This study identified missense mutations in the ligand binding domain of the oncoprotein PML-RARα in 5 of 8 patients with acute promyelocytic leukemia (APL) with 2 or more relapses and 2 or more previous courses of all-trans retinoic acid (RA)–containing therapy. Four mutations were novel (Lys207Asn, Gly289Arg, Arg294Trp, and Pro407Ser), whereas one had been previously identified (Arg272Gln; normal RARα1 codon assignment). Five patients were treated with repeat RA plus phenylbutyrate (PB), a histone deacetylase inhibitor, and one patient experienced a prolonged clinical remission. Of the 5 RA + PB-treated patients, 4 had PML-RARα mutations. The Gly289Arg mutation in the clinical responder produced the most defective PML-RARα function in the presence of RA with or without sodium butyrate (NaB) or trichostatin A. Relapse APL cells from this patient failed to differentiate in response to RA but partially differentiated in response to NaB alone, which was augmented by RA. In contrast, NaB alone had no differentiation effect on APL cells from another mutant case (Pro407Ser) but enhanced differentiation induced by RA. These results indicate that PML-RARα mutations occurred with high frequency after multiple RA treatment relapses, indicate that the functional potential of PML-RARα was not correlated with clinical response to RA + PB treatment, and suggest that the response to RA + PB therapy in one patient was related to the ability of PB to circumvent the blocked RA-regulated gene response pathway.
Introduction
Despite major advances in the treatment of acute promyelocytic leukemia (APL) with all-trans retinoic acid (RA) in combination with chemotherapy, relapse occurs in approximately 30% of patients who achieve clinical remission (CR). Most of these patients are either already refractory to or soon become refractory to retreatment with RA.1-3 Among these relapse patients, there is a high incidence of reduced APL cell sensitivity to RA-induced terminal differentiation in vitro.2,4,5 In a study of de novo APL patients treated on intergroup protocol 0129 with sequential RA and chemotherapy, we found PML-RARα mutations in 25% to 30% of patients at first relapse5 (Gallagher, Slack, Willman, et al, unpublished results, September 2001). Others have also reported the finding of RARα-region missense mutations in first or second relapse, RA-treated APL patients.6 7 One objective of the current study was to assess whether multiple relapses from RA therapy is associated with an increased incidence of PML-RARα mutations.
The recruitment of excessive amounts of corepressor proteins to retinoic acid response element (RARE)–regulated gene promoters by a multimeric complex of PML-RARα plays a crucial role in the genesis and maintenance of APL.8-10 The therapeutic response of APL cells to RA depends on the ability of pharmacologic RA concentrations (≥ 100 nM) to dissociate the corepressor complex and to recruit an alternative set of coactivator proteins, all of which depends on binding of RA to the ligand-binding domain (LBD) of the RARα region of PML-RARα. Essential components of the corepressor complex are histone deacetylase (HDAC) enzymes that foster chromatin condensation and reduced gene promoter activity (reviewed in McKenna et al11). Trichostatin A (TSA) is one of a variety of compounds that can inhibit HDAC enzymes, reversing their effect on chromatin.12 TSA was reported to partially overcome resistance to RA-induced differentiation in a subline of NB4 APL cells containing a missense mutation in the LBD of PML-RARα that markedly reduces RA binding and corepressor dissociation.13 In that study, the HDAC inhibitor (HDI) was ineffective alone but positively interacted with RA to induce partial differentiation. Similarly, HDI antileukemic activity in murine leukemias generated in PML-RARα transgenic mice was RA dependent.14 These results suggest that HDI differentiation activity is mediated by modulation of RA/RARE-regulated gene transcription. The results, however, do not exclude the possibility that HDIs might independently modulate the transcription of alternative genes that complement RA/RARE-modulated genes leading to terminal APL cell differentiation. This possibility seems consistent with observations that HDIs alone can modulate the transcription of a limited range of genes that affect cell growth, death, and differentiation in non–RA-dependent leukemias and cancers.12 15
On the basis of such background information, one of us (R.P.W.) and colleagues initiated a pilot clinical trial to test the potential interactive therapeutic effect of the HDI sodium phenylbutyrate (PB) and graded doses of RA in multiple-relapse APL patients clinically resistant to RA and chemotherapeutic agents. Remarkably, the first patient treated with this regimen, dubbed “targeted transcription therapy,” achieved a fourth CR that was sustained for several months.16 Four clinically similar patients, however, did not respond to the same treatment regimen.17 In the current study, we sought to determine if the differential clinical response of these 5 patients might be related to the functional properties of their PML-RARα fusion genes.
Patients, materials, and methods
Patients and treatments
Bone marrow specimens from 8 multiple-relapse APL patients were used for the present studies under a protocol approved by the institutional review board under Helsinki protocol guidelines. Table1 briefly summarizes the treatment histories of these patients. Case numbers were assigned based on the order of laboratory specimen receipt and analysis. All patients had relapsed at least twice (range, 2-4) and had received 2 or more (range, 2-4) courses of therapy containing RA. Additionally, all patients had received 3 or more (range, 3-8) different agents. Two patients had been treated with the anti-CD33 monoclonal antibody HUM-195, and 2 patients had relapsed after allogeneic bone marrow transplantation. All patients had been treated with arsenic trioxide (ATO).
Patients with multiple relapse of acute promyelocytic leukemia: treatment history and response to RA + PB therapy
| Case no. . | No. of relapses . | Courses of RA . | Other agents† . | Response to RA + PB treatment . |
|---|---|---|---|---|
| 1 | 4 | 3 | CTA (4), ATO | NT |
| 2 | 2 | 2 | CTA (2), ATO | NT |
| 3 | 2 | 3 | CTA (5), ATO | No |
| 4 | 3 | 4 | CTA (3), ATO | No |
| 5 | 2 | 2 | CTA (3), ATO, Ab | NT |
| 6 | 3 | 3 | CTA (8), BMT, ATO | Yes |
| 7 | 4 | 3 | CTA (5), BMT, ATO, PB | No |
| 8 | 2 | 3 | CTA (3), Ab, ATO, PB | No |
| Case no. . | No. of relapses . | Courses of RA . | Other agents† . | Response to RA + PB treatment . |
|---|---|---|---|---|
| 1 | 4 | 3 | CTA (4), ATO | NT |
| 2 | 2 | 2 | CTA (2), ATO | NT |
| 3 | 2 | 3 | CTA (5), ATO | No |
| 4 | 3 | 4 | CTA (3), ATO | No |
| 5 | 2 | 2 | CTA (3), ATO, Ab | NT |
| 6 | 3 | 3 | CTA (8), BMT, ATO | Yes |
| 7 | 4 | 3 | CTA (5), BMT, ATO, PB | No |
| 8 | 2 | 3 | CTA (3), Ab, ATO, PB | No |
Treatment history before the detection of PML-RARα mutations.
RA indicates all-trans retinoic acid; PB, sodium phenylbutyrate; CTA, chemotherapeutic agent; ATO, arsenic trioxide; NT, not treated; Ab, HUM-195 antibody; BMT, allogeneic bone marrow transplant.
Numbers in parentheses indicate number of different, previously used chemotherapeutic agents.
Five patients (cases 3, 4, and 6-8) were treated with RA + PB, and more detailed clinical accounts of these patients have been presented elsewhere.16,17 Treatment consisted of escalating doses of RA (30-90 mg/m2 per day) and PB (150-400 mg/kg per day) for 25 days per course up to 5 treatment courses. The first patient treated (case 6 of this report) responded to treatment with a CR of 7 months' duration attended by molecular remission, as determined by negative reverse transcriptase–polymerase chain reaction (RT-PCR) tests for PML-RARα messenger RNA (mRNA).16 17 Three patients (cases 6-8) were treated with PB + RA before initial mutational analysis, but in cases 6 and 7 RNA from second bone marrow specimens were retrospectively available before PB + RA therapy. In cases 3 and 4, mutational analysis was initially performed before RA + PB therapy. In case 8, the only earlier sample was obtained at initial diagnosis. No further stored materials were available on these patients.
Mutation analysis
RT-PCR and DNA sequence analysis were performed as previously described.5 This analysis included 2 PCR amplification procedures in preparation for DNA sequence analysis: (1) direct PCR amplification from complementary DNA with primer pairs covering 2 overlapping sequences of the RARα LBD, which permitted biallelic analysis of both normal RARα and PML-RARα mRNAs and (2) initial first-round PCR amplification with a PML-RARα–specific primer pair, followed by nested or heminested secondary PCR amplification using the RARα LBD primer pairs, which permitted monoallelic assignment of mutations found in this study to PML-RARα. All mutations were confirmed by bidirectional sequencing and repeat analysis from RNA.
RA-binding analysis
COS-1 cells in exponential growth were collected by trypsinization and washed twice with phosphate-buffered saline (Mg++/Ca++ free), and 108 cells were resuspended in 5 mL phosphate-buffered saline. pSG5 expression vector DNA (400 μg), containing either wild-type or mutant PML-RARα, was mixed with the cell suspension for 10 minutes at room temperature. Electroporation was done by using a Gene Pulser II (BioRad, Hercules, CA) at 250 μF and 350 V. After 3 days' culture in Dulbecco modification of Eagle medium (DMEM) with 10% fetal bovine serum (FBS), the cells were harvested for nuclear protein extraction, as described.18 To test RA binding, 0.2 mL nuclear extract was incubated for 15 hours at 4°C with 10 nmol/L [3H]RA (30 Ci/mmol [1.11 × 1012 Bq]; DuPont-NEN, Boston, MA) in the absence or presence of 200-fold excess of unlabeled RA. The unbound RA was removed by incubation with dextran-coated charcoal for 15 minutes and then centrifuged for 15 minutes at 10 000g. The supernatant was analyzed at 4°C by fast performance liquid chromatography (FPLC), using a Superose 6HR 10/30 size-exclusion column (Pharmacia Biotech, Piscataway, NJ), essentially as described.18
Transfection/transactivation procedures
COS-1 cells in exponential growth were seeded at 2 × 105 cells per well in 6-well plates 1 day before transfection, using DMEM with 10% FBS (without antibiotics). Cells were rinsed with serum-free Opti-MEM I (Gibco BRL, Bethesda, MD) and transfected by using the Lipofectamine Plus kit (Gibco BRL). The transfection mixture (190 μL Opti-MEM I, 0.6 μg of the reporter plasmid DR5-tk-Luc, 0.4 μg pS5 containing either wild- type or mutant PML-RARα, 0.3 μg pCMV-β–Galactosidase, 6 μL PLUS reagent, and 4 μL Lipofectamine reagent) was overlaid onto the cells and incubated for 4 hours at 37°C. The transfection mixture was then replaced by 2 mL DMEM containing 10% FBS and different RA concentrations in the absence or presence of sodium butyrate (NaB) or TSA. After 48 hours' incubation, the transfected cells were lysed in 500 μL Report Lysis Buffer (Promega, Madison, WI). After centrifugation, 20 μL cell lysate supernatant was mixed with 100 μL Luciferase Assay Reagent (Promega), and the luciferase activity was measured by a luminometer and then normalized with β-gal activity.
Cell culture and analysis procedures
Enriched APL cell fractions were prepared from heparinized bone marrow specimens, placed in tissue culture in the presence or absence of experimental agents, and analyzed for evidence of cell proliferation, death, and differentiation after variable culture periods, following slight modifications of previously reported procedures.5,19 The AP-1060 cell culture strain was derived from case 1, as described elsewhere.20
For flow cytometric analysis, cellular Fc receptors were first blocked with 200 μg/mL normal mouse immunoglobulin G (Caltag, Burlingame, CA) for 10 minutes at room temperature. The cells were then incubated with phycoerythrin-conjugated CD11b (clone D12) or an isotype-matched control antibody (Becton Dickinson, San Jose, CA) for 15 minutes at room temperature. After washing and resuspending the cells, 20 000 events were analyzed by using a FACScan flow cytometer and Cell Quest software (Becton Dickinson).
Histone acetylation
APL cells were incubated with different concentrations of RA with or without 1 mM NaB at 37°C for 2 to 6 hours. Total cellular proteins were extracted as described.21 Protein extracts of 4 × 105 cells were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 10% Tris-glycine Novex precast gel (Invitrogen, Carlesbad, CA) and electroblotted to nitrocellulose membrane (Pierce, Rockford, IL). The blot was first incubated with 1 μg/mL antiacetylated histone H3 polyclonal antibody (Upstate, Lake Placid, NY) and then with 0.2 μg/mL peroxidase-labeled goat antirabbit antibody (Pierce) for detection with chemiluminescent substrate (Pierce). After stripping (ImmunoPure IgG Elution Buffer; Pierce), the blot was reprobed with 1 μg/mL antiacetylated histone H4 polyclonal antibody (Upstate) or 1:10 000 anti-β-actin (Sigma, St Louis, MO).
Results
Identification of PML-RARα missense mutations
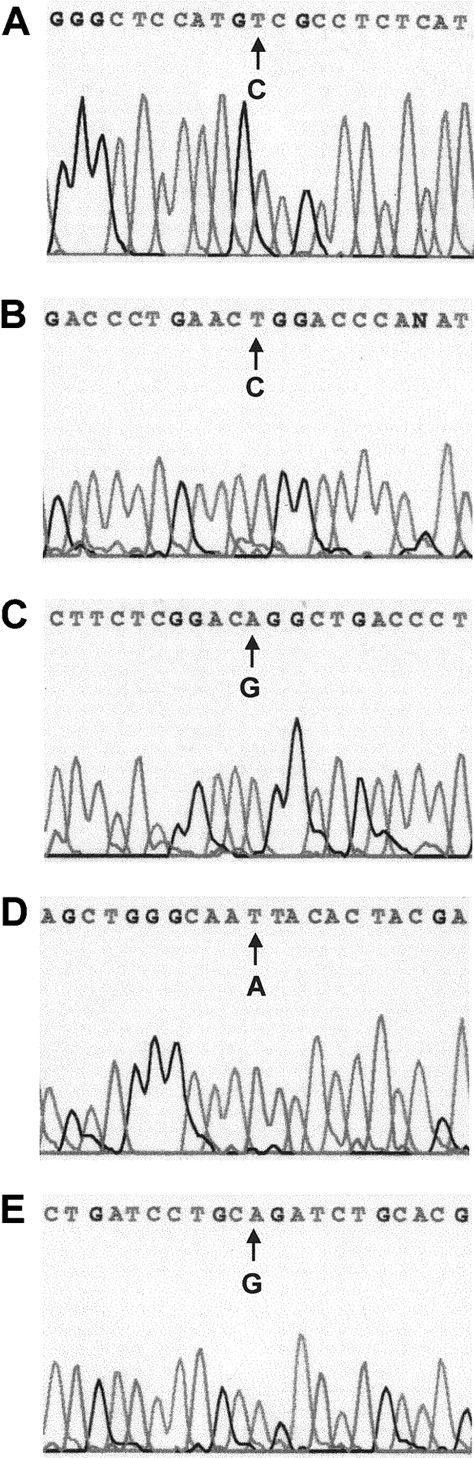
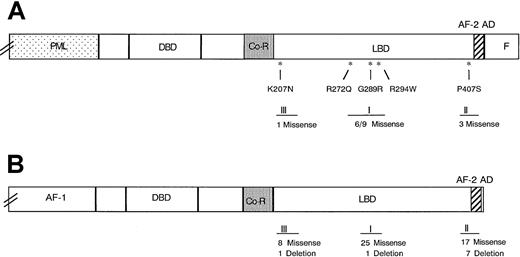
Sequence analysis of the RARα region of PML-RARα mRNA revealed base substitutions in 5 of 8 multiple relapse APL patients (Figure1 and Table2). Each base substitution produced an amino acid codon change characteristic of missense mutations: Pro407Ser (case 1), Arg294Trp (case 4), Gly289Arg (case 6), Lys207Asn (case 7), and Arg272Gln (case 8). No base substitutions were observed in the corresponding sequence of the normal RARα allele (not shown). All 5 mutations occurred in the LBD of the RARα-region of PML-RARα and were distributed in 3 distinct zones (Figure2A,B): 1 near the beginning of the LBD (Lys207Asn), 3 in a central zone (Arg272Gln, Gly289Arg, and Arg294Trp), and 1 in a preterminal zone (Pro407Ser).
Automated DNA sequence analysis of nested, PML-RARα allele-specific PCR products from 5 APL cases with single nucleotide changes.
(A) Case 1, C→T (Pro→Ser). (B) Case 4, C→T (Arg→Trp). (C) Case 6, G→A (Gly→Arg). (D) Case 7, A→T, (Lys→Asn). (E) Case 8, G→A (Arg→Gln).
Automated DNA sequence analysis of nested, PML-RARα allele-specific PCR products from 5 APL cases with single nucleotide changes.
(A) Case 1, C→T (Pro→Ser). (B) Case 4, C→T (Arg→Trp). (C) Case 6, G→A (Gly→Arg). (D) Case 7, A→T, (Lys→Asn). (E) Case 8, G→A (Arg→Gln).
PML-RARα mutation analyses in multiple-relapse APL cases
| Case no. . | PML-RARα type . | Base substitution* . | Missense mutation* . |
|---|---|---|---|
| 1 | L | C1322T | Pro407Ser |
| 2 | S | — | — |
| 3 | L | — | — |
| 4 | S | C982T | Arg294Trp |
| 5 | L | — | — |
| 6 | L | G967A | Gly289Arg |
| 7 | V | A723T | Lys207Asn |
| 8 | L | G917A | Arg272Gln |
| Case no. . | PML-RARα type . | Base substitution* . | Missense mutation* . |
|---|---|---|---|
| 1 | L | C1322T | Pro407Ser |
| 2 | S | — | — |
| 3 | L | — | — |
| 4 | S | C982T | Arg294Trp |
| 5 | L | — | — |
| 6 | L | G967A | Gly289Arg |
| 7 | V | A723T | Lys207Asn |
| 8 | L | G917A | Arg272Gln |
AML indicates acute promyelocytic leukemia.
Nucleotide and amino acid numbers correspond to that for normal RARα1.44
Position of naturally occurring missense mutations in 3 zones of the ligand-binding domain of the RARα region.
Panel A shows the RARα region of PML-RARα, and Panel B 3 zones of clustered mutations in the homologous TRβ defined in extensive studies of RTHS.27,28 Asterisks indicate the positions of the 5 missense mutations found in this report. Roman numerals I to III designate the 3 clustered mutation zones. The numbers below the zones indicate the number of mutations identified in each of the 3 zones, as follows: (A) unique (numerator) and total (denominator) naturally occurring PML-RARα mutants and (B) the number of unique mutations in TRβ in the RTHS in the cited reviews.28 29
Position of naturally occurring missense mutations in 3 zones of the ligand-binding domain of the RARα region.
Panel A shows the RARα region of PML-RARα, and Panel B 3 zones of clustered mutations in the homologous TRβ defined in extensive studies of RTHS.27,28 Asterisks indicate the positions of the 5 missense mutations found in this report. Roman numerals I to III designate the 3 clustered mutation zones. The numbers below the zones indicate the number of mutations identified in each of the 3 zones, as follows: (A) unique (numerator) and total (denominator) naturally occurring PML-RARα mutants and (B) the number of unique mutations in TRβ in the RTHS in the cited reviews.28 29
The mutations were discovered in specimens obtained after 2 to 4 relapses and 3 or 4 previous courses of RA therapy (Table 1). In 2 patients the mutations were found in specimens obtained before the initiation of RA + PB therapy (cases 4 and 6), and, in case 6, a specimen obtained after relapse from RA + PB therapy contained the same mutation. In 2 other RA + PB-treated patients (cases 7 and 8), the mutations were only documented after failure of RA + PB therapy. In case 7, the mutation was not present in a specimen obtained 16 months before RA + PB therapy, and, in case 8, the mutation was not detected in a specimen before any leukemic therapy.
RA-binding activity of mutant PML-RARα proteins
As assessed by FPLC of nuclear extracts from transfected COS-1 cells incubated with 10 nM [3H]RA, 2 mutants, Gly289Arg and Arg294Trp, completely lacked RA-binding activity (Figure3E,H). Two mutants, Lys207Asn and Pro407Ser, bound the ligand primarily in a monomeric/dimeric configuration (Figure 3C,F), in contrast to the wild type L-form control, which expressed more characteristic high molecular mass, multimeric complexes (Figure 3B).8,22 The Arg272Gln L-form mutant bound ligand in a pattern that resembled the wild-type pattern, although reduced in amount (Figure 3D). Because this binding was apparently greater than previously reported for an Arg272Gln S-form mutant,23 we prepared this mutant in an S-form context; ligand binding also showed a modest reduction of multimeric complexes compared with the wild type S-form control (Figure 3G and not shown).
Size-exclusion FPLC analysis of nuclear RA-binding activity in COS-1 cells.
Cells are transfected with pSG5 vector (A), wild-type PML-RARα L form (B), mutant Lys207Asn (C), mutant Arg272Gln (D), mutant Gly289Arg (E), mutant Pro407Ser (F), wild-type PML-RARα S form (G), and mutant Arg294Trp (H). Nuclear extracts were incubated with 10 nmol/L [3H]-RA in the absence (●) or presence (○) of 200-fold excess of unlabeled RA for 15 hours at 4°C. The samples were fractionated over a Superose 6HR 10/30 column at 0.4-mL intervals. Arrows indicate the fraction numbers of marker proteins (in kd) used to calibrate the FPLC column.
Size-exclusion FPLC analysis of nuclear RA-binding activity in COS-1 cells.
Cells are transfected with pSG5 vector (A), wild-type PML-RARα L form (B), mutant Lys207Asn (C), mutant Arg272Gln (D), mutant Gly289Arg (E), mutant Pro407Ser (F), wild-type PML-RARα S form (G), and mutant Arg294Trp (H). Nuclear extracts were incubated with 10 nmol/L [3H]-RA in the absence (●) or presence (○) of 200-fold excess of unlabeled RA for 15 hours at 4°C. The samples were fractionated over a Superose 6HR 10/30 column at 0.4-mL intervals. Arrows indicate the fraction numbers of marker proteins (in kd) used to calibrate the FPLC column.
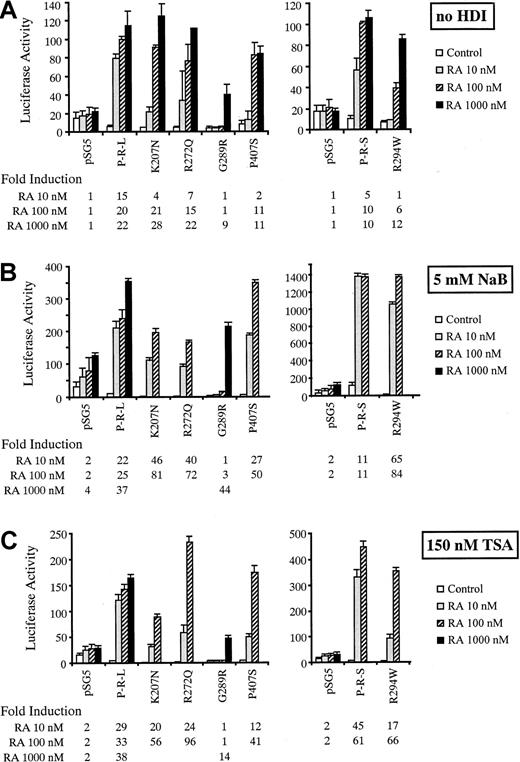
Effect of RA with or without NaB or TSA on mutant PML-RARα transcriptional transactivation activity
All 5 mutant PML-RARαs showed reduced transcriptional transactivation activity at 10 nM RA compared with wild-type L- or S-form PML-RARαs (Figure 4A). The Arg294Trp and Gly289Arg mutants were totally devoid of activity at 10 nM RA, and, the latter, also at 100 nM RA. The activity of the Arg294Trp S-form mutant was also decreased at 100 nM RA. The activity of all mutants was markedly stimulated at 1 μM RA, although this stimulation was much less for the Gly289Arg mutant. Also, notably, the depression of baseline activity from that of the pSG5 vector control in the absence of drug was greater for the mutant PML-RARαs compared with the wild-type PML-RARαs with the exception of the Pro407Ser mutant (Figure 4A).
Transcriptional activity of wild-type and mutant PML-RARα fusion proteins in variable RA concentrations with or without NaB or TSA.
The DR5-tk-luc reporter was cotransfected with L-form PML-RARα (left panel) or S-form PML-RARα (right panel) in the absence of HDI (A), in the presence of 5 mM NaB (B) or 150 nM TSA (C). pSG5 represents the vector alone. Luciferase activity with different concentrations of RA is shown after normalization with β-gal activity with the corresponding, calculated fold-induction value indicated below.
Transcriptional activity of wild-type and mutant PML-RARα fusion proteins in variable RA concentrations with or without NaB or TSA.
The DR5-tk-luc reporter was cotransfected with L-form PML-RARα (left panel) or S-form PML-RARα (right panel) in the absence of HDI (A), in the presence of 5 mM NaB (B) or 150 nM TSA (C). pSG5 represents the vector alone. Luciferase activity with different concentrations of RA is shown after normalization with β-gal activity with the corresponding, calculated fold-induction value indicated below.
The transcriptional transactivation activity induced by 10 nM RA was markedly stimulated in the presence of 5 mM NaB for 4 of the mutant PML-RARαs compared with the stimulation by 10 nM RA alone (Figure4B): Lys207Asn, 46- versus 4-fold; Arg272Gln, 40- versus 7-fold; Pro407Ser, 27- versus 2-fold; and Arg294Trp, 65- versus 1-fold. Conversely, combination NaB had no effect on Gly289Arg mutant activity until a concentration of 1 μM RA, which produced a strong interactive effect (44- versus 9-fold). A more modest stimulatory effect of 5 mM NaB in the presence of 10 nM RA was observed for the wild-type PML-RARαs: L-form, 15- versus 22-fold, and S-form, 5- versus 11-fold. The latter was partly related to the fact that 5 mM NaB produced some augmentation of baseline transcriptional activity in both pSG5 and the wild-type transfectants (but not the mutant transfectants) in the absence of RA supplementation, which likely is related to low levels of endogenous RA and RARs in the COS-1 cell culture system.
TSA (150 μM), a more specific HDI, had the same effects as 5 mM NaB with some differences in detail (Figure 4C). TSA had a lesser effect on baseline pSG5 activity and did not significantly relieve the transcriptional repressive effect of the wild-type PML-RARαs. This effect contributed to the greater stimulatory effect of TSA than NaB on wild-type PML-RARαs in combination with 10 nM RA: L-form, 29-versus 22-fold, and S-form, 45- versus 11-fold. However, the stimulatory interaction between 150 μM TSA and various RA concentrations was generally less than that with 5 mM NaB on the mutant PML-RARαs. These minor differences may reflect quantitative variations from the activity optimum for each agent or could reflect qualitative differences related to alternative, non-HDI activities of NaB.15
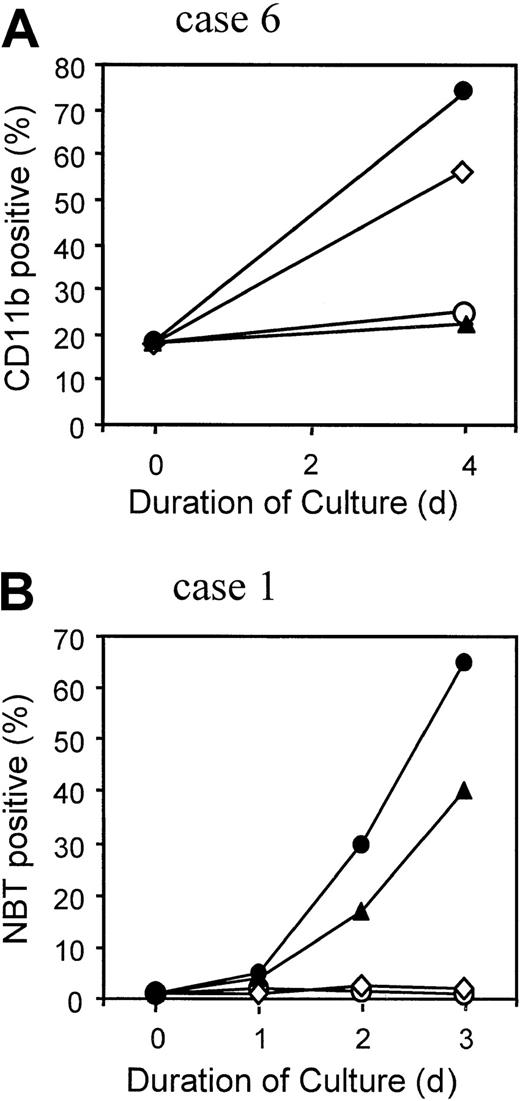
Differentiation response of APL cells harboring PML-RARα Gly289Arg and Pro407Ser mutations
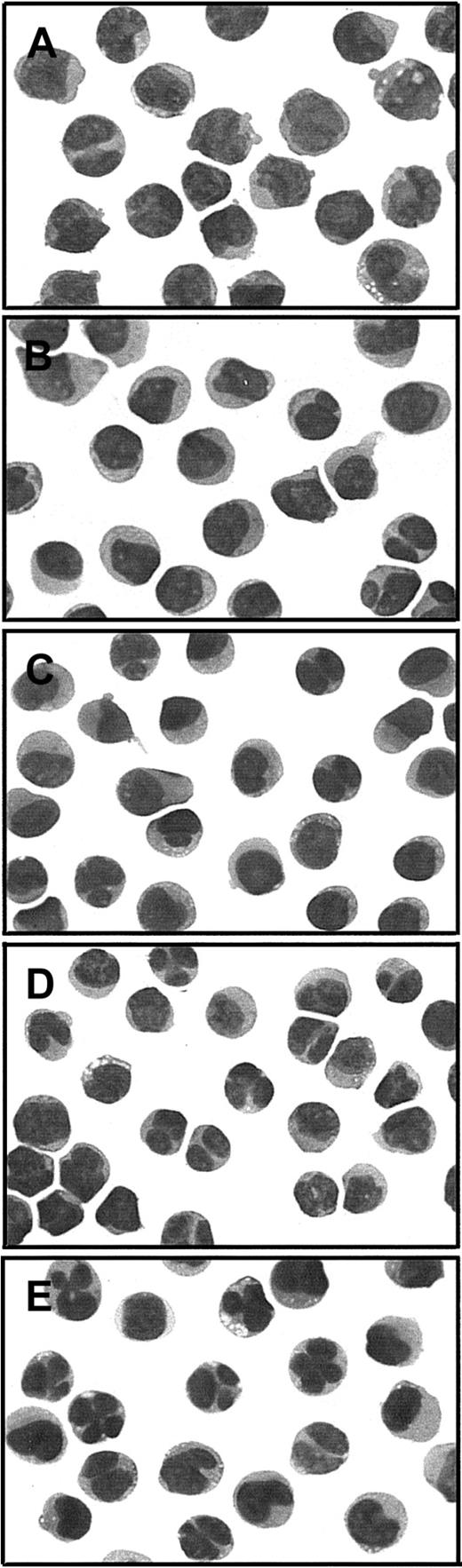
APL cells obtained from clinically responsive case 6 after relapse from RA + PB therapy were tested for in vitro sensitivity to differentiation induction by the 2 therapeutic agents. Up to 1 μM RA, there was little or no evidence of cellular differentiation compared with untreated cells (Figure 5B,C). However, 1 mM NaB alone produced increased nuclear segmentation in a significant proportion of the cells (Figure 5D), and this number was increased in the presence of 1 μM RA (Figure 5E). The impression that these nuclear changes reflected increased differentiation in the presence of NaB, augmented by RA, was confirmed by measurement of the myeloid differentiation marker CD11b. After culture for 4 days, the percentage of CD11b+ cells increased to 58% in the presence of 1 mM NaB alone and 75% in 1 mM NaB combined with 1 μM RA, compared with no increase from the baseline value of 18% in the absence or presence of 1 μM RA alone (Figure6A). Under none of these conditions was the nitroblue tetrazolium dye reduction test positive, suggesting a defect in superoxide production in these multiple relapse APL cells, which had typical cytologic features of the M3 variant form of APL (Figure 5A).24
Cytologic evaluation of APL cells from PML-RARα Gly289Arg mutant case 6 in the absence or presence of RA or NaB alone and in combination.
Photographs of cytospin slide preparations of sodium metrizoate–selected low-density (P ≤ 1.077 g/mL) bone marrow cells were taken at × 1000 magnification of modified Wright stain (Dif-Quik, Sigma, St Louis, MO). (A) Prior to culture; (B-E) cultured × 4 days with (B) no added drug, (C) 1 μM RA, (D) 1 mM NaB, or (E) 1 μM RA + 1 mM NaB.
Cytologic evaluation of APL cells from PML-RARα Gly289Arg mutant case 6 in the absence or presence of RA or NaB alone and in combination.
Photographs of cytospin slide preparations of sodium metrizoate–selected low-density (P ≤ 1.077 g/mL) bone marrow cells were taken at × 1000 magnification of modified Wright stain (Dif-Quik, Sigma, St Louis, MO). (A) Prior to culture; (B-E) cultured × 4 days with (B) no added drug, (C) 1 μM RA, (D) 1 mM NaB, or (E) 1 μM RA + 1 mM NaB.
Differentiation response of APL cells.
APL cells from case 6 (A) and from AP-1060 cells derived from case 1 (B) were cultured for the indicated time in the absence of agents (○), in 1 μM RA (▴), in 1 mM NaB (⋄), or in 1 μM RA + 1 mM NaB (●). The percentage of terminally differentiated APL cells was measured in case 6 by the expression of CD11b surface antigen detected by flow cytometry (A) or in AP-1060 cells by the nitroblue tetrazolium dye reduction test (B).
Differentiation response of APL cells.
APL cells from case 6 (A) and from AP-1060 cells derived from case 1 (B) were cultured for the indicated time in the absence of agents (○), in 1 μM RA (▴), in 1 mM NaB (⋄), or in 1 μM RA + 1 mM NaB (●). The percentage of terminally differentiated APL cells was measured in case 6 by the expression of CD11b surface antigen detected by flow cytometry (A) or in AP-1060 cells by the nitroblue tetrazolium dye reduction test (B).
APL cells were available for in vitro testing from one other patient with a PML-RARα mutation (Pro407Ser; case 1) in the form of a culture strain, called AP-1060.20 In contrast to the Gly289Arg mutant cells, NaB alone had no discernible effect on AP-1060 cell differentiation, as measured cytologically (not shown) or by the nitroblue tetrazolium test (Figure 6B). Similarly, no differentiation was observed with NaB or a subinducing RA concentration (10 nM RA; not shown). A strong positive interactive effect was, however, observed with 100 nM RA, an effective inducing RA concentration (Figure 6B).
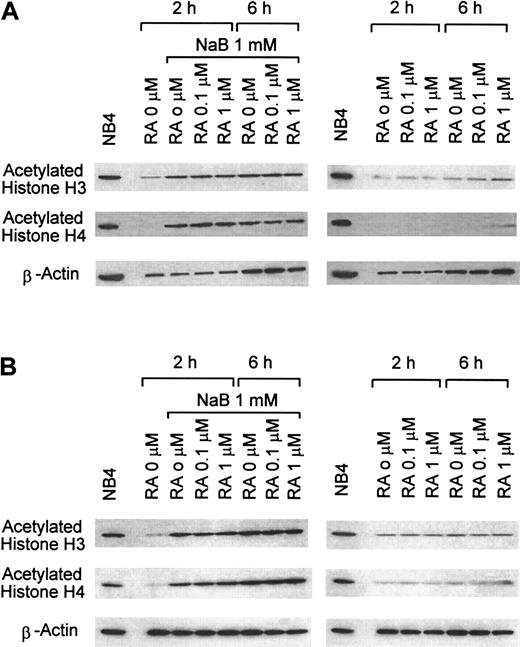
Acetylation of H3 and H4 histones
Exposure to 1 mM NaB for 2 to 6 hours resulted in effective hyperacetylation of H3 and H4 histones in APL cells from the Gly289Arg mutant case and in AP-1060 cells (Figures7A,B), as well as in transfected COS-1 cells (not shown). RA alone (1 μM) produced trace histone acetylation, which only minimally increased the level of histone acetylation produced by 1 mM NaB alone in either cell type.
Western blot analysis of H3 and H4 histone acetylation in APL cells.
APL cells from case 6 (A) and AP-1060 cells (B) were treated with 1 mM NaB, 1 μM RA, or 1 mM NaB + 1 μM RA for the indicated periods of time in tissue culture.
Western blot analysis of H3 and H4 histone acetylation in APL cells.
APL cells from case 6 (A) and AP-1060 cells (B) were treated with 1 mM NaB, 1 μM RA, or 1 mM NaB + 1 μM RA for the indicated periods of time in tissue culture.
Discussion
This study found that 5 (62.5%) of 8 APL patients who had relapsed after 2 or more courses of RA-containing therapy had missense mutations in the RARα-region of PML-RARα. This finding compares with the finding of such mutations in 3 (25%) of 12 first- relapse RA-treated APL patients.5 Although the case numbers are small, these results strongly suggest that repeated and longer exposure to RA-containing therapy is associated with increased risk of developing PML-RARα mutations. It seems improbable that any of these mutations were present in the APL cells before initial therapy, as demonstrated in case 8 in this study, because such mutations have not been observed in more than 30 de novo APL cases, including 5 published cases positive for mutations at relapse.5,7,25 In one patient (case 7), the mutation may have been late emerging, possibly during RA + PB therapy, because no mutation was detected in a specimen obtained 16 months previously during a transient CR on ATO therapy. Conversely, in another patient (case 6), the same mutation was detected before initiating and after relapse from a 7-month remission achieved on RA + PB therapy.16 The latter observation suggests that, once established, the mutant clone may persist, but, clearly, more data on serial specimens and in successive relapses are needed to assess possible subclonal variation. Further studies are also required to determine when the mutations arise and/or become detectable in the APL cell population during the course of therapy and to determine what role the coadministration of chemotherapy or other agents may have in the acquisition of mutations. The emergence of mutant subclones could also affect the rate of clinical recurrence from minimal residual disease levels, a factor to consider in the use of PML-RARα mRNA measurements by RT-PCR to assess relapse risk.26 27
This report adds 4 novel missense mutations to 6 previously identified unique PML-RARα missense mutations, all in the LBD of the RARα region of the molecule (Table 3). A fifth missense mutation (Arg272Gln) was identical to 2 previously reported point mutations,6,7 suggesting that this site could be a hot spot related to RA resistance in APL. The position of the mutations in the RARα region LBD in 13 positive APL relapse cases, counting the redundant cases (Table 3), appears to be segregated into the same 3 zones of clustered mutations identified in the homologous thyroid hormone receptor-β (TRβ) in the more extensively studied resistance to thyroid hormone syndrome (RTHS).28,29 As shown in Figure 2A,B, 9 of 13 PML-RARα mutations occurred in central LBD zone I, corresponding to the site of the most numerous mutations of TRβ in RTHS; 3 of 13 PML-RARα mutations occurred in the near carboxy-terminal activator function-2 region, corresponding to TRβ mutational zone II; and 1 of 13, the Lys207Asn mutant, occurred in a more proximal zone of the RARα-region LBD, corresponding to a recently defined, third zone of clustered mutations in the LBD of TRβ.29 The uniform finding of missense mutations in the PML-RARα LBD contrasts with the finding of some deletion mutations in the TRβ LBD,28 suggesting different mutational mechanisms. Similarly, the finding of a common nonsense mutation in PML-RARα or RARα in RA-sensitive human myeloid leukemia cell lines selected for RA resistance in vitro (NB4 and HL-60, respectively),30-32 suggests that mutation mechanisms may differ in vitro and in vivo.
PML-RARα missense mutations identified in relapse, RA-treated APL patients
| Mutation3-150 . | Missense identified . | References . |
|---|---|---|
| Lys207Asn | 1 | This report |
| Arg272Gln | 3 | Imaizumi et al,6 Marasca et al7 and this report |
| Arg276Trp | 2 | Marasca et al7 and Takayama et al45 |
| Gly289Arg | 1 | This report |
| Leu290Val | 1 | Ding et al5 |
| Arg294Trp | 1 | This report |
| Met297Leu | 1 | Imaizumi et al6 |
| Arg394Trp | 1 | Ding et al5 |
| Pro407Ser | 1 | This report |
| Met413Thr | 1 | Ding et al5 |
| Mutation3-150 . | Missense identified . | References . |
|---|---|---|
| Lys207Asn | 1 | This report |
| Arg272Gln | 3 | Imaizumi et al,6 Marasca et al7 and this report |
| Arg276Trp | 2 | Marasca et al7 and Takayama et al45 |
| Gly289Arg | 1 | This report |
| Leu290Val | 1 | Ding et al5 |
| Arg294Trp | 1 | This report |
| Met297Leu | 1 | Imaizumi et al6 |
| Arg394Trp | 1 | Ding et al5 |
| Pro407Ser | 1 | This report |
| Met413Thr | 1 | Ding et al5 |
RA indicates all-l retinoic acid; APL, acute promyelocytic leukemia.
Nucleotide and amino acid numbers correspond to that for normal RARα1.44
The common positional mutations in the LBD of the TRβ and PML-RARα imply common functional defects as well. Studies of numerous TRβ mutants from all 3 clustered mutation zones indicate that these mutations result in defective activation of thyroid hormone response genes by reducing TR-ligand binding, increasing TR-corepressor binding, and, in some cases, decreasing TR-coactivator binding.28,29,33 A recent study of 5 naturally occurring PML-RARα mutants demonstrated similar RARα-region binding defects.23 In the current study, 2 PML-RARα mutants (Arg294Trp and Gly289Arg) showed a marked reduction in RA binding (Figure 3), and all 5 mutants showed reduced RA transactivation of an RARE-regulated reporter gene compared with wild-type PML-RARα controls (Figure 4). Although more detailed molecular studies are required to understand the heterogeneity of PML-RARα mutant molecular mechanisms, the disproportionate involvement of the basic amino acids, arginine and lysine, that were converted to hydrophobic (tryptophan) or neutral (glutamine and asparagine) amino acids in 50% or more of overall mutations is notable (5 of 10 unique mutations and 8 of 13 total mutations; Table 3). Such alterations, which also might apply to the acquisition of arginine in the Gly289Arg mutation, have been related to changes in interactions with negatively charged residues in the carboxylate moiety of RA, in DNA, and in other proteins, possibly including acetyl and phosphate groups.34-38
The current study clearly demonstrates that there was no relationship between the functional status of PML-RARα and clinical outcome on RA + PB therapy. This finding poses 2 essential questions: Why did case 6 with the most dysfunctional PML-RARα mutation respond to RA + PB treatment, and why did the other 4 patients with either wild-type PML-RARα or less dysfunctional PML-RARα mutations fail to respond? Regarding case 6, there seems little doubt that the Gly289Arg mutation produced severe RA resistance, because the APL cells failed to show any signs of differentiation after treatment with 1 μM RA. At this relatively high concentration, a synergistic interaction between RA and NaB was observed in a transactivation transcription reporter assay (Figure 4B). Although this interaction could be an important element in the clinical response of case 6 to RA + PB therapy, this in vitro response required at least a 10-fold higher RA concentration to elicit compared with the other PML-RARα mutants. These considerations suggest that our additional observation that NaB alone was able to induce partial differentiation of the APL cells from case 6 in short-term culture (Figures 5 and 6) might signify a contributory factor to this patient's clinical response. That this might have been a discriminatory factor is enhanced by the observation that the APL cells from an alternative PML-RARα mutant case (case 1) did not differentiate in response to NaB as a single agent (Figure 6), which is in accord with a reported mutant NB4 subline.13 Thus, NaB may have contributed to the exceptional clinical response of case 6 by modulating critical differentiation-response genes in a non–RA-dependent manner. If this suggestion is valid, it would not have been apparent from the global histone H3 and 4 acetylation studies performed to compare cases 6 and 1, which showed no apparent intercase variation in histone acetylation pattern (Figure 7).
Regarding the failure of the other 4 RA + PB–treated patients to respond, insufficient information is available to offer any specific hypotheses. In our previous study of de novo APL, more first-relapse patients were resistant to RA-induced differentiation than harbored PML-RARα mutations (8 of 10 versus 3 of 12, respectively).5 This finding indicates the involvement of alternative defects in APL cellular RA resistance that might reasonably be expected to be compounded in the cells of patients who have experienced further treatments and relapses. Thus, it would not be surprising if the differentiation response of the APL cells from the 4 nonresponder RA + PB–treated patients did not parallel the PML-RARα transfection transactivation activity, as it did in both case 6 with Gly289Arg and case 1 with Pro407Ser mutant cells. One possible alternative mechanism of RA resistance in these patients, who had previously been extensively treated with both RA and ATO, is hypercatabolism of PML-RARα protein, as reported in NB4 sublines intensively selected with these agents in vitro.39-41Although the ATO-induced hypercatabolism depended on the continuous presence of ATO,40,41 which did not pertain in the current clinical cases, this and other potential mechanisms of clinical APL cellular RA resistance require further study.42Additionally, the 4 nonresponder patients might have had precellular aberrations of RA pharmacology that prevented adequate leukemic cell RA delivery.43 From these considerations, we conclude that neither the presence nor the nature of PML-RARα mutations is predictive for responsiveness to RA + PB therapy.
Supported by grants from the National Institutes of Health (CA56771 [R.E.G.] and CA73136 [R.P.W.]) and from the Lymphoma Foundation (R.P.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robert E. Gallagher, Department of Medical Oncology, Montefiore Medical Center, Rm 601, Hofheimer Bldg, 111 E 210th St, Bronx, NY 10467; e-mail: rgallagh@aecom.yu.edu.

![Fig. 3. Size-exclusion FPLC analysis of nuclear RA-binding activity in COS-1 cells. / Cells are transfected with pSG5 vector (A), wild-type PML-RARα L form (B), mutant Lys207Asn (C), mutant Arg272Gln (D), mutant Gly289Arg (E), mutant Pro407Ser (F), wild-type PML-RARα S form (G), and mutant Arg294Trp (H). Nuclear extracts were incubated with 10 nmol/L [3H]-RA in the absence (●) or presence (○) of 200-fold excess of unlabeled RA for 15 hours at 4°C. The samples were fractionated over a Superose 6HR 10/30 column at 0.4-mL intervals. Arrows indicate the fraction numbers of marker proteins (in kd) used to calibrate the FPLC column.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/4/10.1182_blood.v99.4.1356/6/m_h80422151003.jpeg?Expires=1766956673&Signature=YjHd2cPlIKwvhdMrEJrvyQQuYLSBijkw~Ps7GPjjmnVoB7pmsf8p6OYR3yMUaHNEJRhO59AnG5CcYj~~PpvEsZ1LtFKN3O-1P4OUymdNrxKTbC3thxP42-1aBwerRz9Re8bH76pc9caQl~Gz~WLyVm4axbjg2wxpcfao5bzFxl64m~mfuc7WfuuNQtvYQPgVhDQsTW7KPHWlt1IiJFFD7FWpRn12tpH5aXUaPLearXNJObmu-U93P~veTGm7BBrgl93ZryYX0HAH1jJScJaP1KBZQjAloedGHachELVuT09dg7izlF15i9jhgDvebWtz74zPXWMMTuZvfVIOgA3q6Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal