 0.01 μg/mL,P < .003; 055:B5 0.01 μg/mL, P < .005; 055:B5 0.1 μg/mL, P < .001.; 055:B5 1.0 μg/mL,P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/5/10.1182_blood.v99.5.1699/5/m_h80522217007.jpeg?Expires=1765893535&Signature=tRdUq42iZvfGE56dAvaFh0NrGI2Hr5XG1WJ3DwftZy-HJuPFothoZPK1rxTC4173U57TC8gY3AJ9LiCgt2Z0ZV~tX3O1f1xRGtpetXBhVgaU7fCrYMnA7PhbSup9jFeoReduKKdA3LQW0Z~cc~4fI~UleLsO2OS6tEqBU66PTOcz3dHjVRI8aXaKnCIl0Bo37~9hwtQiZdDI8h02d7yv~jpWGejet7EasLbLHIYiKBgpma27AwAffLt3MxbYIEW3WllENaEKDdtMTkXkGOZkQozvooODbcezGq4VJh10sTAjKQlcnx22stixnch7YGbihAYRuoav0Yhl6jLtK58-0Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal