Abstract
CD150 signaling lymphocytic activation molecule (SLAM), a T/B/dendritic cell surface glycoprotein, is a costimulatory receptor involved in T-cell activation and is also a receptor for measles virus. CD150-induced signal transduction is controlled bySAP/SH2D1A, the gene that is aberrant in X-linked lymphoproliferative disease and familial hemophagocytic lymphohistiocytosis. This report shows that CD150 colocalizes with the T-cell receptor (TCR) following CD3 triggering in human peripheral blood T cells and is rapidly and reversibly tyrosine phosphorylated on TCR cross-linking. The Src-like kinases Lck and Fyn phosphorylate tyrosine residues in the cytoplasmic tail of CD150. The results demonstrate that the SAP protein has 2 modes of binding to CD150. Binding to the motif Thr-Ile-Tyr281Ala-Gln-Val occurs in a phosphotyrosine-independent fashion and to the motif Thr-Val-Tyr327Ala-Ser-Val in a phosphotyrosine-dependent manner. Within both SAP binding motifs the threonine residue at position −2 to tyrosine is essential to stabilize the interaction irrespective of tyrosine phosphorylation, a feature unique to the SAP SH2 domain. A leucine residue, Leu278, further stabilizes nonphospho binding of SAP to Tyr281 of CD150. SAP blocking of the tyrosine phosphatase SHP-2 occurs primarily on Tyr281 of CD150 because SHP-2 requires both Tyr281 and Tyr327 for binding to CD150, and SAP binds to nonphosphorylated Tyr281. CD150 exhibits lateral mobility, segregating into intercellular contacts. The lateral mobility and homophilic clustering of CD150 between neighboring cells is not dependent on SAP/CD150 interaction.
Introduction
CD150 signaling lymphocytic activation molecule (SLAM) is a cell surface glycoprotein of relative mass 70 kd found on activated/memory T cells, B cells, and dendritic cells.1,2It is a member of the immunoglobulin superfamily and shares homology with CD84, CD229, CD244, CD48, CD2, 19A, and Ly108.3 CD150 was recently shown to be the second receptor for measles virus in addition to CD46.4-7 CD150 is a self-ligand8,9with diverse immunologic functions including T/B-cell costimulation,1,10,11 induction of interferon γ (IFN-γ) in Th1 T-cell clones, redirection of Th2 clones to a Th1 or Th0 phenotype,12-14 and inhibition of apoptosis in B cells.15 Despite the importance of CD150 in these processes and its newly described function as the second measles virus receptor, little is known of the signaling properties of this receptor. CD150 on T cells binds to SAP/SH2D1A, the gene product that is absent or aberrant in X-linked lymphoproliferative disease16,17,18-20 and familial hemophagocytic lymphohistiocytosis.21 In B cells, CD150 binds to EAT-2, a molecule similar to SAP.22 Both molecules are small “floating” SH2 domains with short C-terminal tails. SAP blocks the binding to CD150 of the ubiquitous tyrosine phosphatase SHP-2 in T cells16 and has been shown to bind the guanine exchange factor p62dok1.23 The crystal structure of SAP in complex with a peptide of CD150 was recently solved.24This study revealed that SAP has novel requirements for binding to CD150 in a “3-pronged interaction” involving the central tyrosine, a valine residue at +3 to tyrosine (typically required for many SH2 domains), and a threonine residue at position −2 unique for SAP binding. The optimal motif suggested by studies with peptide libraries and fluorescence polarization is T (I/V) YxxV.24The cytoplasmic tail of CD150 has this optimal motif surrounding Tyr281 (TIYAQV; single-letter amino acid codes) and Tyr327 (TVYASV) in addition to a motif surrounding Tyr307, which lacks the valine at position +3 (TIYVAA).
Although crystal structure and biacore data have shed light on the mode of SAP binding to CD150, many fundamental questions remain to be addressed regarding CD150/SAP binding and the role of CD150/SAP binding in T-cell activation. It is unclear whether SAP binds to the motifs on CD150 surrounding Tyr307 and Tyr327 in T cells. It is also unclear which tyrosine kinases are able to phosphorylate CD150, how stringent the SAP binding motif requirement is for SAP/CD150 interaction in T cells, or the exact mode of binding of SHP-2 to CD150. Additionally, the requirement for CD150/SAP interaction in CD150 lateral mobility and homophilic interactions between cells is unknown. These questions are addressed in the present study.
Materials and methods
Cells and antibodies
COS-7 cells (American Type Culture Collection [ATCC], Rockville, MD) were grown in Dulbecco modified Eagle medium (DMEM; Gibco, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 2 mM glutamine, 1 mM sodium pyruvate, 100 IU/mL penicillin, and 100 g/mL streptomycin. Jurkat and Jurkat TAg T cells and EL4 mouse thymoma cells (ATCC) stably transfected with CD150 or CD8-SLAM4 chimeras were grown in RPMI supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 1 mM sodium pyruvate, 100 IU/mL penicillin, and 100 μg/mL streptomycin. Human peripheral blood mononuclear cells (PBMCs) were collected from healthy volunteers in the laboratory.
CD150 was immunoprecipitated using mouse monoclonal anti-hCD150 (2E7). Cell surface staining of CD150 was performed using mouse monoclonal anti-hCD150 (A12). Horseradish peroxidase (HRP)–conjugated antiphosphotyrosine monoclonal antibody cocktail (PY-7E1, PY-1B2, PY20) and HRP-conjugated streptavidin were from Zymed (San Francisco, CA). Rabbit polyclonal anti-SHP2, anti-Fyn, and anti-Lck antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-FLAG monoclonal M5 was obtained from Sigma Chemical (St Louis, MO). Anti–mouse IgG1, IgG2a antisera conjugated to Texas red or fluorescein isothiocyanate (FITC) were obtained from Southern Biotechnology (Birmingham, AL).
Polymerase chain reaction–mediated mutagenesis
A polymerase chain reaction (PCR) strategy was adopted to introduce point mutations into the cytoplasmic tail of CD150 (Figure 2contains the nomenclature of these constructs). Overlapping 5′ and 3′ PCR primers were designed that encoded the desired mutations. These were used to create 2 overlapping fragments of CD150 for each mutation constructed (Figure 2B). Overlapping PCR fragments were run on 1% agarose gels, cut from the gel, and purified using Qiagen gel purification columns (Qiagen, Valencia, CA). Fragments were then annealed to each other in a PCR and reamplified using 5′ and 3′ primers to make full-length CD150 with the required point mutations. Primers for full-length CD150, SAP-FLAG, and cytoplasmic CD150 for yeast 2-hybrid assays had restriction sites incorporated (EcoR1 5′BamH1 3′) to facilitate cloning into pCDNA3 (Invitrogen, Carlsbad, CA), pCMV-FLAG (Eastman Kodak, New Haven, CT), and pGAD424 (Clontech, Palo Alto, CA), respectively. For transient transfection in Jurkat TAg, CD150, Tyr13Phe, and Thr13Ala constructs were subcloned into the EcoRI and SalI sites of the mammalian expression vector pCI (Promega, Madison, WI).
The sequences of the PCR primers are listed in Table1.
PCR primers used in this study
| CD150 full-length 5′ | CGCGAATTCATGCAGTTGAGAAGAAGAGGTAA |
| CD150 cytoplasmic 5′ | CCCGAATTCCAGTTGAGAAGAAGAGGT |
| CD150 full length 3′ | CGCGGATCCTCAGCTCTCTGGAAGTGTCAC |
| CD150 Y281F 5′ | AAAAGCCTTACGATCTTTGCCCAAGTC |
| CD150 Y281F 3′ | TGGTTTCTGGACTTGGGCAAAGATCGTAAG |
| CD150 Y307F 5′ | GACCCTTGCACCACCATATTTGTTGCTGCC |
| CD150 Y307F 3′ | AGGCTCTGTGGCAGCAACAAATATGGTGG |
| CD150 Y327F 3′ | CCGCTCGAGTCAGCTCTCTGGAAGTGTCACACTAGCAAAGACTGT |
| CD150 T279A 5′ | GTGGAAAAAAAAAGCCTTGCGATCTATGCC |
| CD150 T279A 3′ | CTGGACTTGGGCATAGATCGCAAGGCTTTT |
| CD150 T305A 5′ | GCTCAGGACCCTTGCACCGCCATATATGTT |
| CD150 T305A 3′ | TGTGGCAGCAACATATATGGCGTGCAAGG |
| CD150 T325A 5′ | CAGGAAACAAATTCCATCGCAGTCTATGCT |
| CD150 T325A 3′ | TGTCACACTAGCATAGACTGCGATGGAATT |
| CD150 K276N 5′ | ACAACAGTGGAAAAAAATAGCCTTACGATC |
| CD150 K276N 3′ | GATCGTAAGGCTATTTTTTTCCACTGTTGT |
| CD150 L278I 5′ | GAAAAAAAAAGCATTACGATCTATGCCCAA |
| CD150 L278I 3′ | TTGGGCATAGATCGTAATGCTTTTTTTTTC |
| SAP-FLAG 5′ | CGGGAATTCAATGGACGCAGTGGCTGTGTA |
| SAP-FLAG 3′ | CGCGGATCCTCATGGGGCTTTCAGGCAGAC |
| CD150 full-length 5′ | CGCGAATTCATGCAGTTGAGAAGAAGAGGTAA |
| CD150 cytoplasmic 5′ | CCCGAATTCCAGTTGAGAAGAAGAGGT |
| CD150 full length 3′ | CGCGGATCCTCAGCTCTCTGGAAGTGTCAC |
| CD150 Y281F 5′ | AAAAGCCTTACGATCTTTGCCCAAGTC |
| CD150 Y281F 3′ | TGGTTTCTGGACTTGGGCAAAGATCGTAAG |
| CD150 Y307F 5′ | GACCCTTGCACCACCATATTTGTTGCTGCC |
| CD150 Y307F 3′ | AGGCTCTGTGGCAGCAACAAATATGGTGG |
| CD150 Y327F 3′ | CCGCTCGAGTCAGCTCTCTGGAAGTGTCACACTAGCAAAGACTGT |
| CD150 T279A 5′ | GTGGAAAAAAAAAGCCTTGCGATCTATGCC |
| CD150 T279A 3′ | CTGGACTTGGGCATAGATCGCAAGGCTTTT |
| CD150 T305A 5′ | GCTCAGGACCCTTGCACCGCCATATATGTT |
| CD150 T305A 3′ | TGTGGCAGCAACATATATGGCGTGCAAGG |
| CD150 T325A 5′ | CAGGAAACAAATTCCATCGCAGTCTATGCT |
| CD150 T325A 3′ | TGTCACACTAGCATAGACTGCGATGGAATT |
| CD150 K276N 5′ | ACAACAGTGGAAAAAAATAGCCTTACGATC |
| CD150 K276N 3′ | GATCGTAAGGCTATTTTTTTCCACTGTTGT |
| CD150 L278I 5′ | GAAAAAAAAAGCATTACGATCTATGCCCAA |
| CD150 L278I 3′ | TTGGGCATAGATCGTAATGCTTTTTTTTTC |
| SAP-FLAG 5′ | CGGGAATTCAATGGACGCAGTGGCTGTGTA |
| SAP-FLAG 3′ | CGCGGATCCTCATGGGGCTTTCAGGCAGAC |
Plasmid construction
For expression in COS-7 and Jurkat-TAg cells, CD150 or CD150 tail mutants were cloned into the multiple cloning sites of pCDNA3 (Invitrogen), pCI (Promega), or enhanced green fluorescent protein (pEGFP; Clontech), and sequenced to confirm their identity. CD150 mutants encompassing the 77-amino acid tail of CD150 were also cloned into the EcoR1 and BamH1 sites of the yeast Gal-4 activation domain vector pGAD424 (Clontech). Human SAP was cloned into the first multiple cloning site of the yeast Gal4 binding domain vector pBRIDGE (Clontech) and also into pCDNA3 and pCMV-FLAG.
Yeast 2-hybrid system and β-galactosidase assay
Human CD150 mutants were subcloned in the vector pGAD424; SAP complimentary DNA (cDNA) was cloned in the vector pBRIDGE (Clontech). Competent yeast cells (strain Y190) were cotransformed using the standard LiAc/TE/PEG transformation protocol and transformants were selected in media lacking tryptophan and leucine. Liquid culture assay using ONPG as the substrate was used to measure β-galactosidase activity (yeast protocols handbook, Clontech). For phosphotyrosine-dependent binding analysis an altered Fyn construct was inserted into the second multiple cloning site of pBRIDGE as previously described.25
Immunoprecipitation and Western blotting
COS-7 cells (107) were transfected by the dimethylaminoethyl (DEAE)–dextran method and biotinylated with sulfo-NHS-LC-Biotin (Pierce, Rockford, IL) as described previously.16 Lysis was carried out with 1% Triton X-100 as described before.16 Cell lysates were clarified by centrifugation at 14 000g for 15 minutes at 4°C and the crude lysate was precleared using 30 μL protein G-agarose beads (Gibco) and 3 μg pooled mouse IgG for 1 hour. Immunoprecipitations were carried out using 3 μg of the indicated antibody and 30 μL protein G-agarose beads for 3 hours at 4°C. Beads were then washed 3 times with decreasing concentrations of Triton X (1%, 0.1%, 0%). Crude lysates and immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride filters (Millipore, Bedford, MA). Filters were blocked for 1 hour with 5% skim milk (or 3% bovine serum albumin) and then probed with the indicated antibodies. Bound antibody was revealed using HRP-conjugated secondary antibodies using enhanced chemiluminescence (Supersignal, Pierce). For antiphosphotyrosine blotting we used a directly HRP-conjugated antibody cocktail (Pierce).
Immunofluorescent staining
Peripheral blood was collected from healthy volunteers and PBMCs were isolated by centrifugation over a Ficoll gradient (Amersham Pharmacia Biotech, Piscataway, NJ). PBMCs were activated for 24 hours to up-regulate cell surface CD150 expression (data not shown) in RPMI supplemented with 10% fetal calf serum (FCS) with phytohemagglutinin (PHA) at 5 μg/mL plus interleukin-2 (IL-2) at 100 U/mL. Cells were then washed twice in ice-cold phosphate-buffered saline (PBS) prior to CD150 cross-linking experiments. CD150 or CD3 were cross-linked using 10 μg anti-CD150 A12 (mouse IgG1) or anti-CD3 OKT3 (mouse IgG2a) respectively, incubated on ice for 20 minutes followed by addition of FITC-conjugated anti-IgG1 or IgG2a with incubation at 37°C for 10 minutes. Cross-linking was terminated with 3 washes of ice-cold PBS before the cells were mounted on coverslips for microscopy. In some experiments cells were permeabilized with methanol and nuclei were stained with 1 mM Hoescht dye. Immunofluorescence was visualized with a Nikon Optiphot II microscope connected to a SPOT (Diagnostic Instruments, Sterling Heights, MI) digital camera.
Transient transfection in Jurkat-TAg cells
Jurkat TAg cells (20 × 106) were transfected by electroporation using a Biorad Genepulser apparatus using the settings 250 V, 960 μF, 200 Ω, and 100 μg DNA resuspended in RPMI with 100 mM HEPES in 0.4 cm gap cuvettes.
Results
CD150 is rapidly and reversibly tyrosine phosphorylated following TCR cross-linking
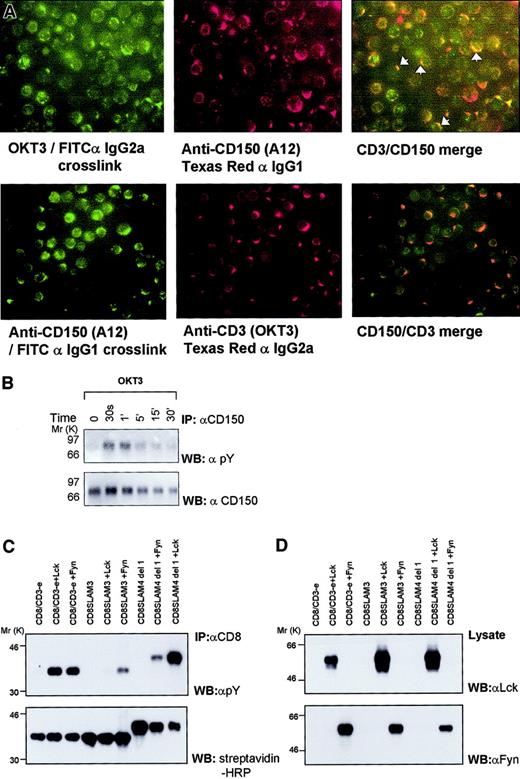
Because CD150 has previously been shown to have T-cell costimulation function1 in experiments using monoclonal antibodies, we investigated whether this effect could in part be due to an ability of CD150 to cluster in close proximity with the TCR at the T-cell plasma membrane. Human T cells were preactivated for 48 hours with PHA and IL-2 to induce high levels of surface CD150 expression (data not shown). After a “rest” period of 24 hours, anti-CD3 (OKT3) or anti-CD150 (A12) with an isotype-specific secondary antibody was used to cross-link one of the cell surface receptors. Next, the cells were stained for the second receptor (CD150 or CD3). Cross-linking of either CD150 or CD3 results in partial colocalization of both molecules on activated T cells; colocalization of CD3 and CD150 is more apparent when CD150 is cross-linked first (Figure1A, lower panel). Cross-recognition of the first antibodies by the second antibodies was unlikely because isotype-specific reagents were used (A12 is IgG1, OKT3 is IgG2a). Moreover, FITC-labeled CD150+ B cells did not label with Texas red anti-IgG2a. Thus, CD150 appears to cluster in close proximity to the TCR/CD3 complex on the T-cell plasma membrane. Antibody-induced lateral mobility of CD150 may be analogous to CD150 movement into the interface of 2 Jurkat T cells transfected with CD150 shown below.
CD150/TCR colocalization, CD3 triggering-mediated CD150 tyrosine phosphorylation, and identification of c Fyn/Lck phosphorylation sites in CD150.
(A) Cross-linking CD3 on normal human peripheral blood T cells with OKT3 results in colocalization of CD150 and CD3 on the T-cell plasma membrane. The top left panel shows FITC staining with capping of CD3 on PBMCs. Top middle panel is the same field as top left panel showing CD150 localization with Texas red following CD3 capping. Top right panel shows the top panels merged to reveal colocalization of CD150 and CD3 following CD3 cross-linking. White arrowheads highlight selected areas of colocalization. Lower right panel is FITC staining showing capping of CD150 on PBMCs following CD150 capping; bottom middle panel is same field as bottom left panel showing localization of CD3 following CD150 capping. Bottom right panel is the bottom panels merged to show colocalization of CD150 and CD3. (B) Cross-linking of CD3 in the human T-cell line Jurkat, stably transfected with CD150 results in rapid, reversible tyrosine phosphorylation of CD150; results are representative of 3 separate experiments. (C-D) Phosphorylation of chimeric CD8-truncated CD150 (CD8-SLAM4 del1, CD8-SLAM3) constructs by the Src kinases Fyn and Lck in COS-7 cells.
CD150/TCR colocalization, CD3 triggering-mediated CD150 tyrosine phosphorylation, and identification of c Fyn/Lck phosphorylation sites in CD150.
(A) Cross-linking CD3 on normal human peripheral blood T cells with OKT3 results in colocalization of CD150 and CD3 on the T-cell plasma membrane. The top left panel shows FITC staining with capping of CD3 on PBMCs. Top middle panel is the same field as top left panel showing CD150 localization with Texas red following CD3 capping. Top right panel shows the top panels merged to reveal colocalization of CD150 and CD3 following CD3 cross-linking. White arrowheads highlight selected areas of colocalization. Lower right panel is FITC staining showing capping of CD150 on PBMCs following CD150 capping; bottom middle panel is same field as bottom left panel showing localization of CD3 following CD150 capping. Bottom right panel is the bottom panels merged to show colocalization of CD150 and CD3. (B) Cross-linking of CD3 in the human T-cell line Jurkat, stably transfected with CD150 results in rapid, reversible tyrosine phosphorylation of CD150; results are representative of 3 separate experiments. (C-D) Phosphorylation of chimeric CD8-truncated CD150 (CD8-SLAM4 del1, CD8-SLAM3) constructs by the Src kinases Fyn and Lck in COS-7 cells.
Triggering of the TCR with antibodies directed at CD3 partially mimics activation induced by major histocompatibility complex-peptide, resulting in phosphorylation of CD3 ITAMS and recruitment and activation of a variety of tyrosine kinases including the src-like tyrosine kinases Lck and Fyn [AI11] (see Clements et al26 for review). Because of the inducible proximity of CD150 and TCR/CD3, we next studied the effect of anti-CD3 triggering on CD150 phosphorylation. To this end a Jurkat transfectant was used. Following anti-CD3 treatment of Jurkat T cells stably transfected with CD150, we indeed observed rapid transient tyrosine phosphorylation of CD150 (Figure 1B). The intensity of phosphorylation peaked between 30 seconds and 1 minute and returned to basal levels after 5 minutes. These data suggest that proximal tyrosine kinases activated by TCR stimulation also function to phosphorylate tyrosine residues in the cytoplasmic tail of CD150. Tyrosine phosphorylation of CD150 did not occur in response to cross-linking of CD150 alone indicating that CD150 capping, and concomitant TCR cocapping (Figure 1A), is insufficient in itself to activate CD150-phosphorylating kinases consistent with the role of CD150 as a costimulatory molecule.
All 3 tyrosines of CD150 (Tyr281, Tyr307, Tyr327) are phosphorylated by the src kinase Fyn
To identify possible TCR-proximal tyrosine kinases that might be responsible for the rapid phosphorylation of CD150 following CD3 cross-linking, we focused on the 2 prominent kinases in TCR signaling Fyn and Lck. We have previously shown that the src kinase Fyn is able to phosphorylate CD150 in COS-7 cells.16 Chimeric constructs (Figure 2D) encoding the ectodomain of CD8 fused to the truncated cytoplasmic tail of CD150 encompassing the first SAP motif and Tyr307 (CD8-SLAM4 del1) or the first SAP binding motif alone (CD8-SLAM3) were therefore coexpressed with Fyn in COS-7 cells. COS-7 cells were used to avoid interference by endogenous Fyn. Immunoprecipitation of CD8 followed by Western blotting against phosphotyrosine demonstrates that Fyn is able to phosphorylate both CD8-SLAM3 (which contains the first SAP motif, Tyr281 only) and CD8-SLAM4 del1 (which contains Tyr281 and Tyr307 but not Tyr327) indicating that it phosphorylates Tyr281 and Tyr307 (Figure 1C). Coexpression of Lck with the chimeric constructs results in phosphorylation of CD8-SLAM4 del1 but not CD8 SLAM3, thus indicating that Lck phosphorylates Tyr307 but not Tyr281 (Figure 1C). Because Fyn phosphorylates both CD8-SLAM3 and CD8-SLAM4 del1, which contain Tyr281 or Tyr281 plus Tyr307, respectively, we further mapped CD150 phosphorylation using a series of CD150 constructs incorporating tyrosine-phenylalanine mutations (Figure 2B shows the nomenclature of these constructs). It is clear that Fyn is able to phosphorylate CD150 at position Tyr281 and Tyr327, but phosphorylates Tyr307 to a slightly lesser extent (Figure3A, second panel).
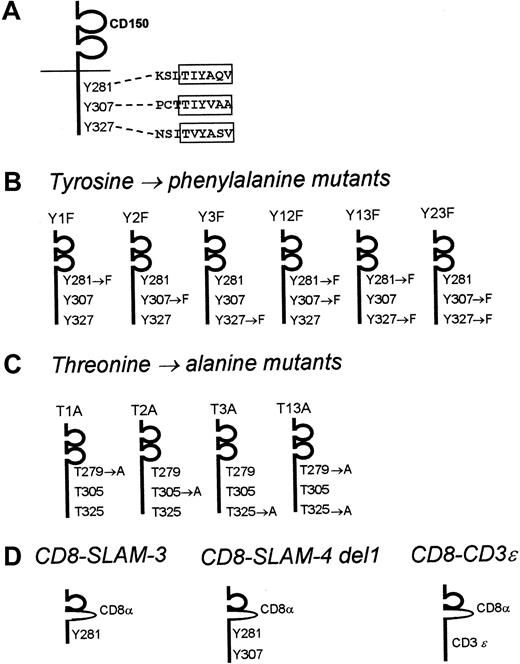
CD150 mutation/deletion constructs used in this study.
(A) CD150. The 3 SAP consensus sequences surrounding Tyr281, Tyr307, and Tyr327 are indicated. (B) CD150 point mutants, containing tyrosine-phenylalanine substitutions in the SAP-binding motifs. (C) CD150 point mutants, containing threonine-alanine substitutions at position −2 to the central tyrosine in the SAP binding motif. (D) Chimeric CD8-CD150/CD3ε constructs encoding the ectodomain of CD8 and the cytoplasmic and membrane domains of CD150.
CD150 mutation/deletion constructs used in this study.
(A) CD150. The 3 SAP consensus sequences surrounding Tyr281, Tyr307, and Tyr327 are indicated. (B) CD150 point mutants, containing tyrosine-phenylalanine substitutions in the SAP-binding motifs. (C) CD150 point mutants, containing threonine-alanine substitutions at position −2 to the central tyrosine in the SAP binding motif. (D) Chimeric CD8-CD150/CD3ε constructs encoding the ectodomain of CD8 and the cytoplasmic and membrane domains of CD150.
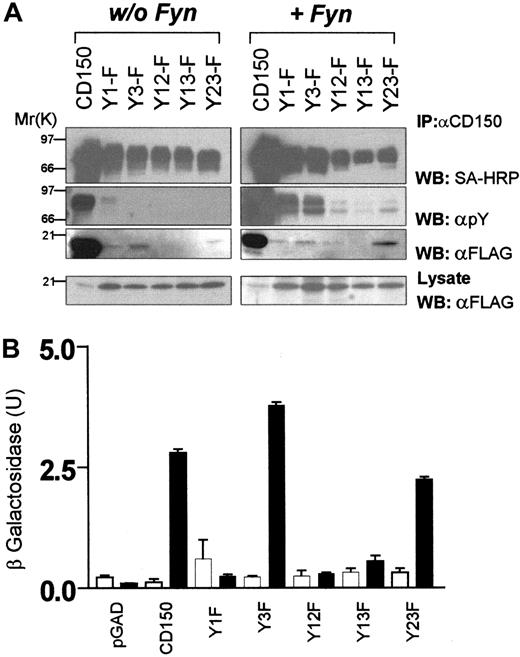
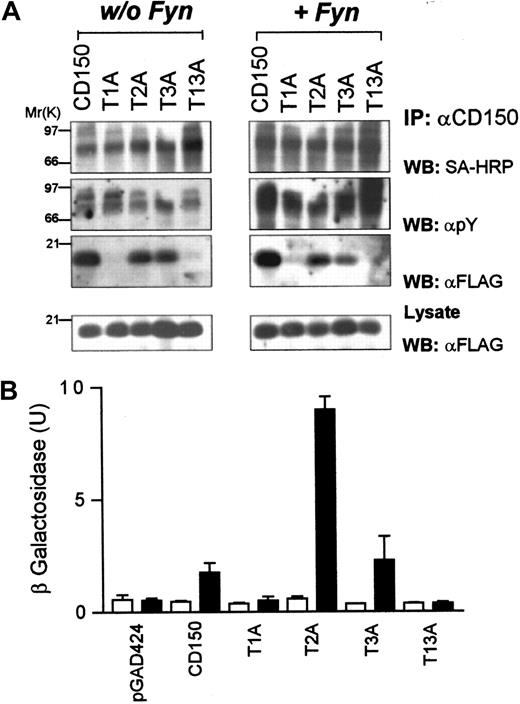
Identification of phosphotyrosine-dependent and -independent SAP binding motifs in CD150.
(A) COS-7 cells transiently transfected with CD150, or CD150 constructs with SAP motif tyrosine mutations (Figure 2 lists nomenclature) plus SAP-FLAG with or without Fyn coexpression. CD150 immunoprecipitation followed by antiphosphotyrosine, streptavidin-HRP, and anti-FLAG Western blot. (B) Yeast 2-hybrid analysis of SAP binding to non–tyrosine-phosphorylated CD150 or CD150 with tyrosine-phenylalanine mutations. ▪, +SAP; ■, +pBridge. Quantitation of β-galactosidase activity was performed as described in “Materials and methods.”
Identification of phosphotyrosine-dependent and -independent SAP binding motifs in CD150.
(A) COS-7 cells transiently transfected with CD150, or CD150 constructs with SAP motif tyrosine mutations (Figure 2 lists nomenclature) plus SAP-FLAG with or without Fyn coexpression. CD150 immunoprecipitation followed by antiphosphotyrosine, streptavidin-HRP, and anti-FLAG Western blot. (B) Yeast 2-hybrid analysis of SAP binding to non–tyrosine-phosphorylated CD150 or CD150 with tyrosine-phenylalanine mutations. ▪, +SAP; ■, +pBridge. Quantitation of β-galactosidase activity was performed as described in “Materials and methods.”
SAP binds to amino acid residues on CD150 embedding Tyr281 or Tyr327
SAP binding to the membrane-proximal Tyr281 motif of CD150 has been described previously.16 As shown in Figure 2A, CD150 also has a perfect SAP binding motif surrounding Tyr327, but it is unclear whether SAP can indeed bind to this motif in a physiologic manner. We decided to use a point mutational mapping approach to address this question. CD150 constructs encoding various combinations of tyrosine-phenylalanine mutations in the cytoplasmic tail were generated for expression in mammalian cells (Figure 2B has details).
Coexpression of CD150 with SAP in COS-7 cells leads to constitutive weak tyrosine phosphorylation of CD150 on all 3 SAP binding sites (Figure 3A, left panel). A large increase of phosphorylation of each construct was observed following cotransfection of Fyn. Predictably the single tyrosine mutants (Tyr1Phe, Tyr3Phe) were phosphorylated to a greater extent than the double mutants (Tyr12Phe, Tyr13Phe, Tyr23Phe; Figure 3A, right panel). Following immunoprecipitation of CD150, coimmunoprecipitation of SAP was observed on all the mutants except Tyr13Phe, which has Tyr281 and Tyr327 mutated to phenylalanine. This result demonstrates that SAP binds to Tyr281 and, following phosphorylation, Tyr327. SAP cannot bind to Tyr307 under these conditions.
Because expression of CD150 and SAP in COS-7 cells leads to constitutive weak tyrosine phosphorylation, dissection of the phosphotyrosine-independent binding of SAP to CD150 was performed using the yeast 2-hybrid system. Yeast cells cannot accomplish tyrosine phosphorylation unless they are transfected with tyrosine-kinase DNA. They therefore provide a suitable expression system for nonphosphotyrosine mapping of the SAP/CD150 interaction.25CD150 Gal-4 activation domain and SAP Gal-4 binding domain chimeric constructs (see “Materials and methods”) were transformed into the yeast strain Y190. SAP/CD150 interaction was then measured with a liquid β-galactosidase reporter assay. Mutation of Tyr281 in CD150 leads to total abrogation of SAP binding in the yeast 2-hybrid system (Figure 3B, “Tyr1Phe”). Mutation of Tyr327 (Tyr3Phe) or Tyr307 plus Tyr327 (Tyr23Phe) has little effect on nonphospho binding of SAP to CD150 (Figure 3B) showing that in the absence of Src-like kinase-mediated phosphorylation, CD150 binds SAP only via Tyr281. In conclusion, the yeast experiments confirm the data obtained in COS-7 cells: SAP binds Tyr281 independently of phosphorylation, does not bind Tyr307, and binds Tyr327 only after phosphorylation.
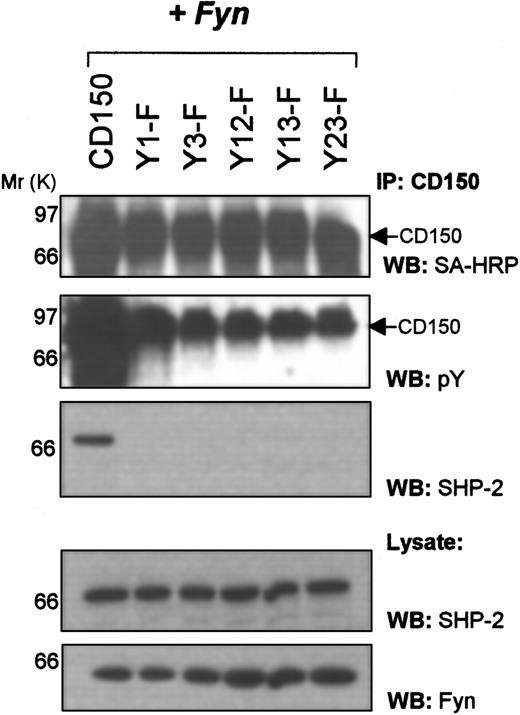
SHP-2 binding to CD150 involves both SAP binding consensus sites at Tyr281 and Tyr327
One major proposed function of the SH2D1A gene product SAP is that of blocking recruitment of other signal transduction molecules to CD150 and related molecules (CD84, CD229, CD244).22 One such signaling molecule, SHP-2 (a ubiquitous tyrosine phosphatase with tandem SH2 domains), is known to bind CD150, which event can be blocked by SAP.16 Because SAP can bind to the motifs surrounding Tyr281 and Tyr327 of CD150 (Figure 3), we further investigated the binding requirements for SHP-2 to CD150. SHP-2 exhibits a requirement for both tyrosines, Tyr281 and Tyr327, to bind CD150 (Figure 4). Mutation of Tyr281 (Tyr1Phe) or Tyr327 (Tyr3Phe) alone or in combination results in abrogation of SHP-2 binding (Figure 4, third panel). This result suggests that SAP binding to nonphosphorylated Tyr281 alone would be sufficient to block the SHP-2/CD150 interaction.
The tyrosine phosphatase SHP-2 requires both Tyr281 and Tyr327 of CD150 to interact.
COS-7 cells transfected with the indicated CD150 constructs andfyn were surface biotinylated and immunoprecipitated with anti-CD150 followed by Western blotting with the indicated antibodies.
The tyrosine phosphatase SHP-2 requires both Tyr281 and Tyr327 of CD150 to interact.
COS-7 cells transfected with the indicated CD150 constructs andfyn were surface biotinylated and immunoprecipitated with anti-CD150 followed by Western blotting with the indicated antibodies.
The threonine residue at position −2 to Tyr281 and Tyr327 is critical for the SAP/CD150 interaction
Amino acid residues N-terminal to the central tyrosine of the SAP binding motifs could function to stabilize the SAP/CD150 complex in the non–tyrosine-phosphorylated state.24 27 Indeed, the crystal structure of SAP in complex with a peptide of CD150 encompassing the first SAP motif shows that Thr279 hydrogen bonds with Glu17 and a buried water molecule of SAP. Here we show that the threonine residue at position −2 is in fact critical for SAP binding independently of tyrosine phosphorylation on the motifs surrounding Tyr281 and Tyr327. Introduction of Thr>Ala point mutations in the SAP motifs completely abrogates binding to CD150, expressed in COS-7 cells (Figure 5A). The majority of SAP binding occurs on motif number 1 although in the presence offyn some SAP binding is observed at motif 3 (Figure 5A, right panel, Thr1Ala mutation). In the absence of fyn the CD150 constructs are constitutively tyrosine phosphorylated at low levels (Figure 5A, left and right panels). To measure SAP binding to nonphosphorylated CD150 incorporating threonine mutations we used the yeast 2-hybrid system (Figure 5B). Mutation of the threonine residue in either the first SAP motif (Thr1Ala), or the first and third motif in combination (Thr13Ala) completely abrogates SAP binding (Figure 5B), whereas mutation of Thr305 (Thr2Ala) or Thr325 (Thr3Ala) alone results in strong β-galactosidase activity (Figure 5B). These results show that without tyrosine phosphorylation, SAP binding to Tyr281 is dependent on the threonine residue at position −2. This confirms the observations in COS-7 cells, that following tyrosine phosphorylation byfyn the threonine residue stabilizes binding of SAP to motifs surrounding TyrY281 and Tyr327. Thus, the threonine in the SAP motif T(I/V)YxxV is essential for binding CD150 at each motif.
SAP requires a threonine residue at position −2 in the consensus motif T(I/V)pYxx(V/I) to stabilize both phosphotyrosine- and non–phosphotyrosine-dependent binding to CD150.
(A) COS-7 cells transiently transfected with CD150 or CD150 with the indicated threonine-alanine substitutions plus SAP-FLAG with or without Fyn coexpression. CD150 was immunoprecipitated followed by Western blotting with the indicated antibodies. (B) Yeast 2-hybrid analysis of SAP binding to non–tyrosine-phosphorylated CD150 T-A constructs. ▪, +SAP; ■, +pBridge. Quantitation of β-galactosidase activity was performed as described in “Materials and methods.”
SAP requires a threonine residue at position −2 in the consensus motif T(I/V)pYxx(V/I) to stabilize both phosphotyrosine- and non–phosphotyrosine-dependent binding to CD150.
(A) COS-7 cells transiently transfected with CD150 or CD150 with the indicated threonine-alanine substitutions plus SAP-FLAG with or without Fyn coexpression. CD150 was immunoprecipitated followed by Western blotting with the indicated antibodies. (B) Yeast 2-hybrid analysis of SAP binding to non–tyrosine-phosphorylated CD150 T-A constructs. ▪, +SAP; ■, +pBridge. Quantitation of β-galactosidase activity was performed as described in “Materials and methods.”
The nonphosphotyrosine interaction of SAP with Tyr281 is partially stabilized by Leu278 of CD150
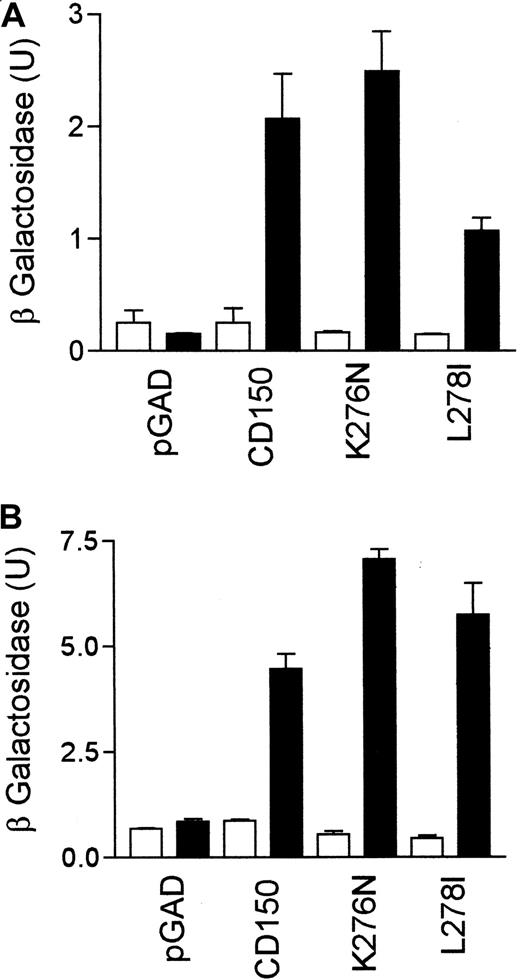
The presence of a threonine residue at position −2 to the central tyrosine in the SAP binding motif of CD150 is critical for stabilizing the interaction in the presence or absence of tyrosine phosphorylation. However, this does not explain the unique affinity of SAP for nonphosphorylated Tyr281. For example, Tyr327 of CD150 is preceded by a threonine residue Thr325, but SAP does not bind CD150 at this position in the absence of tyrosine phosphorylation (Figures 3 and 4).We sought to identify candidate amino acid residues N-terminal to the Try281 SAP motif that might contribute to the nonphospho binding. Amino acid residues surrounding the Tyr281 motif of human and mouse CD150 were compared with those surrounding all the other SAP motifs of the human and mouse CD150 family members (Table 2). CD84, CD229, and CD244 are known to require tyrosine phosphorylation for SAP binding.25 28 The plasma membrane–proximal SAP consensus regions of human, mouse, and monkey CD150 are unique in having a lysine residue at position −5, and a leucine residue at position −3 to the central tyrosine (Table 2, residues denoted with an asterisk, and Figure 6). Lys276 and Leu278 were mutated to the corresponding residues found at the third SAP consensus motif N-terminal to Tyr327, which binds SAP only following tyrosine phosphorylation. Yeast 2-hybrid analysis was performed with CD150 mutants encoding point mutations of Lys276Asn and Leu278Ile (Figure 7). Mutation of Lys276Asn had no effect on the ability of SAP to bind nonphosphorylated CD150 (Figure 7A). In contrast mutation of Leu278Ile resulted in a marked reduction in nonphosphotyrosine binding of SAP to CD150 (Figure7A). On introduction of fyn into the yeast this reduction in binding was reversed (Figure 7B), showing that Leu278 contributes to the phosphotyrosine-independent binding of SAP with CD150. Therefore binding of SAP to CD150 is dependent on a number of residues N-terminal and C-terminal to the central tyrosine in the motifs (Table 2 and Figure 6). The unique blocking function of SAP binding to nonphosphorylated motif number 1 is partially explained by a stabilizing effect of Leu278.
Amino acid comparison of the SAP motifs in SLAM-related receptors
| Receptor motif number . | Amino acid position . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| − 5 . | − 4 . | − 3 . | − 2 . | − 1 . | 0 . | + 1 . | + 2 . | + 3 . | |
| hSLAM-1 | K* | S | L* | T | I | Y | A | Q | V |
| hSLAM-2 | P | C | T | T | I | Y | V | A | A |
| hSLAM-3 | N | S | I | T | V | Y | A | S | V |
| mSLAM-1 | K* | S | L* | T | I | Y | A | Q | V |
| mSLAM-2 | P | C | T | T | I | Y | V | A | A |
| mSLAM-3 | N | P | T | T | V | Y | A | S | V |
| hLy9-1 | D | P | V | T | P | Y | V | T | E |
| hLy9-2 | G | E | N | T | V | Y | A | Q | V |
| hLy9-3 | S | S | A | T | I | Y | C | S | I |
| mLy9-1 | E | A | I | T | P | Y | D | K | V |
| mLy9-2 | D | S | N | T | I | Y | C | S | V |
| hCD84 | P | V | N | T | V | Y | S | E | V |
| mCD84-1 | S | K | K | T | V | Y | A | V | V |
| mCD84-2 | P | V | T | T | I | Y | S | S | V |
| h2B4-1 | G | G | S | T | I | Y | S | M | I |
| h2B4-2 | P | A | Y | T | L | Y | S | L | I |
| h2B4-3 | F | N | S | T | I | Y | E | V | I |
| m2B4-1 | D | R | G | T | M | Y | S | M | I |
| m2B4-2 | E | K | C | T | V | Y | S | V | V |
| m2B4-3 | L | S | C | T | V | Y | E | E | V |
| Ly108-1 | P | G | N | T | V | Y | A | Q | V |
| Ly108-2 | D | S | M | T | I | Y | S | I | V |
| Receptor motif number . | Amino acid position . | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| − 5 . | − 4 . | − 3 . | − 2 . | − 1 . | 0 . | + 1 . | + 2 . | + 3 . | |
| hSLAM-1 | K* | S | L* | T | I | Y | A | Q | V |
| hSLAM-2 | P | C | T | T | I | Y | V | A | A |
| hSLAM-3 | N | S | I | T | V | Y | A | S | V |
| mSLAM-1 | K* | S | L* | T | I | Y | A | Q | V |
| mSLAM-2 | P | C | T | T | I | Y | V | A | A |
| mSLAM-3 | N | P | T | T | V | Y | A | S | V |
| hLy9-1 | D | P | V | T | P | Y | V | T | E |
| hLy9-2 | G | E | N | T | V | Y | A | Q | V |
| hLy9-3 | S | S | A | T | I | Y | C | S | I |
| mLy9-1 | E | A | I | T | P | Y | D | K | V |
| mLy9-2 | D | S | N | T | I | Y | C | S | V |
| hCD84 | P | V | N | T | V | Y | S | E | V |
| mCD84-1 | S | K | K | T | V | Y | A | V | V |
| mCD84-2 | P | V | T | T | I | Y | S | S | V |
| h2B4-1 | G | G | S | T | I | Y | S | M | I |
| h2B4-2 | P | A | Y | T | L | Y | S | L | I |
| h2B4-3 | F | N | S | T | I | Y | E | V | I |
| m2B4-1 | D | R | G | T | M | Y | S | M | I |
| m2B4-2 | E | K | C | T | V | Y | S | V | V |
| m2B4-3 | L | S | C | T | V | Y | E | E | V |
| Ly108-1 | P | G | N | T | V | Y | A | Q | V |
| Ly108-2 | D | S | M | T | I | Y | S | I | V |
Residues unique to the plasma membrane–proximal SAP consensus regions in SLAM.
Diagrammatic representation of critical amino acid residues in the cytoplasmic tail of CD150 and alignment with mouse and monkey CD150 sequences.
The primary SAP binding motif is boxed. (1) Leu278 partially stabilizes the nontyrosine phosphorylation-dependent SAP/CD150 interaction. (2) Thr279 is critical in the SAP motif for phospho- and non–phosphotyrosine-dependent binding. (3) Tyr281 is phosphorylated byfyn and lck and binds SAP and SHP-2. It is essential for SHP-2 binding. (4) Val284 interacts with the hydrophobic SH2 domain pocket of SAP.24 (5) Tyr307 is phosphorylated by fyn and lck but does not interact with SAP. (6) Thr325 stabilizes phosphotyrosine-dependent binding of SAP to the third motif in CD150. (7) Tyr327 is phosphorylated by fyn,and weakly binds SAP. (8) Val330 interacts with the hydrophobic SH2 domain pocket of SAP.24
Diagrammatic representation of critical amino acid residues in the cytoplasmic tail of CD150 and alignment with mouse and monkey CD150 sequences.
The primary SAP binding motif is boxed. (1) Leu278 partially stabilizes the nontyrosine phosphorylation-dependent SAP/CD150 interaction. (2) Thr279 is critical in the SAP motif for phospho- and non–phosphotyrosine-dependent binding. (3) Tyr281 is phosphorylated byfyn and lck and binds SAP and SHP-2. It is essential for SHP-2 binding. (4) Val284 interacts with the hydrophobic SH2 domain pocket of SAP.24 (5) Tyr307 is phosphorylated by fyn and lck but does not interact with SAP. (6) Thr325 stabilizes phosphotyrosine-dependent binding of SAP to the third motif in CD150. (7) Tyr327 is phosphorylated by fyn,and weakly binds SAP. (8) Val330 interacts with the hydrophobic SH2 domain pocket of SAP.24
Leu278 at position −3 in the first SAP binding motif of CD150 partially stabilizes non–phosphotyrosine binding but is not required for phosphotyrosine-dependent interaction.
(A) Yeast 2-hybrid β-galactosidase assay for SAP/CD150 non–tyrosine phosphorylation–dependent interaction with CD150 or CD150 Lys276Asn or CD150 Leu278Ile. ▪, +SAP; ■, +pGBT9. (B) Altered yeast 2-hybrid using a modified Fyn (Fyn2YF R-Q, see “Materials and methods” for details) to phosphorylate CD150. β-Galactosidase assay to measure strength of the SAP/CD150 interaction data are representative of 3 separate experiments. ▪, +SAP + Gyn2YF R-Q; ■, +pBridge.
Leu278 at position −3 in the first SAP binding motif of CD150 partially stabilizes non–phosphotyrosine binding but is not required for phosphotyrosine-dependent interaction.
(A) Yeast 2-hybrid β-galactosidase assay for SAP/CD150 non–tyrosine phosphorylation–dependent interaction with CD150 or CD150 Lys276Asn or CD150 Leu278Ile. ▪, +SAP; ■, +pGBT9. (B) Altered yeast 2-hybrid using a modified Fyn (Fyn2YF R-Q, see “Materials and methods” for details) to phosphorylate CD150. β-Galactosidase assay to measure strength of the SAP/CD150 interaction data are representative of 3 separate experiments. ▪, +SAP + Gyn2YF R-Q; ■, +pBridge.
CD150 exhibits lateral mobility in Jurkat T cells, forming adhesion plaques between neighboring cells; SAP is recruited to these contacts, but SAP-CD150 interaction is not required for their formation
CD150 is proposed to function as a T-cell costimulatory receptor and we have observed that CD150 and CD3 colocalize on CD150 or CD3 triggering. We asked whether one mechanism of CD150 costimulation might be to facilitate the formation of homophilic adhesion contacts between adjacent cells.
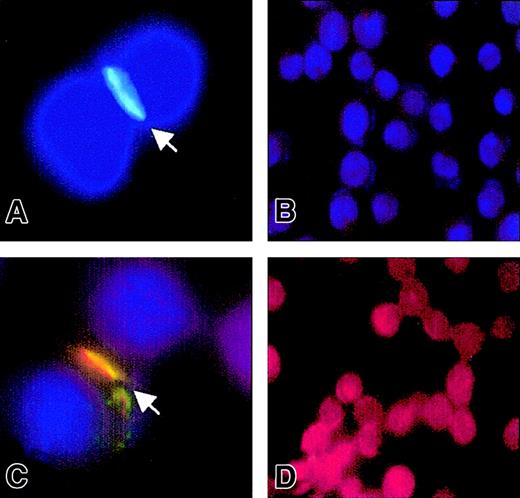
As a first approach to investigating the ability of CD150 to migrate to cell contacts, we used Jurkat cells transiently transfected with CD150-EGFP. The use of the EGFP tag on the C-terminus of CD150 allows visualization of its location either within the cell or on the plasma membrane with fluorescence microscopy. Following transfection of Jurkat-TAg T cells with CD150-EGFP we observed aggregation of CD150 at the plasma membrane contacts between neighboring cells (Figure8A). Clustering of CD150 at the cellular junctions results in recruitment of SAP to the same region (Figure 8C) as visualized with an anti–SAP-Cy3 monoclonal antibody. We conclude that CD150 has a lateral mobility in the plasma membrane and forms adhesion contacts between cells, presumably due to homophilic interaction. Within these adhesion contacts there is a marked accumulation of SAP.
CD150 displays lateral mobility and spontaneously segregates into intercellular contacts with SAP.
Jurkat-Tag cells were transiently transfected with CD150-EGFP. After 24 hours the cells were fixed in methanol and stained with anti–SAP-Cy3 or Hoescht or both to visualize the nuclei. (A) Jurkat-Tag transfected with CD150-EGFP, counterstained with Hoescht. Arrow points to cell-cell contact rich in CD150. (B) Jurkat-Tag untransfected. (C) SAP colocalizes at the cell interface with CD150. Jurkat-Tag transfected with CD150-EGFP, stained with anti–SAP-Cy3 and Hoescht. Arrow points to cell interface. (D) Jurkat-Tag untransfected, stained with anti–SAP-Cy3. In the absence of CD150, SAP is expressed homogeneously throughout the cytoplasm.
CD150 displays lateral mobility and spontaneously segregates into intercellular contacts with SAP.
Jurkat-Tag cells were transiently transfected with CD150-EGFP. After 24 hours the cells were fixed in methanol and stained with anti–SAP-Cy3 or Hoescht or both to visualize the nuclei. (A) Jurkat-Tag transfected with CD150-EGFP, counterstained with Hoescht. Arrow points to cell-cell contact rich in CD150. (B) Jurkat-Tag untransfected. (C) SAP colocalizes at the cell interface with CD150. Jurkat-Tag transfected with CD150-EGFP, stained with anti–SAP-Cy3 and Hoescht. Arrow points to cell interface. (D) Jurkat-Tag untransfected, stained with anti–SAP-Cy3. In the absence of CD150, SAP is expressed homogeneously throughout the cytoplasm.
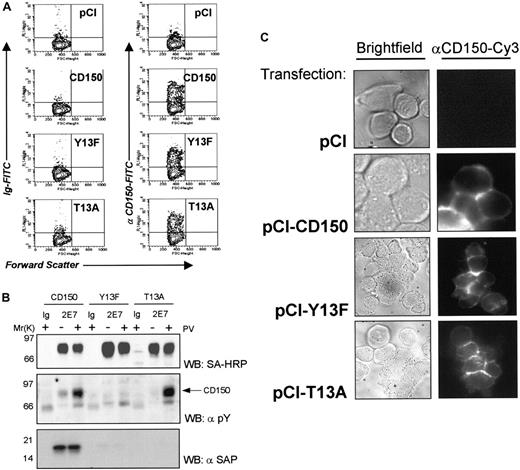
We next asked whether SAP/CD150 interaction was necessary for the formation of the CD150 adhesion contacts. We transfected Jurkat-TAg T cells with CD150-Tyr13Phe and Thr13Ala, both of which are impaired in their ability to bind SAP, and used fluorescence microscopy to visualize the cellular location of CD150. On transient transfection, 30% to 45% cell surface expression of CD150 was obtained (Figure 9A). SAP was able to bind only to the intact CD150 construct (Figure 9B) as predicted from the experiments in COS-7 cells and yeast. SAP binding to the mutant CD150 constructs Tyr13Phe and Thr13Ala was undetectable in the presence or absence of tyrosine phosphorylation of the CD150 tail, induced with 1 mM sodium pervanadate treatment (Figure 9B). Cell clusters of Jurkat-TAg expressing these CD150 constructs show clear CD150 concentration on the plasma membrane at the interface between neighboring CD150+ cells, with no differences between wild-type CD150 or mutated CD150 (Figure 9C). This result demonstrates that CD150 lateral mobility and formation of intercellular contacts on T cells is not dependent on SAP binding.
Segregation of CD150 into cell-cell contacts is not dependent on SAP binding.
(A) Jurkat T cells stably transfected with SV40 large T antigen (Jurkat-TAg) were transiently transfected with CD150 or the 2 non–SAP-binding mutants Tyr13Phe or Thr13Ala. Expression levels were consistently 30% to 45%. (B) SAP binds to CD150, but not Tyr13Phe or Thr13Ala mutants in Jurkat TAg cells. (C) CD150 is concentrated on the plasma membrane at the cell-cell contact of adjacent Jurkat-TAg cells transfected with CD150 or Thr>Ala/Tyr>Phe mutants.
Segregation of CD150 into cell-cell contacts is not dependent on SAP binding.
(A) Jurkat T cells stably transfected with SV40 large T antigen (Jurkat-TAg) were transiently transfected with CD150 or the 2 non–SAP-binding mutants Tyr13Phe or Thr13Ala. Expression levels were consistently 30% to 45%. (B) SAP binds to CD150, but not Tyr13Phe or Thr13Ala mutants in Jurkat TAg cells. (C) CD150 is concentrated on the plasma membrane at the cell-cell contact of adjacent Jurkat-TAg cells transfected with CD150 or Thr>Ala/Tyr>Phe mutants.
Discussion
CD150 was the first member of a growing family of lymphocyte cell surface receptors (CD84, CD229, CD244) that have been shown to bind to the signaling molecule SAP. This receptor/signaling molecule pair has recently acquired increased significance with the discovery that CD150 is the most commonly used receptor for measles virus entry into lymphocytes. Additionally, SAP is mutated in 3 immune diseases: X-linked lymphoproliferative disease, combined variable immunodeficiency disease, and familial hemophagocytic lymphohistiocytosis.
CD150 has been reported to function as a costimulatory receptor on activated T cells, although the mechanism for this is unknown. In this study, we observed close physical proximity of the TCR with CD150 following cross-linking of either molecule. In addition, following triggering of CD3, CD150 was rapidly and transiently tyrosine phosphorylated. These results are consistent with a major signaling function of CD150 during costimulation. It is likely that tyrosine phosphorylation of CD150 in the immune synapse functions to recruit SH2 domain-containing signaling molecules, including SAP to the supramolecular activation cluster (SMAC). Indeed, in preliminary experiments we observe CD150 in the “immune synapse” formed between antigen-specific cytotoxic lymphocyte (CTL) clones and B cells pulsed with their cognate antigen (data not shown). CD150 is unique among its homologues in the immunoglobulin superfamily in that it is able to bind SAP, a floating SH2 domain, in the absence of tyrosine phosphorylation. In this study, using a detailed mutagenesis mapping approach we have shown that SAP binding to CD150 is in fact bimodal. Prior to tyrosine phosphorylation, SAP binds the membrane-proximal motif surrounding Tyr281. Following tyrosine phosphorylation by tyrosine kinases such as Fyn, SAP binds additionally to the distal motif surrounding Tyr327. SAP has been proposed to have 2 nonmutually exclusive functions. First, it functions to block recruitment to CD150 of SH2 domain-containing signaling molecules, the prototypic example being SHP-2.16 Second, it has been demonstrated to act as an adapter molecule, binding the guanine exchange factor p62dok1.23 It is reasonable to predict that immediately following TCR engagement, and subsequent CD150 phosphorylation, SAP is recruited to both SAP motifs at Tyr281 and Tyr327, to block or adapt at these positions.
Physicochemical analyses of SAP binding to peptides spanning Tyr281 of CD150 have suggested that amino acid residues N-terminal to the central tyrosine of the SAP binding site strengthen the interaction.24 27 Our findings extend these observations to binding analysis in cells. The threonine residue at position −2 serves to stabilize the binding of SAP to CD150 in both modes (phospho/nonphospho) and on both SAP motifs. We investigated the mechanism of nonphospho binding to the first tyrosine by mutational analysis in the yeast 2-hybrid system. The Leu278 residue at position −3 contributes to nonphosphotyrosine binding but is not sufficient alone to explain the affinity of SAP to this region.
A major proposed function of adhesion molecule pairs such as leukocyte function-associated antigen 1 (LFA-1; CD11a/CD18)/intercellular adhesion molecule (ICAM-1) is to strengthen the interaction between T cells and antigen-presenting cells (APCs). Additionally, these interactions help to sequester TCR into the immune synapse. Here we demonstrate that CD150 has lateral mobility in the plasma membrane and readily clusters at the interface between 2 transfected cells. SAP is recruited into these CD150-rich regions, although SAP/CD150 interaction is not required for their formation. It is therefore likely that an important function of CD150 is to increase the affinity of memory T cells (which are CD150+) with APCs during the anamnestic response. In support of this concept, professional APCs (dendritic cells and activated macrophages) are highly positive for CD150 surface expression.
The authors would like to thank Drs Bruce and Barbara Furie for the use of their microscopy equipment, and Glenn Merrill-Skoloff for his expert assistance with digital microscopy. The authors are also indebted to Dr Charles Gullo, Dr Massimo Morra, and Dr William Faubion for critical reading of the manuscript and valuable discussions.
Supported by a grant from the National Foundation March of Dimes. D.H. is supported by a fellowship from the Leukemia and Lymphoma Society. M.S. is supported by a fellowship from Ministerio de Educacion y Ciencia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Duncan Howie, Division of Immunology, RE-204, Beth Israel Deaconess Medical Center, Harvard Medical School, 330 Brookline Ave, Boston, MA 02215; e-mail:dhowie@caregroup.harvard.edu or terhorst@caregroup.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal