MLL (also known as ALL1, HTRX, or HRX) gene translocations are among the most common chromosomal abnormalities in both B-lineage acute lymphoblastic leukemia (B-ALL) and acute myeloid leukemia (AML). This MLLinvolvement means that these leukemias constitute a distinct disease1 with a particularly poor prognosis, which needs to be more easily identified and better characterized if therapeutic regimens are to be improved. MLL gene rearrangements have been sporadically reported in acute lymphoblastic T-cell leukemia (T-ALL),2 3 but the incidence of MLLinvolvement in T-ALL could be consistently underestimated becauseMLL rearrangements would seem to be due to translocations that are difficult to detect by classic cytogenetic methods. Studying the MLL gene could lead to the identification of a new group of T-ALL, especially if this group also turned out to have a poor clinical outcome. Here we present evidence that the MLL gene could be in fact involved in more than 8% of adult T-ALL.
We looked for MLL abnormalities in bone marrow samples from 47 adults and 34 children with unselected T-ALL. Immunophenotyping and the assignment of the leukemias to the T lineage were based on the classification of the European Group for the Immunological characterization of Leukemias. No correlation was observed between the maturation of T-ALL and the involvement of the MLL gene. All the patients were screened by Southern blot analysis with various probes encompassing exon 3 to exon 22 of theMLL gene, and also, where possible, by fluorescence in situ hybridization (FISH). None of the children showed MLLinvolvement, but in 4 adults a rearranged MLL gene was found by at least 2 different methods: Southern blot analysis (Figure1), reverse transcriptase–polymerase chain reaction (RT-PCR), and/or FISH analysis. Southern blot analysis showed that another adult (P5) had rearranged bands corresponding in size to the site-specific DNA cleavage induced by the topoisomerase II inhibitors that have been found in the MLLgene.4 This result was further strengthened by the fact that FISH analysis in this case revealed no abnormalities. Moreover, Southern blot analysis of the P5 relapse sample did not reveal the rearranged bands observed at diagnosis, strongly suggesting that the MLL gene is not implicated in the T-ALL of this patient.
Involvement of the MLL gene in T-ALL.
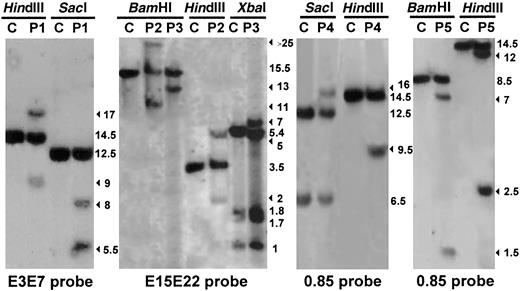
Southern blot analysis performed on genomic DNA extracted from leukemic cells from 5 patients with T-ALL (P1, P2, P3, P4, and P5) compared with human placenta used as a negative control (C). The enzymes used to digest the DNA are indicated. The filter was assayed for hybridization to the MLL PCR-amplified cDNA probes: E3E7 (nucleotides 3137-4125), E15E22 (nucleotides 5171-6130), and 0.85 (nucleotides 3750-4614); nucleotide numbering is with respect to the MLLcDNA sequence (Genbank accession no. NM_005933). The numbers indicate band size (kbp). The rearranged fragments are indicated by arrows. Southern blot analysis of the MLL gene revealed rearranged DNA fragments, with at least 2 restriction endonucleases in 5 of the 47 adults studied.
Involvement of the MLL gene in T-ALL.
Southern blot analysis performed on genomic DNA extracted from leukemic cells from 5 patients with T-ALL (P1, P2, P3, P4, and P5) compared with human placenta used as a negative control (C). The enzymes used to digest the DNA are indicated. The filter was assayed for hybridization to the MLL PCR-amplified cDNA probes: E3E7 (nucleotides 3137-4125), E15E22 (nucleotides 5171-6130), and 0.85 (nucleotides 3750-4614); nucleotide numbering is with respect to the MLLcDNA sequence (Genbank accession no. NM_005933). The numbers indicate band size (kbp). The rearranged fragments are indicated by arrows. Southern blot analysis of the MLL gene revealed rearranged DNA fragments, with at least 2 restriction endonucleases in 5 of the 47 adults studied.
The relevant clinical and biological data for the patients withMLL involvement are given in Table1. An interesting common feature was that, in 3 of the 4 cases (P1, P2, P3), the breakpoint occurred outside of the breakpoint cluster region (bcr) commonly involved inMLL translocations,5 which may in part account for the fact that the incidence of MLL rearrangement has up to now been underestimated in T-ALL. In P1, the breakpoint was located upstream from the usual MLL bcr (between exons 4c and 5), and the fusion partner was the AF10 gene. In P2 and P3, the breakpoint was located downstream from the usual MLL bcr, between exons 20 and 21 and exons 18 and 19, respectively. Only in P4 the breakpoint occurred in the usual MLL bcr (between exon 6 and exon 7). With P2, P3, and P4, the MLL gene fused with the AF6q27 gene. As with all the other 11q23 translocations so far investigated, an RT-PCR on leukemic cells from P1, P2, P3, and P4 revealed an in-frame fusion transcript encoding the amino-terminus of MLL and the carboxy-terminus of either AF10 (P1) or AF6q27 (P2, P3, and P4). The fusion occurred at breakpoints already described for the partner genes AF10 andAF6q27.6,7 It will be noted that the contribution of the MLL protein to the fusion product was larger for P2 and P3 than in other cases described to date. It included, in particular, the major part of the Drosophila trithorax zinc-fingers domain of the MLL protein.8 The deletion of the exon 8 of the MLL gene observed in some cases of T-ALL9 was not found by RT-PCR on leukemic cells from the 5 rearranged cases (P1 to P5). P1 and P2 achieved complete remission (CR) after 2 courses of induction chemotherapy, and they received allogenic bone marrow transplants as consolidation therapy. They were still alive 7 years and 3 years, respectively, after transplantation. P3, P4, and P5 achieved CR after intensive standard chemotherapy but relapsed a few months later and died. These observations suggest that where there is MLL involvement, T-ALL, like other forms of leukemia, responds particularly poorly to standard therapies.
Clinical and molecular characteristics of T-ALL patients with MLL rearrangement
| Patient no., sex/age (y) . | Overall survival (follow up, y) . | Karyotype . | FISH analysis . | Fusion transcript . | Genomic breakpoint accession no. . |
|---|---|---|---|---|---|
| P1, M/19 | Alive (7), allografted | 46,XY[12]/44,XY,t(1;20)(p32;q12),-12,-13, +der(12)t(12;13)(p11-12;q11),der(14q), -21,-21,i(21q)[9] | NA | MLL(nt:3634)/AF10 (nt:1931) | AF490423 |
| P2, M/26 | Alive (3), allografted | 46,XY[8]/46,XY,der(7q),del(11)(q14q24)[18] | Split | MLL(nt:5952)/AF6q27 (nt:106) | AF490424 |
| P3, M/16 | Died (1) | 46,XY[11]/47,XY,del(9)(p22),del(11)(q21q23), del(13)(q14q21),+mar[15] | Split | MLL(nt:5655)/AF6q27 (nt:106) | NA |
| P4, M/24 | Died (2) | 45,XY,del(11)(q21q23-24),add(12)(p12), -18[1]/45,XY,idem,add(17)(p12)[2] | Split | MLL(nt:4086)/AF6q27 (nt:106) | NA |
| P5, M/36 | Died (1) | 46,XY[18]/46,XY,del(4)(q31q34),del(6)(q13q16-21), -15,-22,+mar[9] | Normal | Cleavage (topoisomerase II inhibitor site) |
| Patient no., sex/age (y) . | Overall survival (follow up, y) . | Karyotype . | FISH analysis . | Fusion transcript . | Genomic breakpoint accession no. . |
|---|---|---|---|---|---|
| P1, M/19 | Alive (7), allografted | 46,XY[12]/44,XY,t(1;20)(p32;q12),-12,-13, +der(12)t(12;13)(p11-12;q11),der(14q), -21,-21,i(21q)[9] | NA | MLL(nt:3634)/AF10 (nt:1931) | AF490423 |
| P2, M/26 | Alive (3), allografted | 46,XY[8]/46,XY,der(7q),del(11)(q14q24)[18] | Split | MLL(nt:5952)/AF6q27 (nt:106) | AF490424 |
| P3, M/16 | Died (1) | 46,XY[11]/47,XY,del(9)(p22),del(11)(q21q23), del(13)(q14q21),+mar[15] | Split | MLL(nt:5655)/AF6q27 (nt:106) | NA |
| P4, M/24 | Died (2) | 45,XY,del(11)(q21q23-24),add(12)(p12), -18[1]/45,XY,idem,add(17)(p12)[2] | Split | MLL(nt:4086)/AF6q27 (nt:106) | NA |
| P5, M/36 | Died (1) | 46,XY[18]/46,XY,del(4)(q31q34),del(6)(q13q16-21), -15,-22,+mar[9] | Normal | Cleavage (topoisomerase II inhibitor site) |
FISH analysis using an MLL probe (Vysis) showed that the MLL gene was split between chromosomes 6 and 11 in P2, P3, and P4. The nucleotide number for the fusion transcripts came from the cDNA sequences of MLL (Genbank accession no. NM_005933),AF10 (Genbank accession no. U13948), and AF6q27(Genbank accession no. U02478). For P1 and P2, the breakpoints were cloned and characterized, and the nucleotide sequences were filed at the Data Library of the National Center for Biotechnology Information.
M indicates male; NA, not available; and nt, nucleotide.
To our knowledge, this is the first study of MLLabnormalities in a series of cases of T-ALL. Our findings, like those of previous studies on B-ALL and AML, suggest that the involvement of the MLL gene in adults T-ALL is recurrent, with an incidence of more than 8%. In view of these results and the fact that lymphoblastic leukemia with MLL translocation seems to constitute a distinct disease with a poor prognosis, we would recommend that adults with T-ALL be screened by FISH analysis for MLLabnormalities.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal