The biallelic platelet-specific Gov antigen system—implicated in refractoriness to platelet transfusion, neonatal alloimmune thrombocytopenia, and posttransfusion purpura—is carried by the glycosylphosphatidylinositol (GPI)–linked protein CD109. The recent identification of the human CD109 complementary DNA (cDNA) has allowed the molecular nature of the Gov alleles to be elucidated. By using reverse transcriptase–polymerase chain reaction (RT-PCR) to amplify CD109 cDNAs from 6 phenotypically homozygous Govaa and Govbb individuals, we have determined that the Gov alleles differ by an A to C single nucleotide polymorphism (SNP) at position 2108 of the coding region, resulting in a Tyr/Ser substitution at CD109 amino acid 703. Allele-specific PCR sequence-specific primers (SSP), PCR-restriction fragment length polymorphism, and real-time PCR studies of 15 additional donors (5 Govaa, 5 Govbb, and 5 Govab) confirmed that this SNP correlates with the Gov phenotype. In addition, Chinese hamster ovary cells transiently expressing nucleotide 2108 A>C CD109 cDNA variants were recognized specifically by allele-specific Gov antisera, indicating that this polymorphism defines the Gov alloantigenic determinants. Real-time PCR was then used to genotype 85 additional Gov phenotyped donors. In all but 3 cases, genomic testing concurred with the Gov phenotype. Repeat testing corrected 2 of these discrepancies in favor of the genotyping result. The third discrepancy could not be resolved, likely reflecting low-level CD109 expression below the sensitivity of the phenotyping assay. We conclude that the Gov alleles are defined by a 2108 A>C SNP that results in a Tyr703Ser substitution of CD109 and that genotyping studies are more accurate for Gov alloantigen determination than are conventional serologic methods.
Introduction
The biallelic Gov antigen system is carried by the platelet membrane protein CD109.1-3 The CD109 antigen, first described on activated T cells and platelets,2,4-6is also found on endothelial cells and on a subset of primitive bone marrow progenitor and candidate hematopoietic stem cells.7-9 Although the precise function of CD109, a monomeric glycosylphosphatidylinositol (GPI)–anchored glycoprotein of approximately 170 kD,2 is not known, its recent identification as a novel member of the α2M/C3, C4, C5 family of thioester-containing proteins suggests that it may be capable of mediating covalent cell-substrate and cell-cell interactions (see accompanying article by Lin et al,10 page 1683).
Gov alloantibodies are implicated in refractoriness to platelet transfusion (PR) and can cause neonatal alloimmune thrombocytopenia and posttransfusion purpura.1,11,12 Until recently, however, the importance of the Gov antigens in alloantibody-mediated platelet destruction has not been defined. We have recently shown that the incidence of Gov alloantibodies, especially in PR patients, is high with an incidence exceeded only by those directed against the human platelet antigen 1 (HPA-1) system antigens.11Unfortunately, Gov phenotyping by serologic techniques is problematic for several reasons. First, the polyclonal antisera that are required are in limited supply and of variable quality, and often contain HLA, and sometimes HPA, alloantibodies as well. Second, CD109 also expresses the ABH antigens.13 Third, CD109 expression on platelets is low (approximately 2000 ± 400 molecules per thrombin-activated platelet7,14), and in addition shows significant interindividual variability, such that reliable phenotyping is sometimes precluded by very low-level expression. Finally, as the CD109 protein is readily released from the platelet membrane (presumably by GPI anchor cleavage), phenotyping must be performed on fresh samples.11 The elucidation of the molecular nature of the Gov alleles would permit genotypic Gov determination, thereby obviating the difficulties associated with phenotypic analysis.
As the Gov epitopes are unaffected by deglycosylation but are disrupted by sodium dodecyl sulfate denaturation,3 it is likely that the Gov alloantigens reflect differences in the primary CD109 amino acid sequence. Our recent identification of the human CD109 complementary DNA (cDNA)10 has allowed us to compare the nucleotide sequences of the 2 putative CD109 Gov alleles. Analysis of CD109 cDNAs derived from the platelet messenger RNA of 6 donors of known Gov phenotype (3 Govaa and 3 Govbb) revealed an allele-specific A to C single nucleotide polymorphism (SNP) at nucleotide 2108 of the coding sequence that results in a Tyr703Ser substitution in the CD109 protein. Expression studies using allele-specific CD109 cDNA variants confirmed that this SNP was sufficient for the formation of the Gov alloantigenic determinants. PCR-SSP, PCR-restriction fragment length polymorphism (RFLP), and Taqman real-time PCR genomic DNA analysis were subsequently developed to permit donor genotyping.
Materials and methods
Donor panels
Three donor panels comprising samples from a total of 106 apheresis donors were analyzed: panel 1, 6 donors (3 Govaaand 3 Govbb) whose CD109 cDNAs were sequenced to determine the difference between the 2 Gov alleles; panel 2, 15 blood group O donors (5 Govaa, 5 Govab, and 5 Govbb) with Gov phenotype confirmed by 2 or more monoclonal antibody-specific immobilization of platelet antigens (MAIPA) assays; and panel 3, 85 additional donors whose Gov phenotypes were determined once by MAIPA. Only donors with platelet counts more than 150 × 109/L were included in the 3 panels. All samples were obtained with informed consent and with the approval of the National Blood Service ethics review board.
Gov phenotyping by MAIPA assay
Determination of the Gov phenotype of platelets of all donors was performed by the modified MAIPA assay,15,16 using well-characterized polyclonal Gov-specific antisera and the CD109 monoclonal antibody (mAb) D2 (gift of R. W. Finberg, Boston, MA), as described previously.11 Chinese hamster ovary (CHO) cells transiently expressing Gov allele-specific cDNAs (106cells per assay) were characterized by MAIPA as described11but using the CD109 mAb TEA 2/16 (Pharmingen, San Diego, CA).
RNA extraction and cDNA synthesis
Total platelet RNA was prepared from panel 1 donors by using RNA STAT-60 messenger RNA isolation reagent following the manufacturers protocol (Tel-Test, Friendswood, TX). Briefly, 1 × 1010platelets were homogenized in 1 mL RNA STAT-60, 0.2 volume of chloroform was added, and the mixture was centrifuged at 12 000g for 15 minutes. RNA was precipitated from the aqueous layer by adding 0.5 volume isopropanol and spinning at 12 000g for 15 minutes at 4°C, washed in 70% ethanol, and resuspended in 400 μL of RNAse-free water. cDNA was prepared by incubating 120 μL RNA with 12 μL oligo dT primer at 70°C for 10 minutes and rapidly transferring to 4°C. Next, 6 μL SuperRT reverse transcriptase (HT Biotechnology, Cambridge, United Kingdom; 420 U/mL), 6 μL RNAse inhibitor RNAsin (Promega, Madison, WI; 0.8 U/μL), 30 μL reaction buffer, and 250 μM of each dNTP were added, and the reaction volume was brought to 300 μL with RNAse-free water. cDNA was synthesized at 42°C for 40 minutes.
PCR amplification and DNA sequence analysis of CD109 cDNAs
Platelet cDNA was used as the template for PCR amplification of CD109 sequences in 8 overlapping segments that spanned the entire open reading frame (ORF; Table 1). PCR reactions (50 μL) containing 1 × PCR buffer (Life Technologies, Burlington, ON, Canada), 1.5 mM MgCl2, 200 μM each dNTP, 1 μM each primer, 1.25 U Taq polymerase (Life Technologies), and 3 μL cDNA underwent 40 cycles of 94°C (45 seconds), primer-specific annealing temperature (Table 1; 45 seconds), and 72°C (45-60 seconds), using a Perkin Elmer 2400 thermocycler. PCR products (30 μL) were separated electrophoretically on a 1.2% agarose/TAE gel containing 1 μg/mL ethidium bromide, bands were excised, and DNA was purified by using the QIAquick gel extraction kit (Qiagen, Mississauga, ON, Canada). Sequencing reactions (3-5 μL purified product per reaction) were carried out by using the Thermosequenase Cy5.5 dye terminator sequencing kit (Amersham Pharmacia Biotech) with the use of either the initial amplification primers (Table 1) or selected internal CD109-specific primers as appropriate. Sequences were subsequently analyzed by using the Open Gene automated DNA sequencing system (Visible Genetics, Toronto, ON, Canada). In parallel, PCR products were cloned into PmeI-digested pMAB1, a pBS SK(−) (Stratagene, La Jolla, CA) derivative containing a PmeI restriction site within the polylinker. Resultant plasmid clones were analyzed by alkaline lysis/restriction digestion, and, as appropriate (and following an additional overnight 13% polyethylene glycol/1.6 M NaCl precipitation), by DNA sequence analysis.
CD109-specific oligonucleotide polymerase chain reaction primers
| Fragment . | Sense primer . | Antisense primer . | Size (bp) . | Ta (°C) . | ||
|---|---|---|---|---|---|---|
| 1 | K1-80 | (−24) | K1-650 | (544) | 568 | 59 |
| 5′-GTAGCCCAGGCAGACGCC-3′ | 5′-GTGACAACCACTGTTGGATCAA-3′ | |||||
| 2 | K1-1 | (445) | K1-1120 | (1014) | 570 | 50 |
| 5′-CGCATTGTTACACTCTTCTC-3′ | 5′-TACATTTCTTGAAATACCTG-3′ | |||||
| 3 | K1-1022 | (910) | K1-REV-1 | (1747) | 838 | 50 |
| 5′-GATTCTTCAAATGGACTTT-3′ | 5′-GGCTGTGTCACAGAGATC-3′ | |||||
| 4 | K1-1400 | (1291) | GSP1 | (2165) | 875 | 55 |
| 5′-TGAATTCCCAATCCTGGAGGA-3′ | 5′-GCCACCCAAGAAGTGATAGA-3′ | |||||
| 5 | K1-M43 | (1898) | 6R4N | (2998) | 1101 | 56 |
| 5′-TTCAGGAATGTGGACTCTGG-3′ | 5′-CGGCTTCAAGGAAACATCT-3′ | |||||
| 6 | K1-3080 | (2948) | 1-5N | (3859) | 912 | 56 |
| 5′-CTGGGAGCACTTGGTTGTCA-3′ | 5′-CAGCAACATCTAAATCAAAGGC-3′ | |||||
| 7 | K1-3570 | (3462) | 7U3N | (4337) | 876 | 50 |
| 5′-ACAATTTCAGACTTCTGAGG-3′ | 5′-CACAGCCAAAGTTCCATA-3′ | |||||
| 8 | K1-3920 | (3812) | K1-4600 | (4489) | 678 | 55 |
| 5′-GACGAAGATCTATCCAAAATC-3′ | 5′-GCTAGGACCTGTTGTACACC-3′ | |||||
| Fragment . | Sense primer . | Antisense primer . | Size (bp) . | Ta (°C) . | ||
|---|---|---|---|---|---|---|
| 1 | K1-80 | (−24) | K1-650 | (544) | 568 | 59 |
| 5′-GTAGCCCAGGCAGACGCC-3′ | 5′-GTGACAACCACTGTTGGATCAA-3′ | |||||
| 2 | K1-1 | (445) | K1-1120 | (1014) | 570 | 50 |
| 5′-CGCATTGTTACACTCTTCTC-3′ | 5′-TACATTTCTTGAAATACCTG-3′ | |||||
| 3 | K1-1022 | (910) | K1-REV-1 | (1747) | 838 | 50 |
| 5′-GATTCTTCAAATGGACTTT-3′ | 5′-GGCTGTGTCACAGAGATC-3′ | |||||
| 4 | K1-1400 | (1291) | GSP1 | (2165) | 875 | 55 |
| 5′-TGAATTCCCAATCCTGGAGGA-3′ | 5′-GCCACCCAAGAAGTGATAGA-3′ | |||||
| 5 | K1-M43 | (1898) | 6R4N | (2998) | 1101 | 56 |
| 5′-TTCAGGAATGTGGACTCTGG-3′ | 5′-CGGCTTCAAGGAAACATCT-3′ | |||||
| 6 | K1-3080 | (2948) | 1-5N | (3859) | 912 | 56 |
| 5′-CTGGGAGCACTTGGTTGTCA-3′ | 5′-CAGCAACATCTAAATCAAAGGC-3′ | |||||
| 7 | K1-3570 | (3462) | 7U3N | (4337) | 876 | 50 |
| 5′-ACAATTTCAGACTTCTGAGG-3′ | 5′-CACAGCCAAAGTTCCATA-3′ | |||||
| 8 | K1-3920 | (3812) | K1-4600 | (4489) | 678 | 55 |
| 5′-GACGAAGATCTATCCAAAATC-3′ | 5′-GCTAGGACCTGTTGTACACC-3′ | |||||
Primer pairs used for the polymerase chain reaction (PCR) amplification of 8 overlapping CD109 complementary DNA (cDNA) fragments spanning the entire open reading frame (ORF) are shown. The position of the 5′ end of each oligonucleotide with respect to the published CD109 cDNA sequence10 is noted in parentheses. The CD109 ORF encompasses nucleotides 1 to 4335. The size of each PCR product (in base pairs) and the annealing temperature (Ta) used for the corresponding primer pair are listed.
By combining direct PCR sequencing and the analysis of subcloned fragments, we ensured that the DNA sequence of each PCR-derived cDNA fragment was obtained independently at least twice. Each fragment was sequenced fully in both directions.
Expression of Gova and Govb CD109 cDNAs in CHO cells
On the basis of the presence of an A at position 2108 of the coding region, our recently identified human CD109 cDNA (clone K1)10 was shown to correspond to the putativeGova allele. To construct a full-lengthGovb cDNA, a 572-base pair (bp) fragment encompassing nucleotide 2108 was generated by PCR from platelet cDNA from one of the original Govbb donors, using oligonucleotide primers K1-2019 (5′-GTGGACTCTGGGTATTGACAGATGC-3′) and K1-2591R (5′-CCGTTGGATTTCTGATGTCC-3′). Digestion with BstXI and SacI (MBI Fermentas, Burlington, ON, Canada) produced a 147-bp fragment that was then inserted into SacI (partial)/BstXI-digested pBS/K110, yielding pBS/K1b. The fidelity of the PCR-derived portion of the resultantGovb cDNA and the presence of a C at position 2108 were then verified by DNA sequence analysis. A 2088-bpBsmBI/BstEII (MBI Fermentas) fragment of pBS/K1b was then inserted into the BsmBI/BstEII-digested expression vector pK1/YFP10, yielding pK1b/YFP. The presence of the Govb allele in the final construct was confirmed by restriction digestion withBstNI.
CHO cells were grown, plated, and transfected with 10 μg pK1/YFP, pK1b/YFP, or control pIRES-EYFP (Clontech, Palo Alto, CA) plasmid DNA, using Lipofectamine and OPTI-MEM I medium (Life Technologies) as described.10 After 40 to 45 hours, transfected cells were harvested and washed as described10 and analyzed by MAIPA in parallel with untransfected control CHO cells.
DNA sequence analysis of introns flanking Gova/bpolymorphism-bearing CD109 exon
To facilitate genomic DNA analyses of the Govalleles, the intron/exon junctions of the exon bearing the putative Gov-specific nucleotide substitution, as well as the DNA sequence of the flanking introns, were determined. Specifically, CD109 cDNA-specific oligonucleotides binding in the vicinity of this substitution were used for the direct sequencing of p4L10, a pCYPAC-1–derived PAC genomic clone bearing the human CD109 locus (J.Y.A.P. and A.C.S., unpublished observation, August 2000), followed by the alignment of cDNA and corresponding genomic sequences.
PCR-SSP analysis of Gova/b alleles
Gova/b allele-specific antisense oligonucleotide primers differing by a single 3′ nucleotide (5′-TTCAAATTCTTGGTAAATCCTGT-3′ [Gova]; 5′-TTCAAATTCTTGGTAAATCCTGG-3′ [Govb]) were combined with a common sense primer binding in the adjacent intron (5′-ATGACCTTATGATGACCTATTC-3′), yielding a 225-bp product. Oligonucleotide primers that amplify a 429-bp fragment of the human growth hormone gene (sense, 5′-GCCTTCCCAACCATTCCCTTA-3′; antisense, 5′-TCACGGATTTCTGTTGTGTTTC-3′) were used as control. To each PCR tube were added 2 μL of 100 ng/μL genomic DNA in distilled H2O, 1 μL 10 × PCR buffer (160 mM [NH4]2S04, 15 mM MgCl2, 670 mM Tris-HCl pH 8.8, and 0.1% wt/vol Tween 20), 0.2 μL of a mixture of dNTPs (2.5 mM each of dATP, dCTP, dGTP, and dTTP; Perkin Elmer, Cambridge, United Kingdom), 1 μL each of allele-specific primer and common primer (0.5 μM final concentration), 1 μL each of control primers (0.1 μM final concentration), 2.8 μL distilled H20, and 0.35 U Taq DNA polymerase (Sigma, Poole, United Kingdom). Thirty-three amplification cycles (96°C for 60 seconds; 96°C for 25 seconds, 70°C for 45 seconds, 72°C for 30 seconds (5 cycles); 96°C for 25 seconds, 65°C for 45 seconds, 72°C for 30 seconds (20 cycles); 96°C for 25 seconds, 51°C for 45 seconds, 72°C for 30 seconds (8 cycles); and 72°C for 3 minutes) using a Hybaid Omnigene thermocycler with heated lid. PCR products were size-separated electrophoretically in a 2.5% agarose/tris borate EDTA (TBE) gel containing 0.2 μg/mL ethidium bromide.
PCR-RFLP analysis of Gova/b alleles
A 448-bp fragment containing the Gov SNP was obtained by PCR amplification of genomic DNA using the oligonucleotide primers 5′-TTTAGATTATTTTGGCTT-3′ (sense) and 5′-ATGGTTAGTTCAGGTCAA-3′ (antisense). To each PCR tube were added 4 μL of 100 ng/μL genomic DNA in distilled H2O, 2 μL of 10 × PCR buffer, 0.4 μL of a mixture of dNTPs (2.5 mM of each), 2 μL of each primer (0.5 μM final concentration), 9.2 μL distilled H20, and 0.7 U Taq DNA polymerase. Reactions subsequently underwent 30 amplification cycles (96°C for 60 seconds; 96°C for 25 seconds, 53°C for 45 seconds, 72°C for 30 seconds [30 cycles]; and 72°C for 3 minutes). PCR product (15 μL), without further purification, was digested with 5 U BstNI (New England BioLabs, Hitchin, United Kingdom) for 90 minutes at 60°C as recommended by the manufacturer. The restriction pattern was subsequently analyzed in a 2.5% agarose/TBE gel containing 0.2 μg/mL ethidium bromide.
Allele-specific real-time PCR analysis
Genomic primers and probes were designed for genotyping using Taqman real-time PCR technology. Reactions were performed with 100 nM each of the oligonucleotide primers 5′-TGTATCAGTTCTTGGTTTTGTGATGTT-3′ (sense) and 5′-CCAAGAAGTGATAGAATCAGGTACAGTTAC-3′ (antisense) in a total volume of 25 μL containing 1 μL genomic DNA. In addition, each reaction contained 100 nM of the FAM-labeled Gova-specific probe 5′-TATTATCTTGACTTCAGTTACAGGATTTAC CAAGAATTTG-3′ and 200 nM of the VIC-labeled Govb-specific probe 5′-TATTATCTTGACTTCAGTTCCAGGATTTACCAAGAAT-3′. Reactions were incubated at 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C (15 seconds) and 64°C (60 seconds). Allelic discrimination was determined by using a post-PCR plate reader (PE 7700, Perkin Elmer).
Results
Gov phenotyping
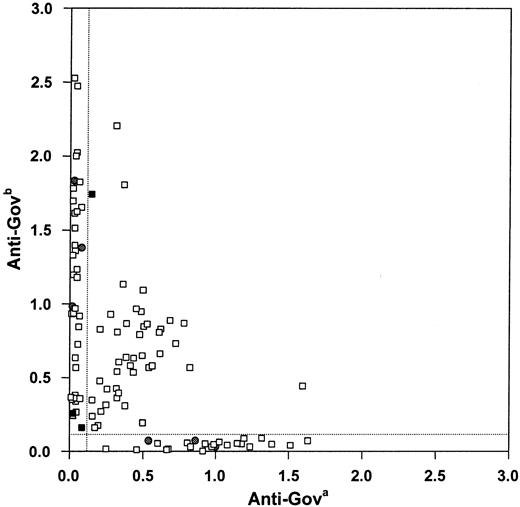
The Gov phenotyping by MAIPA of the 106 donors tested is shown in Figure 1. Interestingly, the strength of reactivity with the Gov-specific antisera was found to vary considerably among donors, especially in Gov homozygotes. Such variability underscores the present difficulties in Gov phenotyping. Indeed, in 3 of the 106 cases analyzed, the initial reaction with the Gov-specific antisera was so weak that erroneous phenotyping results were obtained (see below).
Gov phenotyping of the 106 donors analyzed.
Platelets from 106 donors were tested for Gov phenotype by MAIPA using well-characterized Gova and Govb antisera. Antiserum-specific absorbance values (490 nm) are shown. Panel 1 donors, filled circles; panels 2 and 3 donors, squares. Vertical and horizontal dotted lines indicate Gova and Govbpositivity, respectively, as defined by mean reactivity (± 3 SD) with control AB sera in the 106 samples examined. The 3 panel 3 donors not exhibiting concordance of phenotype and genotype are shown as filled squares.
Gov phenotyping of the 106 donors analyzed.
Platelets from 106 donors were tested for Gov phenotype by MAIPA using well-characterized Gova and Govb antisera. Antiserum-specific absorbance values (490 nm) are shown. Panel 1 donors, filled circles; panels 2 and 3 donors, squares. Vertical and horizontal dotted lines indicate Gova and Govbpositivity, respectively, as defined by mean reactivity (± 3 SD) with control AB sera in the 106 samples examined. The 3 panel 3 donors not exhibiting concordance of phenotype and genotype are shown as filled squares.
PCR amplification and sequence analysis of CD109 cDNAs
The complete CD109 ORF was amplified from the platelet cDNA of the 6 donors in panel 1 by using the oligonucleotide primers listed in Table 1. PCR products were sequenced directly or after subcloning, as appropriate. Overall, the sequence of each fragment was obtained independently at least twice in both directions, such that the entire CD109 ORF was assessed unambiguously for all 6 donors. Our strategy was expected to amplify cDNA sequences corresponding to both chromosomal copies of CD109. We anticipated, therefore, that putative Gov allele-specific sequence differences would not only correlate with the phenotyping results and would result in an amino acid substitution, but also that Govaa and Govbb individuals would be homozygous for such differences, whereas they could potentially be heterozygous for other unrelated CD109 polymorphisms. Consistent with this notion, a single homozygous SNP was found. This polymorphism showed concordance with the Gov phenotyping and resulted in an amino acid substitution. Although the CD109 cDNA sequences of the 3 Govbb donors corresponded to that previously isolated in our laboratory from a KG1a library,10 the 3 cDNAs derived from Govaa donors contained an adenine at nucleotide 2108 instead of a cytosine (Figure 2). Codon 703 of CD109 therefore changes from TAC (Gova) to TCC (Govb), resulting in a Tyr/Ser substitution.
An A/C SNP at position 2108 of the CD109 cDNA defines the Gov alloantigens.
CD109 cDNA sequences flanking nucleotide 2108 and derived from a Govaa (top panel) or a Govbb (bottom panel) panel 1 donor are shown. Nucleotide coordinates correspond to the CD109 cDNA sequence of Lin et al.10 The nucleotide 2108 SNP is concordant with the Gov phenotyping and results in an amino acid substitution.
An A/C SNP at position 2108 of the CD109 cDNA defines the Gov alloantigens.
CD109 cDNA sequences flanking nucleotide 2108 and derived from a Govaa (top panel) or a Govbb (bottom panel) panel 1 donor are shown. Nucleotide coordinates correspond to the CD109 cDNA sequence of Lin et al.10 The nucleotide 2108 SNP is concordant with the Gov phenotyping and results in an amino acid substitution.
Transient expression of Gova- and Govb-specific CD109 cDNAs
To confirm that the Gov alloantigens were defined by the Tyr703Ser polymorphism of CD109, we expressed full-length CD109 cDNAs containing either A (Gova) or C (Govb) at nucleotide 2108 in CHO cells and analyzed the transfected cells by MAIPA. As shown in Figure 3, although anti-Govaserum reacted with cells expressing the putativeGova allele but not theGovb allele, anti-Govb serum reacted only with cells expressing Govb.
MAIPA analysis of
Gova andGovb transfectants. CHO cells were transfected with Gova/YFP-,Govb/YFP-, or control YFP-expressing plasmid vectors and analyzed by MAIPA by using well-characterized Gova and Govb antisera and control blood group AB sera. Anti-Gova and anti-Govb sera react only with Gova- andGovb-expressing cells, respectively. The increased signal produced with the anti-Govb serum reflects the known higher titer of this reagent.11 Three independent AB sera produced results similar to the AB serum data shown. CHO, untransfected cells; CHO YFP, cells transfected with the empty vector pIRES-EYFP; CHO Gova/YFP, cells transfected with the Gova-expressing pIRES-EYFP derivative pK1/YFP; CHO Govb/YFP cells transfected with theGovb-expressing pIRES-EYFP derivative pK1b/YFP. Error bars indicate ± 2 SD from the mean, based on 2 experiments each performed in duplicate.
MAIPA analysis of
Gova andGovb transfectants. CHO cells were transfected with Gova/YFP-,Govb/YFP-, or control YFP-expressing plasmid vectors and analyzed by MAIPA by using well-characterized Gova and Govb antisera and control blood group AB sera. Anti-Gova and anti-Govb sera react only with Gova- andGovb-expressing cells, respectively. The increased signal produced with the anti-Govb serum reflects the known higher titer of this reagent.11 Three independent AB sera produced results similar to the AB serum data shown. CHO, untransfected cells; CHO YFP, cells transfected with the empty vector pIRES-EYFP; CHO Gova/YFP, cells transfected with the Gova-expressing pIRES-EYFP derivative pK1/YFP; CHO Govb/YFP cells transfected with theGovb-expressing pIRES-EYFP derivative pK1b/YFP. Error bars indicate ± 2 SD from the mean, based on 2 experiments each performed in duplicate.
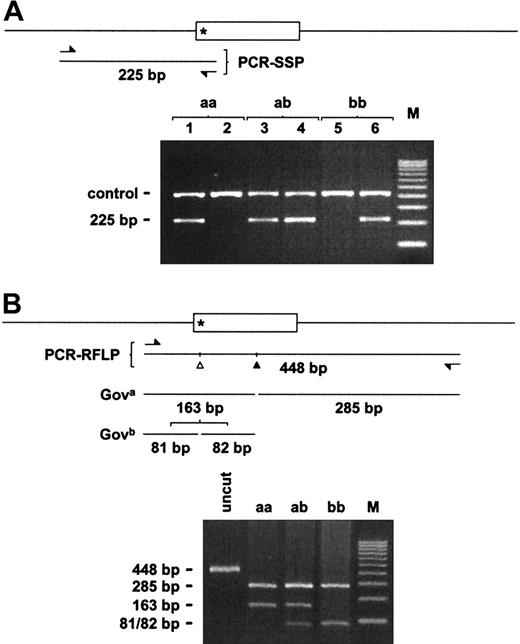
PCR-SSP analysis
To facilitate subsequent genotyping of the Gov alleles, the exon bearing the putative Gov-specific nucleotide substitution identified above and the flanking introns were defined by genomic DNA sequencing. Nucleotide 2108 was found to lie 3 nucleotides from the 5′ end of the corresponding exon (subsequently identified as exon 19 [J.Y.A.P. and A.C.S., unpublished observation, January 2001]; Figure4). On the basis of these sequence data, a pair of allele-specific antisense primers and a third common sense primer binding in the adjacent intron were designed to permit the detection of the nucleotide 2108 A>C SNP in genomic DNA by SSP analysis. As illustrated in Figure 5A, these primers amplified a 225 bp product in an allele-specific manner. Of a total of 21 phenotyped samples analyzed (panels 1 and 2), complete concordance between PCR-SSP analysis and phenotyping was observed.
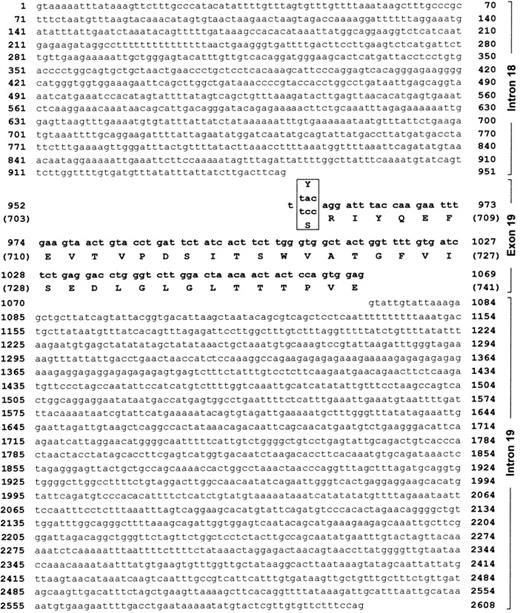
DNA sequence of CD109 exon 19 and flanking introns.
The CD109 nucleotide 2108 A>C SNP was found to lie 3 nucleotides from the 5′ end of CD109 exon 19. The DNA sequence of exon 19 and of flanking introns 18 and 19 is shown (GenBank accession no. AF410460). Exon 19 sequence and the corresponding predicted peptide sequence are shown in bold. Nucleotides are numbered sequentially, beginning at the 5′ end of intron 18. Amino acid coordinates (parentheses) correspond to the CD109 numbering scheme of Lin et al.10 Both allele-specific codon 703 variants (Gova, TAC;Govb, TCC) are shown (box). The nucleotide 2108 A>C SNP results in a Tyr to Ser amino acid substitution at position 703 of CD109.
DNA sequence of CD109 exon 19 and flanking introns.
The CD109 nucleotide 2108 A>C SNP was found to lie 3 nucleotides from the 5′ end of CD109 exon 19. The DNA sequence of exon 19 and of flanking introns 18 and 19 is shown (GenBank accession no. AF410460). Exon 19 sequence and the corresponding predicted peptide sequence are shown in bold. Nucleotides are numbered sequentially, beginning at the 5′ end of intron 18. Amino acid coordinates (parentheses) correspond to the CD109 numbering scheme of Lin et al.10 Both allele-specific codon 703 variants (Gova, TAC;Govb, TCC) are shown (box). The nucleotide 2108 A>C SNP results in a Tyr to Ser amino acid substitution at position 703 of CD109.
Gov genotyping by PCR-SSP and PCR-RFLP analyses.
CD109 exon 19 (open box) and flanking introns are diagrammed. The Gov SNP (*) lies at the 5′ end of exon 19. Relative positions of the PCR primers (arrows) used to generate the 225 and 448 bp PCR products used for genomic SSP and RFLP analyses are shown. (A) Allele-specific antisense oligonucleotide primers differing by a single 3′ nucleotide corresponding to the Gov SNP yield allele-specific 225 bp bands. All reactions also contained control HGH primers that resulted in a 429 bp product (control). In addition, although the reactions shown in lanes 1, 3, and 5 containedGova-specific primers, those in lanes 2, 4, and 6 contained Govb-specific primers. (B) TheGova allele contains a single BstNI site (filled triangle) that is common to both alleles. The Gov SNP results in an additional BstNI site (open triangle) that is specific to the Govb allele. As a result,BstNI digestion of the 448 bp Gova-specific PCR product yields 2 fragments of 163 and 285 bp. In contrast, digestion of the Govb-specific product yields 3 fragments of 285, 81, and 82 bp. M, 100 bp DNA ladder; uncut, no BstNI added; aa/ab/bb, Gov genotype.
Gov genotyping by PCR-SSP and PCR-RFLP analyses.
CD109 exon 19 (open box) and flanking introns are diagrammed. The Gov SNP (*) lies at the 5′ end of exon 19. Relative positions of the PCR primers (arrows) used to generate the 225 and 448 bp PCR products used for genomic SSP and RFLP analyses are shown. (A) Allele-specific antisense oligonucleotide primers differing by a single 3′ nucleotide corresponding to the Gov SNP yield allele-specific 225 bp bands. All reactions also contained control HGH primers that resulted in a 429 bp product (control). In addition, although the reactions shown in lanes 1, 3, and 5 containedGova-specific primers, those in lanes 2, 4, and 6 contained Govb-specific primers. (B) TheGova allele contains a single BstNI site (filled triangle) that is common to both alleles. The Gov SNP results in an additional BstNI site (open triangle) that is specific to the Govb allele. As a result,BstNI digestion of the 448 bp Gova-specific PCR product yields 2 fragments of 163 and 285 bp. In contrast, digestion of the Govb-specific product yields 3 fragments of 285, 81, and 82 bp. M, 100 bp DNA ladder; uncut, no BstNI added; aa/ab/bb, Gov genotype.
PCR-RFLP analysis
The CD109 nucleotide 2108 A>C substitution introduces aBstNI restriction site on the putativeGovb allele. By using genomic DNA from panel 1 and 2 donors, we amplified by PCR a 448 bp CD109 genomic fragment containing the nucleotide 2108 SNP and digested the PCR products withBstNI. As illustrated in Figure 5B, BstNI digestion resulted in allele-specific restriction fragment patterns:Govaa, 285 and 163 bp;Govbb, 285, 81, and 82 bp; andGovab, 285, 163, 81, and 82 bp. The sense PCR primer was chosen so that the Govb-specific digestion of the 163 bp band would yield 2 unresolvable bands of 81 and 82 bp, thereby maintaining the intensity of the smaller product under UV illumination. Concordance between PCR-SSP and PCR-RFLP analysis was observed in all cases.
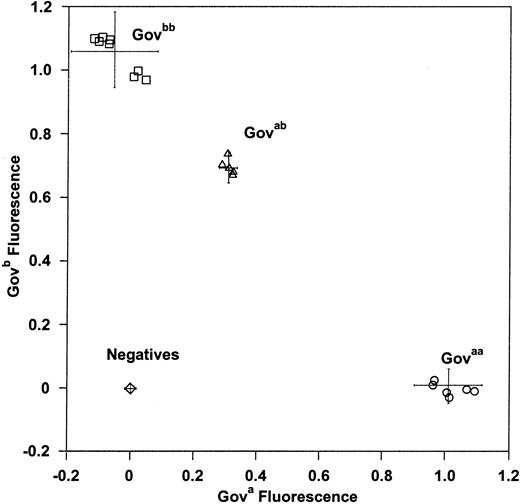
Allele-specific real-time PCR analysis
To facilitate high throughput Gov genotyping without the requirement for post-PCR manipulation, CD109 nucleotide 2108 A- and C-specific probes were designed for Taqman-based allele-specific real-time PCR. The analysis of panel 1 and 2 samples by this approach (Figure 6) showed complete concordance with Gov phenotype and with the results of PCR-SSP and PCR-RFLP analysis.
Gov genotyping by Taqman-based real-time PCR.
Samples from 21 donors in panel 1 and 2 were genotyped by Taqman real-time PCR by using conditions optimized to discriminate betweenGova and Govb alleles. Allele-specific fluorescent signals are shown. Three distinct groups, corresponding to Govaa (open circles), Govab(open triangles), and Govbb (open squares), are observed. Negatives (open diamonds), no template control PCR reactions. Error bars indicate ± 2 SD from the mean.
Gov genotyping by Taqman-based real-time PCR.
Samples from 21 donors in panel 1 and 2 were genotyped by Taqman real-time PCR by using conditions optimized to discriminate betweenGova and Govb alleles. Allele-specific fluorescent signals are shown. Three distinct groups, corresponding to Govaa (open circles), Govab(open triangles), and Govbb (open squares), are observed. Negatives (open diamonds), no template control PCR reactions. Error bars indicate ± 2 SD from the mean.
Real-time PCR-based genotyping agreed with the known Gov phenotype in 82 of 85 subsequently tested additional samples (panel 3). Discordance between genotype and phenotype was observed in the 3 remaining cases, which were then analyzed further (Figure 1; Table2): Genotyping by PCR-SSP and PCR-RFLP confirmed the results of the initial Taqman analyses. In addition, as the Gov phenotypes of panel 3 samples had been determined only once by MAIPA using single Gova and Govb antisera, Gov phenotyping was repeated by using fresh platelets and additional Gov antisera. Repeat samples were also reanalyzed by real-time PCR. Overall, repeat testing resolved the discrepancies in favor of the genotype in all but one case. In the remaining case, no significant signal was produced with any of the Gova antisera, even when twice as many platelets were used in the MAIPA assay.
Discrepancies between Gov phenotyping and genotyping
| Donor no. . | Phenotype by MAIPA . | Real-time PCR . | PCR-SSP . | PCR-RFLP . | ||
|---|---|---|---|---|---|---|
| Initial . | Repeat . | Initial . | Repeat . | Initial . | Initial . | |
| 30 | bb | bb | ab | ab | ab | ab |
| 84 | ab | bb | bb | bb | bb | bb |
| 95 | bb | ab | ab | ab | ab | ab |
| Donor no. . | Phenotype by MAIPA . | Real-time PCR . | PCR-SSP . | PCR-RFLP . | ||
|---|---|---|---|---|---|---|
| Initial . | Repeat . | Initial . | Repeat . | Initial . | Initial . | |
| 30 | bb | bb | ab | ab | ab | ab |
| 84 | ab | bb | bb | bb | bb | bb |
| 95 | bb | ab | ab | ab | ab | ab |
Three donors from panel 3 were identified whose initial phenotypes did not agree with the genotypes determined by real-time polymerase chain reaction (PCR). In all 3 cases, the PCR-SSP and PCR-restriction fragment length polymorphism (RFLP) analyses confirmed the genotype determined by Taqman. Phenotyping and genotyping were subsequently repeated by using fresh samples and additional Gov antisera. Gov typing discrepancies were thereby resolved in favor of the genotypes in 2 (donors 84 and 95) of the 3 cases. Initial, results obtained from the first blood sample investigated; Repeat, results of repeat monoclonal antibody-specific immobilization of platelet antigens (MAIPA) assay and real-time PCR using fresh blood samples.
Taken together, the results of Gov phenotyping by MAIPA; the expression of Gov allele-specific CD109 cDNAs; and the PCR-SSP, PCR-RFLP, and real-time PCR analyses confirm that the Gova and Govb alleles are defined by the CD109 nucleotide 2108 A>C SNP.
Discussion
The biallelic Gov platelet antigen system is known to be carried by CD109, an approximately 170-kd GPI-anchored membrane glycoprotein that is expressed by a subset of hematopoietic progenitor and candidate stem cells, and by activated platelets and T cells.2,7-9Gov alloantibodies are implicated in refractoriness to platelet transfusion and can cause neonatal alloimmune thrombocytopenia and posttransfusion purpura.3,11 Whether CD109 may additionally play a role in organ or bone marrow transplantation by functioning as a minor histocompatibility antigen is not known. The importance of the Gov antigens in alloantibody-mediated platelet destruction has only recently been defined. We have recently demonstrated that the immunogenicity of the Gov alloantigens is similar to that of the HPA-5 alloantigens and is exceeded only by that of the HPA-1 alloantigens.11 In light of this information, the difficulties of conventional Gov phenotyping by serologic methods are particularly frustrating, underscoring the need for an alternative approach to determining Gov type.
In this report, we have determined the molecular basis of the Gov platelet antigen system. As the Gov epitopes are unaffected by deglycosylation, but are sensitive to sodium dodecyl sulfate denaturation,3 we reasoned that the Gov alleles would reflect differences in the primary CD109 amino acid sequence. Furthermore, and by analogy with all but one of 19 characterized platelet antigen systems,17 we anticipated that the Gov allelic differences would most likely arise from a SNP. Our recent identification of the human CD109 cDNA10, together with the ability to perform Gov phenotyping by MAIPA11 (Figure 1), has allowed us to test this hypothesis. Beginning with platelet RNA from 6 donors of known Gov phenotype, we used CD109-specific RT-PCR and sequence analysis to determine that the Gova andGovb alleles differ by a single A/C polymorphism at nucleotide 2108 of the CD109 ORF that results in a Tyr to Ser substitution at position 703 in the CD109 protein (Figures 2, 4). Expression studies indicated that this polymorphism is indeed responsible for the formation of the Gov alloantigenic determinants (Figure 3). Confirmatory PCR-SSP, PCR-RFLP, and Taqman-based allele-specific PCR genotyping studies using genomic DNA from a core panel of 21 donors of known phenotype demonstrated absolute concordance with Gov phenotype, as determined by MAIPA (Figures 5, 6).
On genotyping an additional 85 phenotyped donors, although 82 samples were concordant, in 3 cases the Taqman-based genotype disagreed with the phenotype as determined by MAIPA (Figure 1; Table 2). Notably, in all 3 discrepant cases, phenotyping had been based on low absorbance values (Figure 1). As the real-time PCR-based genotype could be confirmed by alternative genotyping methods, it appeared that the MAIPA assay had yielded discrepant results in all 3 cases. Indeed, repeat phenotyping using larger numbers of freshly obtained platelets, as well as additional typing antisera, revealed that the original typing studies had been in error in 2 of 3 cases (Table 2). In the final case, however, even additional modified testing did not resolve the discrepancy, likely reflecting very low-level CD109 expression below the detection limit of the MAIPA assay.
Platelet CD109 expression is known to be low, with previous studies reporting only about 2000 ± 400 molecules detectable after thrombin activation.7,14 Recent studies from our group have detected a similar number of molecules on weakly activated, but P-selectin (CD62P)–negative platelets.11 In addition, in the course of our Gov phenotyping studies, we detected considerable interindividual variability in reactivity with Gov antisera (Figure 1). Indeed, in the present study, the Gov phenotype of a significant number of donors was based on very low absorbance readings with one or both of the Gova or Govb antisera. Thus, it was not unexpected that we observed Gov typing discrepancies. The ability of the Gov genotyping studies described here to resolve such discrepant or ambiguous cases underscores the greater sensitivity and accuracy of these approaches. Whether the observed variability in reactivity with Gov antisera reflects interindividual variation in CD109 expression or is paralleled by altered platelet survival is unknown and requires further study.
Although the precise function of CD109 remains obscure, its recent identification as a novel member of the α2M/C3, C4, C5 family of thioester-containing protease inhibitors and complement proteins10 suggests that it may be capable of mediating covalent cell-substrate and/or cell-cell interactions. Whether the Tyr703Ser polymorphism reported here has any functional consequences is presently not known. Although the effect of this substitution (small polar to large polar amino acid) on CD109 structure is unknown, computer modeling would suggest that such an effect, if any, is minimal. Consistent with the observation that there is no difference in the apparent molecular weight of the Gova or Govb CD109 variants, and that deglycosylation of CD109 does not affect the binding of Gov alloantibodies, the amino acid 703 substitution is not predicted to change the number of potential CD109 glycosylation sites. In addition, sequence comparison of CD109 with other members of the α2M/C3, C4, C5 family suggests that the Tyr703Ser polymorphism should not alter the substrate specificity or the thioester reactivity of CD109. Nevertheless, as our understanding of CD109 function becomes clearer, it will be of interest to determine whether the Gova and Govb CD109 variants are functionally distinct.
In this report, we have elucidated the molecular basis of the Gov alloantigen system. On the basis of these data, we have demonstrated the feasibility of Gov genotyping by 3 different techniques and have shown that this approach is superior to conventional serologic methods for determining Gov phenotype. Such DNA-based methods will allow the reliable typing of donors and patients and will also facilitate studies on the potential role(s) of the Gov alloantigens in other clinical settings such as organ and bone marrow transplantation. Although serologic methods will continue to be required to determine if immunization against a Gov alloantigen has occurred, the elucidation of the molecular basis of the Gov alloantigen system will also aid in the detection of Gov alloantibodies. Current approaches to alloantibody detection— MAIPA, for example—are cumbersome and are hampered by both low levels of CD109 expression and the instability of the GPI anchor. These problems would be obviated by the availability of Gov alloantigen-specific peptide reagents that could be used for antibody detection. Experiments to identify the minimal recombinant CD109 fragments required for Gov alloantibody detection are currently under way.
We thank Ed Conway and David Spaner for helpful comments during the course of this work and for critically reviewing the manuscript, as well as Xiang-Fu Wu and Rakash Nayar for technical assistance. In addition, we gratefully acknowledge the support of the apheresis clinic staff and the technical assistance of the Platelet Immunology Laboratory of the National Blood Service–East Anglia.
Supported by grants from the Medical Research Council of Canada and the National Cancer Institute of Canada (ACS/DRS) and from National Blood Service England (WHO/KC).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andre C. Schuh, Rm 7366, Medical Sciences Bldg, University of Toronto, 1 King's College Circle, Toronto, ON, Canada, M5S 1A8; e-mail: andre.schuh@utoronto.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal