The postinduction level of minimal residual disease (MRD) was quantified with a competitive polymerase chain reaction (PCR) technique in 104 children with acute lymphoblastic leukemia (ALL) diagnosed between June 1993 and January 1998 and followed for a median of 4.2 years. A significant correlation was found between the MRD level on day 15 (D15) and day 29 (D29) after the start of induction therapy (rs = 0.70, P < .0001). The 15 patients with T-cell disease had higher D29 MRD than those with B-lineage ALL (P = .01). Age was positively related to D29 MRD (rs = 0.32, P = .001). The 16 patients who had a relapse had higher D15 and D29 MRD levels than the patients who stayed in remission (median levels D15, 1% versus 0.1%,P = .03; D29, 0.4% versus 0.01%,P = .0001). No patients with a MRD level less than 0.01% on D29 have so far had a relapse, whereas the 7-year probability of event-free survival for patients with higher MRD levels was 0.52 (P = .0007). The group of patients with a D29 MRD less than 0.01% included patients with T-cell disease, white blood cell count more than 50 × 109/L at diagnosis, or age 10 years or older, and could not be identified by up-front criteria. The best-fit Cox model to predict the risk of relapse included D29 MRD (P = .004) and age (P = .009). These findings indicate that with the present treatment protocol MRD quantification at an early stage of therapy identifies patients with a very low risk of relapse. Further trials are needed to reveal whether such patients with D29 MRD less than 0.01% can be cured with less intensive chemotherapy, which would reduce the risk of serious late effects as well as the costs of therapy.
Introduction
In the Nordic countries the outcome of acute lymphoblastic leukemia (ALL) in childhood has improved markedly within the last 3 decades from about 20% probability of event-free survival (pEFS) in the early 1970s to currently almost 80% pEFS.1This improvement is a result of a series of protocols in which the treatment has been intensified based on up-front risk criteria.1 This intensification of treatment carries a risk of clinically significant late effects. Thus, new approaches are needed to identify patients who have a high probability of being cured with less toxic therapy.2
Patients with a poor treatment response by morphologic criteria have a high risk of relapse,3,4 but morphologic studies will only identify a minority of those children with ALL who eventually fail. Quantification of minimal residual disease (MRD) has been of prognostic value in children with ALL. Several studies have shown that children with a high leukemic cell burden at the end of induction therapy have an inferior outcome compared to children with a lower leukemic cell burden.5-12 However, most of the assays applied in these studies have been qualitative or at best semiquantitative and have mainly been applied to identify patients with an increased relapse rate. To identify patients with a superior prognosis, a qualitative polymerase chain reaction (PCR) test may not be sufficient, because a large proportion of those patients who are eventually cured will show PCR positivity during and after induction therapy. A recent study indicated that the very good-risk patients may be identified as those with a very low MRD at the end of induction therapy, whereas MRD evaluation at a later time point is needed to distinguish between patients with resistant disease and those with a slowly disappearing but chemosensitive disease.13Consequently, a precise, sensitive, and quantitative method is probably required to stratify treatment according to the postinduction leukemic cell burden.
To explore whether patients with a good prognosis could be identified early during therapy, the Nordic Society of Pediatric Haematology and Oncology (NOPHO) ALL MRD-95 study quantified residual leukemia on treatment days 15 (D15) and 29 (D29) in 104 Nordic children by a recently described precise, competitive PCR method.14 The study was approved by the local ethical committees according to the Helsinki Declaration.
Patients, materials, and methods
Patients
Bone marrow and blood samples were collected at the time of diagnosis, at D15, and at D29 from children with non–B-cell ALL in Denmark, Iceland, and Norway and shipped to the National University Hospital, Rigshospitalet, Copenhagen. The patients were between 1 and 16 years old (median, 4.5 years). The study was initiated for patients diagnosed at the National University Hospital, Rigshospitalet, Copenhagen, June 1, 1993 (n = 34, period I), and since September 1, 1995 extended to include all centers treating childhood ALL in Denmark, Norway, and Iceland (n = 70, period II). The study was closed January 31, 1998. All patients were treated according to the NOPHO ALL-92 protocol.1 We received samples from 197 patients. Of the 104 patients examined for MRD, 66 patients were examined both at D15 and D29, 4 patients were examined only at D15 (including 1 patient with a relapse), and 34 patients were examined only at D29. Of the remaining 93 patients, 90 were excluded due to a too small sample volume, poor sample (ie, DNA) quality, or because samples were not obtained at diagnosis. Three patients were not included because of no clonal rearrangements in TCR or IGH genes. The end of follow-up was September 1, 2000.
Risk classification was determined by age and white blood cell (WBC) counts at diagnosis (standard risk [SR]: 2-10 years and WBCs < 10 × 109/L; intermediate risk [IR]: <2 years or ≥10 years or WBCs = 10-49 × 109/L; and high risk [HR]: WBCs ≥50 × 109/L), the presence of central nervous system (CNS) involvement or testicular leukemia, a mediastinal mass, T-cell disease, certain cytogenetic translocations, or an M3 D15 or an M2 D29 bone marrow sample (all HR criteria).1
Induction therapy consisted of prednisolone (60 mg/m2 per day divided into 3 doses on days 1-36, then tapering), vincristine (2.0 mg/m2 on days 1, 8, 15, 22, 29, and 36), doxorubicin (40 mg/m2 on days 1, 22, and 36 [+day 8 for patients with HR-ALL], Erwinia asparaginase (30 000 IU/m2 on days 37-46), and intrathecal methotrexate (days 1, 8, 15, and 29).1
Mononuclear cells from bone marrow and blood were isolated by density centrifugation (Lymphoprep, Pharmacia, Uppsala, Sweden). DNA was prepared, either from viable cells washed in 0.9% NaCl and cryopreserved in RPMI and dimethyl sulfoxide in liquid nitrogen or from freshly isolated mononuclear cells, by NaCl precipitation15or phenol extraction. The DNA concentration and quality were measured by spectrophotometry and the DNA was stored at −20°C.
Quantification of MRD
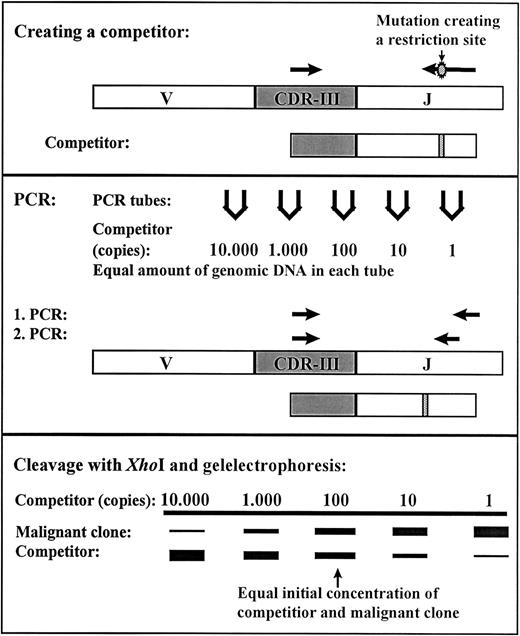
Quantification of MRD for all patients was performed at the National University Hospital, Rigshospitalet, Copenhagen as previously described.14 In short, the rearranged genes of the immunoglobulin heavy chain (IGH), the T-cell receptor γ(TCRG), and the incomplete V2D3 rearrangement of T-cell receptor δ (TCRD) were used as clonal markers. At diagnosis, PCR was performed on genomic DNA using primers located in conserved domains on each side of the CDRIII regions of the rearranged IgH, TCR-γ, and TCR-δ. The PCR products were sequenced and for the majority of patients it was possible to identify a clonal marker in more than one of these 3 systems. The marker TCR-γ was preferentially used, then TCR-δ, and finally IgH for designing of a clone-specific primer. We have previously shown that the determined level of MRD is generally independent of the gene rearrangement used as comparison of 2 or 3 different clonal markers of the clone dominating at diagnosis gave the same level of MRD at D29.14 A competitor was constructed by introducing the restriction site XhoI in a PCR product that was identical to parts of the highly specific rearranged IGH, TCRG, or TCRDgenes of the malignant clone. This PCR product was cloned using the pCR2.1-TOPO kit (Invitrogen, Leek, The Netherlands) and PCR-amplified using vector specific primers to form the competitor. The correct sequence was verified by DNA sequencing and the concentration was determined. Using a clone-specific primer and a consensus primer located externally to the XhoI site the competitor and the DNA from the malignant clone were amplified under identical conditions. The PCR was performed as seminested where the clone-specific primer was reused and the consensus primer moved inward. After cleavage withXhoI, the PCR products originating from the competitor and the malignant clone could be separated by gel electrophoresis and the amount of residual disease could be determined (Figure1). The method is very sensitive with a detection limit of down to one malignant cell in 106 normal cells depending on the amount of mononuclear cells analyzed.
Competitive PCR method for residual disease quantification.
A competitor was constructed by PCR using genomic DNA from the malignant clone as template together with a clone-specific primer and primer-XhoI to introduce a restriction site. The first amplification in the seminested PCR was done with the clone-specific primer and primer-1 and the second amplification with the clone-specific primer and primer-2. The PCR products from the competitor are cleaved by the restriction enzyme XhoI; the PCR products from the malignant clone remain uncleaved. The MRD level can be determined from the lanes with equal amplification of the competitor and malignant clone.14
Competitive PCR method for residual disease quantification.
A competitor was constructed by PCR using genomic DNA from the malignant clone as template together with a clone-specific primer and primer-XhoI to introduce a restriction site. The first amplification in the seminested PCR was done with the clone-specific primer and primer-1 and the second amplification with the clone-specific primer and primer-2. The PCR products from the competitor are cleaved by the restriction enzyme XhoI; the PCR products from the malignant clone remain uncleaved. The MRD level can be determined from the lanes with equal amplification of the competitor and malignant clone.14
Statistical analyses
The Mann-Whitney U test, Kruskal-Wallis test, χ2 test, Fisher exact test, and Spearman rank order correlation analysis were applied to compare distribution of parameters between subgroups and correlation between parameters (rS = correlation coefficient).16 Stepwise Cox multivariate proportional hazards regression analyses were done to detect prognostic factors.17 Parameters were included and excluded from the models at significance limits of 0.05 and 0.10, respectively. The Kaplan-Meier method was applied for estimation of remission duration and for the generation of survival curves.18 Subgroups were compared with the log-rank test. In all analyses 2-sided P < .05 were regarded as being significant. Data were analyzed using the SPSS software (SPSS, Chicago, IL).
Results
The distribution of age, sex, phenotype, risk group stratification, or clinical outcome did not differ significantly between the group of 104 patients who were analyzed for MRD with competitive PCR and those 93 patients who were not included. WBC count at diagnosis was higher in the group of patients that was analyzed for MRD than in the group of patients not included in this study (median, 19 versus 9 × 109/L, P = .03). We found no significant difference in the MRD levels at D29 when comparing samples with a duration of shipment of more than 1 day to samples with a duration of shipment of 1 day or less.
A limiting factor to the sensitivity was a low number of mononuclear cells in the bone marrow aspirates obtained at D15 and D29, when hypoplasia frequently was found. Thus, for 5 patients at D15 and 19 patients at D29, the clonal immune gene rearrangements were not detectable. Prior to the statistical analyses, the MRD level for such patients was set to the reciprocal value of the number of cells examined. None were induction failures.
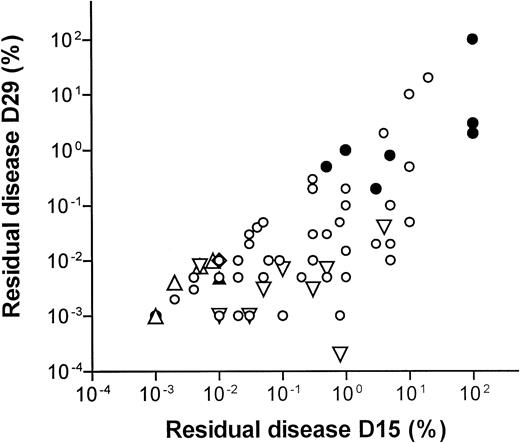
Figure 2 shows a scatter plot of the 66 patients that were examined both at D15 and at D29 after the initial diagnosis. When including all patients, the median D15 and D29 MRD levels were 0.3% and 0.01% (75% ranges, 0.008%-10% and 0.001%-1.2%, respectively). The MRD level on D15 was correlated to the level on D29 (rS = 0.70, P < .0001). This correlation was higher for T-lineage ALL (rS = 0.90,P = .002) compared to B-lineage ALL (rS = 0.61, P < .0001). A similar correlation between the D15 and D29 MRD levels was found if only patients who had detectable MRD levels at D15 and D29 were included (n = 51, rS = 0.74, P < .0001).
D15 versus D29 residual disease levels.
Scatterplot of 66 patients examined both at D15 and at D29 after the start of treatment. Open symbols denote B-lineage leukemia and filled symbols denote T-lineage leukemia. ■ indicates that both D15 and D29 MRD levels are above the limit of detection. ▿ indicates that D15 MRD is detectable, but D29 is below the limit of detection and the level is set to the reciprocal value of the number of cells examined. ⋄ indicates that D29 MRD is detectable, but D15 level is below the limit of detection and the level is set to the reciprocal level of the number of cells examined. ▵ indicates that D15 and D29 levels are both below the limit of detection and the MRD levels are set to the reciprocal values of the number of cells examined. Rank correlations: all patients, n = 66, rS = 0.70, P < .0001; T-lineage ALL, n = 9, rS = 0.90, P = .002; B-lineage ALL, n = 57, rS = 0.61,P < .0001.
D15 versus D29 residual disease levels.
Scatterplot of 66 patients examined both at D15 and at D29 after the start of treatment. Open symbols denote B-lineage leukemia and filled symbols denote T-lineage leukemia. ■ indicates that both D15 and D29 MRD levels are above the limit of detection. ▿ indicates that D15 MRD is detectable, but D29 is below the limit of detection and the level is set to the reciprocal value of the number of cells examined. ⋄ indicates that D29 MRD is detectable, but D15 level is below the limit of detection and the level is set to the reciprocal level of the number of cells examined. ▵ indicates that D15 and D29 levels are both below the limit of detection and the MRD levels are set to the reciprocal values of the number of cells examined. Rank correlations: all patients, n = 66, rS = 0.70, P < .0001; T-lineage ALL, n = 9, rS = 0.90, P = .002; B-lineage ALL, n = 57, rS = 0.61,P < .0001.
Table 1 shows Spearman correlations between MRD and WBC counts and between MRD and age. No significant correlation was found between WBC count and MRD although patients with a WBC count more than 200 × 109/L at diagnosis had higher MRD levels than the remaining patients (median D29, 0.65% versus 0.01%, P = .04). Both on D15 and D29 significant correlations were found between MRD and age (D15, rS = 0.46, P < .0001; D28, rS = 0.32, P = .001).
MRD in relation to clinical parameters
| . | No. D15 . | Median MRD D15, % . | No. D29 . | Median MRD D29, % . |
|---|---|---|---|---|
| WBC count at diagnosis (× 109/L) | ||||
| Less than 10 | 26 | 0.3 | 38 | 0.01 |
| 10 to lower than 50 | 26 | 0.075 | 37 | 0.01 |
| 50 to lower than 100 | 5 | 0.01 | 11 | 0.03 |
| 100 to lower than 200 | 8 | 0.4 | 10 | 0.02 |
| 200 or more | 5 | 0.8 | 4 | 0.65 |
| Subgroup comparisons | rS = −0.13, ns | rS = 0.10, ns | ||
| Age at diagnosis, y | ||||
| 1.0 to younger than 2.0 | 6 | 0.025 | 11 | 0.01 |
| 2.0 to younger than 6.0 | 41 | 0.1 | 54 | 0.009 |
| 6.0 to younger than 10.0 | 14 | 1 | 18 | 0.03 |
| 10.0 or older | 9 | 1 | 17 | 0.2 |
| Subgroup comparisons | rS = 0.46,P < .0001 | rS = 0.32, P = .001 | ||
| Male | 43 | 0.3 | 59 | 0.02 |
| Female | 27 | 0.05 | 41 | 0.01 |
| Subgroup comparisons | ns | ns | ||
| SR | 21 | 0.3 | 29 | 0.01 |
| IR | 30 | 0.095 | 43 | 0.01 |
| HR | 19 | 0.5 | 28 | 0.035 |
| Subgroup comparisons | ns | ns | ||
| B lineage | 60 | 0.15 | 86 | 0.01 |
| T lineage | 10 | 2 | 14 | 0.35 |
| Subgroup comparisons | ns | P= .01 | ||
| Remission | 59 | 0.1 | 85 | 0.01 |
| Relapse | 11 | 1 | 15 | 0.4 |
| Subgroup comparisons | P = .03 | P = .0001 |
| . | No. D15 . | Median MRD D15, % . | No. D29 . | Median MRD D29, % . |
|---|---|---|---|---|
| WBC count at diagnosis (× 109/L) | ||||
| Less than 10 | 26 | 0.3 | 38 | 0.01 |
| 10 to lower than 50 | 26 | 0.075 | 37 | 0.01 |
| 50 to lower than 100 | 5 | 0.01 | 11 | 0.03 |
| 100 to lower than 200 | 8 | 0.4 | 10 | 0.02 |
| 200 or more | 5 | 0.8 | 4 | 0.65 |
| Subgroup comparisons | rS = −0.13, ns | rS = 0.10, ns | ||
| Age at diagnosis, y | ||||
| 1.0 to younger than 2.0 | 6 | 0.025 | 11 | 0.01 |
| 2.0 to younger than 6.0 | 41 | 0.1 | 54 | 0.009 |
| 6.0 to younger than 10.0 | 14 | 1 | 18 | 0.03 |
| 10.0 or older | 9 | 1 | 17 | 0.2 |
| Subgroup comparisons | rS = 0.46,P < .0001 | rS = 0.32, P = .001 | ||
| Male | 43 | 0.3 | 59 | 0.02 |
| Female | 27 | 0.05 | 41 | 0.01 |
| Subgroup comparisons | ns | ns | ||
| SR | 21 | 0.3 | 29 | 0.01 |
| IR | 30 | 0.095 | 43 | 0.01 |
| HR | 19 | 0.5 | 28 | 0.035 |
| Subgroup comparisons | ns | ns | ||
| B lineage | 60 | 0.15 | 86 | 0.01 |
| T lineage | 10 | 2 | 14 | 0.35 |
| Subgroup comparisons | ns | P= .01 | ||
| Remission | 59 | 0.1 | 85 | 0.01 |
| Relapse | 11 | 1 | 15 | 0.4 |
| Subgroup comparisons | P = .03 | P = .0001 |
P values for subgroup comparisons are calculated using either the Mann-Whitney U test or Kruskal-Wallis test. rS indicates Spearman rank correlation coefficient; ns, not significant.
Table 1 shows comparisons of MRD levels within different subgroups. Neither gender nor risk group was significantly related to the D15 or D29 MRD levels. No significant difference in MRD levels was found between patients with B-lineage (n = 60, median = 0.15%, 25%-75% percentile, 0.02%-1%) versus T-lineage disease at D15 (n = 10, median = 2%, 25%-75% percentile, 0.01%-100%,P = .15), whereas at D29 the group of patients with T-lineage disease (n = 14, median = 0.35%, 25%-75% percentile, 0.009%-2.3%) had significantly higher levels of MRD than those with B-lineage disease (n = 86, median = 0.01%, 25%-75% percentile, 0.004%-0.08%, P = .01).
The D29 MRD levels did not differ significantly between patients with SR (median D29 MRD, 0.01%, 1 relapse), IR (median D29 MRD, 0.01%, 6 relapses), and HR ALL (median D29 MRD, 0.04%, 9 relapses). In this study only one patient with SR ALL has so far had a relapse. This patient had the second highest MRD level at D29 (1%) of all the 29 SR patients analyzed on D29 in this study.
Karyotypes by G-banding were available for 76 of the 100 patients who were investigated for MRD at D29. Thirty-five of these patients had an aberrant karyotype. There were no relationships between the modal number and the MRD levels at D29 (rS = −0.01,P = .95). When dividing the patients into groups by the modal number, the median MRD at D29 was 0.01% for patients with modal numbers from 45 to 47, 0.008% for patients with modal numbers from 48 to 51, and 0.01% for patients with a modal number 52 or higher.
After a median length of follow-up for patients still in remission of 4.2 years (75% range, 2.9-6.6 years), 16 patients (7 with T-cell disease) have had a relapse 2 to 80 months from diagnosis (median, 30 months). All T-cell relapses occurred within 3 years from diagnosis. The overall 7-year probability of remaining in remission was 0.75, which is similar to the pEFS of all children more than 1 year of age treated by the NOPHO ALL-92 protocol (0.76 at 8 years).1 The outcome for patients included in period I and period II did not differ significantly. Patients with T-cell disease did worse than those with B-lineage ALL (risk of relapse, 0.47 versus 0.22 at 7 years, P < .0001). Time to relapse was not related to D29 MRD (rS = −0.13, P = .63). Nor did medullary or isolated extramedullary (3 CNS, 1 testicular) differ in the D29 MRD levels (D29, 0.4% versus 2.5%,P = .9).
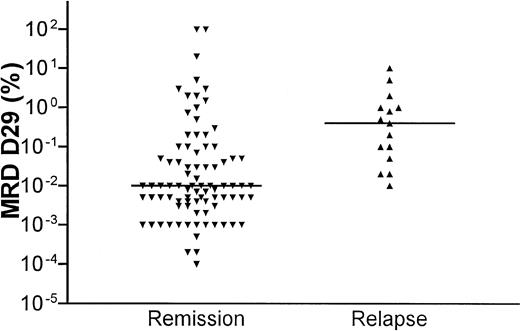
The distribution of MRD levels in relation to clinical outcome is shown in Figure 3. Patients who stayed in continuous clinical remission (CCR) had significantly lower D15 and D29 MRD levels compared with patients who later relapsed (D15 median, 0.1 versus 1%, P = .03; D29 median, 0.01 versus 0.4%,P = .0001). All patients who relapsed had a D29 MRD of 0.01% or higher (the median D29 MRD level for patients who stayed in remission).
D29 residual disease levels in relation to clinical outcome.
Scatterplot of MRD levels at D29 after the start of induction therapy and clinical outcome. The horizontal lines show the median levels at D29 for the patients in remission and the patients who developed a relapse, respectively.
D29 residual disease levels in relation to clinical outcome.
Scatterplot of MRD levels at D29 after the start of induction therapy and clinical outcome. The horizontal lines show the median levels at D29 for the patients in remission and the patients who developed a relapse, respectively.
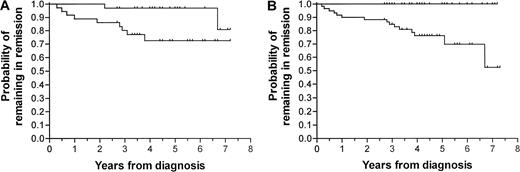
In the survival analyses and the Kaplan-Meier plots, patients with second malignancies (2 patients) or death in remission (1 patient, infection) were censored at the time point of these events. The 70 patients investigated at D15 were divided in 2 groups, one with the MRD level below the median and the other with MRD equal to or higher than the median. Figure 4A shows the Kaplan-Meier estimates based on MRD levels on D15. Only 2 patients in the group of patients with MRD below the median (< 0.3%) suffered a relapse. One patient had a D15 MRD of 0.03% and a relapse 80 months from diagnosis, whereas the other patient had a D15 MRD of 0.01% and a relapse 26 months from diagnosis. Thus, none of the 9 patients (13% of all studied) with a D15 MRD level less than 0.01% have had relapse. The Kaplan-Meier curves for patients with MRD levels less than versus at least 0.01% on D29 are shown in Figure 4B. No relapses were seen among patients with MRD levels below the median of 0.01% compared to a 7-year relapse risk of 0.48 for the patients with higher D29 MRD levels (P = .0007). Only one patient had a D29 MRD of exactly 0.01% and a relapse in the bone marrow 2 months from diagnosis. We found a significant difference in the 6-year risk of relapse for patients with D29 MRD levels 0.01% or more and less than 0.3% versus at least 0.3% (longest follow-up, 6.1 years; 6-year risk of relapse, 0.18 versus 0.51, P = .19). Although it did not reach significance, the differences in relapse risk among these 3 MRD subgroups (D29 MRD < 0.1% versus at least 0.01% and < 0.3% versus at least 0.3%) were most striking among patients with T-lineage ALL (cumulative relapse risk, 0% [0 of 3] versus 25% [1 of 4] versus 71% [5 of 7 patients], P = .15). For B-lineage ALL, the difference in outcome for patients with D29 MRD levels less than 0.01% versus at least 0.01% was significant (relapse risk, 0% versus 50%, P = .005), whereas patients with D29 MRD levels of at least 0.01% and less than 0.3% versus at least 0.3% did not differ significantly (P = .66).
Relapse risk plots according to D15 and D29 residual disease levels.
Kaplan-Meier plots. (A) Patients with D15 MRD levels at least (n = 36, lower curve) or less than (n = 34, upper curve) 0.3% (median for patients in remission), P = .02. (B) Patients with D29 MRD levels at least (n = 60, lower curve) and less than (n = 40, upper curve) 0.01% (median for patients in remission),P = .007.
Relapse risk plots according to D15 and D29 residual disease levels.
Kaplan-Meier plots. (A) Patients with D15 MRD levels at least (n = 36, lower curve) or less than (n = 34, upper curve) 0.3% (median for patients in remission), P = .02. (B) Patients with D29 MRD levels at least (n = 60, lower curve) and less than (n = 40, upper curve) 0.01% (median for patients in remission),P = .007.
Multivariate, stepwise Cox regression analysis was done to evaluate the impact of gender, age, WBC count at diagnosis, immunophenotype, and D15 or D29 MRD, respectively. To approximate a normal distribution, WBC count, and D15 and D29 MRD were logarithmically transformed prior to the regression analyses. In both forward and backward analyses only D29 MRD level (B = 0.59, P = .004) and age (B = 0.17,P = .009) were included in the model. The good prognosis group of patients with a D29 MRD less than 0.01% included patients with T-cell disease (n = 3), WBC count more than 50 × 109/L at diagnosis (n = 8), or age at least 10 years (n = 5), and could not be identified by up-front criteria. When testing the D15 MRD level, the WBC count at diagnosis (B = 1.56,P = .01), D15 MRD level (0.53, P = .03), age (B = 0.18, P = .03), and gender (B = 1.47,P = .09) were included in the model. The immunophenotype had prognostic significance in univariate analysis, but this was lost in the final model when age and D29 MRD were entered into the model.
Discussion
About 50% of the patients diagnosed in the Nordic countries in the early 1980s were cured on less intensive treatment than that of the current protocol.1 If these patients could be identified through MRD analyses at an early stage of therapy, this group of patients could be offered less intensive therapy and late effects as well as treatment costs caused by overtreatment could hereby be reduced. The quantification of MRD with a competitive PCR enabled us to detect a large fraction of patients with an excellent prognosis. Thus, in the present study, no patient with MRD levels below that of the median level for all patients examined on D29 (0.01%) has suffered a relapse so far.
Although a few other studies have addressed the relation between postremission MRD levels and outcome, except for the BFM (Berlin-Frankfurt-Münster) study,13 the present prospective unselected study is the largest that includes both T- and B-lineage ALL and all risk groups and that has been able to identify an almost 50% subgroup of patients with a less than 5% risk of relapse. Thus, the results of the present study support the findings of the International BFM Study Group that showed that patients with undetectable MRD at the end of induction (comparable to MRD < 0.01%-0.001%) have a pEFS of 0.98.13 In that study of van Dongen et al,13 71 of 169 patients studied were MRD negative after 5 weeks of induction therapy, and after a median follow-up of 48 months only 2 have had a relapse. These results have in a case-control study been confirmed for patients with medium-risk features.19 Although a few other MRD studies similarly have identified cohorts of children with ALL and an excellent outcome, these studies were all hampered by being either small, the duration of follow-up being small, or the subgroups with the good outcome being selected: (1) Coustan-Smith et al20 used rapid flow cytometry to detect MRD; 165 patients were examined at the end of induction therapy and 123 of these were found to have an MRD less than 0.01% of which 9 patients relapsed (10% 5-year risk of relapse). When the material was subdivided in several ways, a subgroup of 34 patients with National Cancer Institute high-risk B-lineage ALL and a postinduction MRD level less than 0.01% was found to have an excellent outcome (only a single relapse after 5 years of follow-up). (2) Panzer-Grümayer et al21 quantified the D15 bone marrow MRD levels in 68 patients. Very similar to the present study, 14 patients (21%) had an MRD level of 0.01% or less and none of these have relapsed after a median follow-up of 66 months. However, these patients also were included in the BFM study13 and all patients with D15 MRD levels less than 0.01% also were below 0.01% after induction. Thus, the D15 MRD analysis did not improve the identification of patients with an excellent outcome. (3) In the small studies of Jacquy et al,22 Gruhn et al,23 and Dibenedetto et al,24 it similarly was shown that subgroups of patients could be identified in which no patients had relapse. However, either the number of patients studied was fewer than 30,23,24 the median time of follow-up was less than 222 or 3 years,24 only qualitative techniques were used,24 or only patients with B-lineage23or T-cell ALL were included.24 Thus, it is difficult to draw any general conclusions from these studies. Although T-cell leukemias have higher postinduction MRD levels compared to B-lineage ALL, large studies are needed to explore whether different cut-off points are needed for MRD-based therapy stratification for these subsets of patients.
The explanation for our superior identification of a large subgroup of patients with an excellent outcome (100% at 7 years) compared to most of the above-mentioned studies may partly reflect that we used a truly quantitative method to determine MRD and partly reflect a high antileukemic efficacy for the patients with D29 MRD less than 0.01% with the NOPHO ALL-92 protocol.1 An explanation for the relatively lower frequency of postremission patients with positive PCR results in the studies by Cave et al12 and van Dongen et al13 could partly reflect the PCR method applied and partly be based on their use of (1) a prednisolone prephase before induction therapy and (2) the inclusion of asparaginase during the first 4 weeks of treatment (ie, testing a 5-drug regimen compared to our 4-drug therapy during the first 4 weeks).
The relatively large number of excluded patient samples due to inadequate sampling is probably explained by the character of the present pilot study where MRD results were not revealed to the physicians, and thus would not lead to any treatment consequence for the individual patient. However, apart from the WBC count at diagnosis, no data including the relapse risk, indicated that the group of patients included in the study was selected in any way that could have biased the results.
The competitive PCR method is somewhat time consuming because an individual competitor needs to be constructed for each patient. However, because this can be constructed during the first 4 weeks of therapy, stratification of therapy based on quantification of D29 MRD by the competitive PCR method is clinically feasible. A faster, highly reproducible, and more convenient method with no post-PCR handling could be the real-time quantitative PCR technology (RQ-PCR).25 Still, using TCR and IGHgenes as clonal markers, it remains to be shown that this technique has a sensitivity as high as that of a nested PCR.25-27 Because (1) the present study indicates a cut-off limit of 0.01% to identify the patients with the best outcome, (2) this is the mode of patients in remission, and (3) this is within the detection limit of the RQ-PCR, the RQ-PCR could well be sufficiently fast, sensitive, and precise to be applied in large clinical trials. Depending on the demand for a very high sensitivity and precision, future studies will determine whether RQ-PCR is a useful and fast alternative to the competitive PCR or flow cytometric techniques. Because there were no significant differences between the MRD levels in samples that had been exposed to several days of shipment compared to samples with a mailing time of 1 day or less, the competitive PCR technique is feasible for centralized multicenter studies.
In the present study, the time points for detection of MRD were chosen to be D15 and D29 after start of induction therapy, because the goal was to identify a large cohort of patients who could be candidates for an early reduction of treatment intensity. Other studies have shown that for postinduction patients with positive PCR results later time points are useful to distinguish between patients with resistant disease compared to patients with a chemosensitive but slowly responding leukemia.13
The patients analyzed for MRD in the present study did not differ significantly in distribution of age, sex, phenotype, risk group distribution, and clinical outcome from the total group of patients diagnosed during the same period of time and treated according to NOPHO-92 protocol.1 Thus, it is likely that the cut-off limit (D29 MRD, 0.01%) found in the present study will be applicable for other patients treated by the NOPHO ALL-92 protocol. Whether the addition of other prognostic factors such as cytogenetic aberrations, in vitro sensitivity, and pharmacokinetics will change these critical cut-off limits is not known. Thus, we have recently shown that combining in vitro prednisolone sensitivity testing and D29 MRD monitoring may increase the cut-off limits for both these parameters to identify a subgroup of patients with an excellent outcome.28 To explore the individual and combined impact of such prognostic parameters, the NOPHO ALL-2000 protocol includes G-banding karyotyping as well as direct techniques to identify the t(12;21) translocation and high-risk cytogenetic aberrations, in vitro sensitivity testing, pharmacokinetic studies of asparaginase, methotrexate, and 6-mercaptopurine, and MRD quantification at treatment days 29, 50, and 106.
Neither WBC count nor immunophenotype had significant prognostic influence when the D29 MRD and age were introduced in the Cox model. Although age at diagnosis was significantly correlated to both D15 and D29 MRD, both D29 and age were included in the Cox model, which indicates that age has an impact on outcome that is separate from the early response to induction therapy. This could reflect disease-related prognostic factors related to age such as immunophenotype, chromosomal aberrations, and in vitro sensitivity, differences in drug disposition during later treatment phases, and patient compliance to oral chemotherapy.
There was no significant difference in the MRD levels between patients belonging to the SR, IR, and HR groups. This observation stresses that MRD could be an important supplement to be used together with the other known risk factors and may even replace other risk factors and thus offer a more simple and directly response-related treatment stratification of the patients. Hopefully, a consequence would be that in future protocols a higher fraction of patients could be treated with less intensive chemotherapy, thereby avoiding serious late effects. The present study as well as the BFM study13 indicate that nearly half of all children with ALL could be candidates for such less intensive treatment.
Based on the present and similar studies, patients on the NOPHO ALL-2000 non–high-risk protocol (non-T ALL with WBC count <50 × 109/L at diagnosis, no t(4;11), t(1;19), t(9;22), or hypodiploid ALL; approximately two thirds of all children with ALL) will receive identical therapy for the first 3 months and have their MRD quantified at D29, D50, and at D106. If the distribution of patients at these 2 time points confirms the data from the present and similar studies, the subsequent chemotherapy could for future patients be determined by the MRD levels.
We wish to thank Ewa Szojmer, Ingrid Alsing, Tina Hartvig, Line Brixen, Jannie Gregers, Michael Timm, and Kristine Nielsen for their excellent technical assistance. Furthermore we acknowledge the pediatric oncology departments in Denmark, Iceland, and Norway for sending bone marrow and blood samples for this study.
Supported in part by the Danish Medical Research Council (grant no. 9401011), the Biotechnological Centre for Cellular Communication, the Danish Cancer Society (grants no. 9410028, 9610007), the Kornerup Foundation, the Emil C. Hertz Foundation, the Children's Cancer Foundation, Sweden (grant no. 1996-073), and the Children's Cancer Foundation, Denmark.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kjeld Schmiegelow, Dept of Pediatrics, Section of Clinical Hematology and Oncology, Juliane Marie Center, University Hospital, Rigshospitalet, Blegdamsvej 9, DK-2100 Copenhagen, Denmark; e-mail: kschmiegelow@rh.dk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal