The 5q− syndrome is the most distinct of the myelodysplastic syndromes, and the molecular basis for this disorder remains unknown. We describe the narrowing of the common deleted region (CDR) of the 5q− syndrome to the approximately 1.5-megabases interval at 5q32 flanked by D5S413 and theGLRA1 gene. The Ensembl gene prediction program has been used for the complete genomic annotation of the CDR. The CDR is gene rich and contains 24 known genes and 16 novel (predicted) genes. Of 40 genes in the CDR, 33 are expressed in CD34+ cells and, therefore, represent candidate genes since they are expressed within the hematopoietic stem/progenitor cell compartment. A number of the genes assigned to the CDR represent good candidates for the 5q− syndrome, including MEGF1, G3BP, and several of the novel gene predictions. These data now afford a comprehensive mutational/expression analysis of all candidate genes assigned to the CDR.
Introduction
The 5q− syndrome is a myelodysplastic syndrome (MDS) with the 5q deletion, del(5q), as the sole karyotypic abnormality and is characterized by refractory anemia, hypolobulated megakaryocytes, and a low risk of transformation to acute myeloid leukemia (AML).1,2 The molecular basis for the 5q− syndrome has been the subject of extensive investigation, although the putative tumor suppressor gene for the 5q− syndrome remains unidentified.2
We have previously delineated the common deleted region (CDR) of the del(5q) in the 5q− syndrome, using fluorescence in situ hybridization (FISH) analysis and molecular mapping techniques, as the approximately 3-megabases (Mb) region at 5q31-5q32 flanked by the genes for ADRβ2 andIL12β.3,4 This region is distinct from the CDR of the del(5q) at 5q31 in AML and some other more aggressive forms of MDS.5 6
The aim of this study was to narrow the CDR of the 5q− syndrome and to generate the complete genomic annotation of this interval, thus facilitating the identification of the causative gene.
Study design
Patients
FISH analysis
FISH analysis was carried out on bone marrow metaphases (patients 1-15) as previously described.4 A series of cosmid and yeast artificial chromosome (YAC) clones corresponding to known genes or DNA markers (and, where appropriate, bacterial artificial chromosome [BAC] clones) mapping to 5q31-5q34 were used for FISH analysis (Figure 1A).4,9,10The 5q13-assigned YAC 153A2 was also used for FISH analysis4 of the proximal breakpoints.
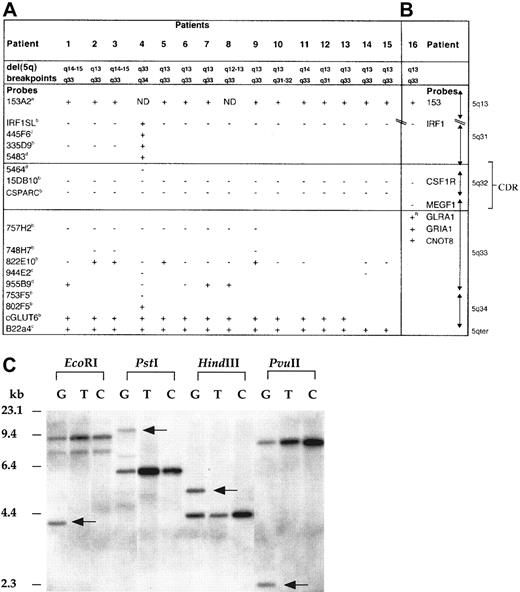
Mapping of the del(5q) in 5q− syndrome patients.
FISH studies (patients 1-15) (A) and gene-dosage studies (patient 16) (B) with the use of probes from 5q31-qter and 5q13. The + indicates retained on del(5q); + R, rearranged on del(5q); −, deleted from del(5q); ND, not determined owing to limited metaphase numbers; boxed area, 5q− syndrome CDR. The following indicate that the probe was donated by the people or institutions listed:a, Kay Davies, Oxford University; b, Jaju et al4; c, Kostrzewa et al9;d, http://www.tree.caltech.edu/ and LBNL; e, National Institutes of Health and Institute of Molecular Medicine Collaboration.10 The 1-16 are 5q− syndrome (MDS-RA) cases studied; patient 4 has been reported previously.4 (C) Southern blot hybridized to a GLRA1 PCR probe showing rearrangement of the GLRA1 gene in patient 16. Previously published primers14 were used for the amplification of theGLRA1 gene, and the PCR-generated probe was sequenced to confirm its identity. The restriction enzymes used are indicated above the lanes. G indicates granulocyte DNA from patient 16; T, T-lymphocyte DNA from patient 16; C, peripheral blood DNA from healthy control. Rearranged fragments are observed in all 4 lanes containing granulocyte DNA from patient 16 (arrows), but not in T-lymphocyte DNA from patient 16, or in DNA from controls. No additional hybridization fragments were observed in the DNA tracks of patient 16 when the same Southern blot was rehybridized with each of the probes for the other 5q-specific genes investigated (see gene-dosage discussion in text).
Mapping of the del(5q) in 5q− syndrome patients.
FISH studies (patients 1-15) (A) and gene-dosage studies (patient 16) (B) with the use of probes from 5q31-qter and 5q13. The + indicates retained on del(5q); + R, rearranged on del(5q); −, deleted from del(5q); ND, not determined owing to limited metaphase numbers; boxed area, 5q− syndrome CDR. The following indicate that the probe was donated by the people or institutions listed:a, Kay Davies, Oxford University; b, Jaju et al4; c, Kostrzewa et al9;d, http://www.tree.caltech.edu/ and LBNL; e, National Institutes of Health and Institute of Molecular Medicine Collaboration.10 The 1-16 are 5q− syndrome (MDS-RA) cases studied; patient 4 has been reported previously.4 (C) Southern blot hybridized to a GLRA1 PCR probe showing rearrangement of the GLRA1 gene in patient 16. Previously published primers14 were used for the amplification of theGLRA1 gene, and the PCR-generated probe was sequenced to confirm its identity. The restriction enzymes used are indicated above the lanes. G indicates granulocyte DNA from patient 16; T, T-lymphocyte DNA from patient 16; C, peripheral blood DNA from healthy control. Rearranged fragments are observed in all 4 lanes containing granulocyte DNA from patient 16 (arrows), but not in T-lymphocyte DNA from patient 16, or in DNA from controls. No additional hybridization fragments were observed in the DNA tracks of patient 16 when the same Southern blot was rehybridized with each of the probes for the other 5q-specific genes investigated (see gene-dosage discussion in text).
Southern analysis: gene dosage
No metaphase cell preparations were available for FISH analysis of case 16, and gene-dosage experiments were therefore designed to allow for a quantitative assessment of the allelic loss of a number of genes assigned to distal 5q and comprising the following:IRF1, CSF1R, MEGF1, GLRA1, GRIA1, and CNOT8(formerly POP2).9,11-14 Gene-dosage experiments14 were also designed for the determination of the allelic loss of the 5q13-assigned probe 153 (subclone of YAC 153A2). Granulocyte and T-lymphocyte cell fractions were separated from peripheral blood samples obtained from patients with the 5q− syndrome by means of Ficoll gradient centifugation and erythrocyte rosetting as previously described.3 High–molecular-weight DNA was obtained from the granulocyte and T-lymphocyte cell fractions from patients with the 5q− syndrome and from the peripheral blood samples of 10 healthy individuals by phenol-chloroform extraction.15 The DNA was digested with the restriction enzyme EcoRI, and Southern blotting was performed according to standard procedures.15 Filters containing patient and control samples were simultaneously hybridized to 2 probes: one of the chromosome 5–specific complementary DNA (cDNA) or genomic probes and the 1.9-kilobase genomic EcoRI-SstI fragment from the renin gene (pHRn ES1.9) (ATCC, Manassas, VA). The renin gene acts as an internal hybridization standard. DNA probe labeling, hybridization, and autoradiography were carried out as previously described.3,14 A comparative densitometric ratio was derived from the 2 hybridization signals in 10 healthy individuals. An approximately 50% reduction in the dosage of the chromosome 5–specific cDNA of interest is consistent with deletion of one allele.3 14 All gene-dosage experiments were carried out on 2 separate occasions.
Genomic annotation of the CDR
Sequence data (comprising draft and finished sequence) were obtained from a total of 37 BAC clones assigned to the CDR of the 5q− syndrome at 5q32 from a contig mapping to this region of chromosome 5 (http://www-gsd.lbl.gov/∼j_martin/ and Lawrence Berkeley National Laboratory [LBNL] [Berkeley, CA]). This contig forms a section of the Ensembl UCSC “golden path” (http://www.ensembl.org/) that is consistent with the National Center for Biotechnology Information database and contains no gaps. Sequence data (Figure 2B for GenBank accession numbers) was analyzed by means of the Ensembl gene prediction program (http://www.ensembl.org/)16 for the presence of coding exons.
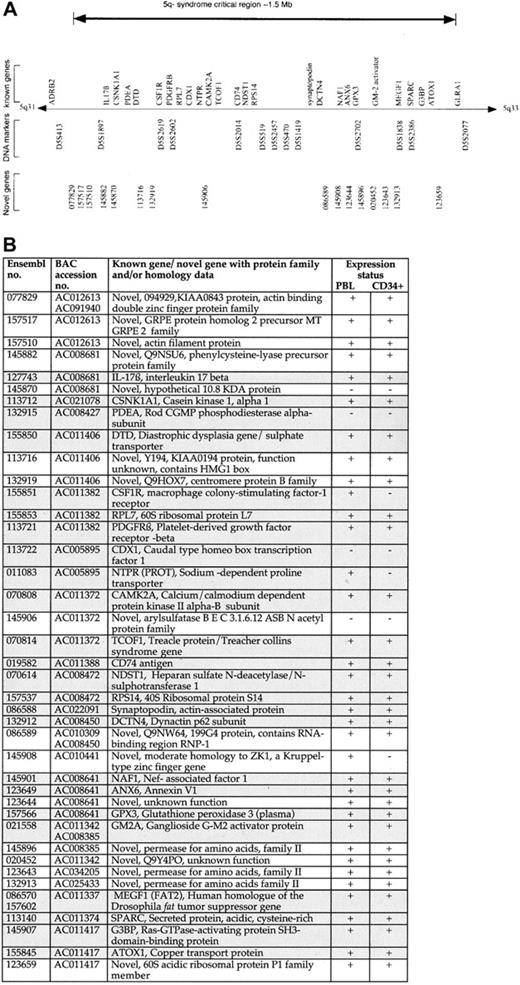
Genomic annotation of the CDR of the 5q− syndrome.
(A) The CDR of the 5q− syndrome showing all known genes9 11-13 and novel genes predicted by Ensembl. (B) Table showing all known genes and novel (predicted) genes assigned to the CDR of the 5q− syndrome with the use of the Ensembl program and their expression profiling. Each of the Ensembl gene numbers shown is preceded by ENSG00000. Sequence data (comprising draft and finished sequence under the GenBank accession numbers shown) was obtained from a complete BAC tiling path from the CDR of the 5q− syndrome (http://www-gsd.lbl.gov/∼j_martin/ and LBNL) and analyzed by means of the Ensembl gene prediction program (http://www.ensembl.org/) for the presence of coding exons. The genes were predicted by the Ensembl analysis pipeline from either a GeneWise or a Genscan prediction followed by confirmation of the exons by comparisons with protein, cDNA, and EST databases (Ensembl v1.2.0 and previous versions). Genes identical to known human genes or protein sequences are referred to as known genes (shaded boxes); genes homologous to, or containing a region of similarity to, gene or protein sequences from human or other species or sequences homologous to only ESTs are referred to as novel genes. Expression profiling was performed on all known and novel genes by means of RT-PCR. Primers were designed to span at least one intron in all cases, and optimal conditions were determined for each primer pair. PBL indicates peripheral blood leukocyte. Expression status is indicated as follows: +, RT-PCR product obtained on at least 2 occasions; −, no RT-PCR product obtained on at least 2 occasions. Note that Ensembl genes 145870 (predicted) and 145906 (predicted) were shown to be positive for expression in human colon and prostate, respectively.
Genomic annotation of the CDR of the 5q− syndrome.
(A) The CDR of the 5q− syndrome showing all known genes9 11-13 and novel genes predicted by Ensembl. (B) Table showing all known genes and novel (predicted) genes assigned to the CDR of the 5q− syndrome with the use of the Ensembl program and their expression profiling. Each of the Ensembl gene numbers shown is preceded by ENSG00000. Sequence data (comprising draft and finished sequence under the GenBank accession numbers shown) was obtained from a complete BAC tiling path from the CDR of the 5q− syndrome (http://www-gsd.lbl.gov/∼j_martin/ and LBNL) and analyzed by means of the Ensembl gene prediction program (http://www.ensembl.org/) for the presence of coding exons. The genes were predicted by the Ensembl analysis pipeline from either a GeneWise or a Genscan prediction followed by confirmation of the exons by comparisons with protein, cDNA, and EST databases (Ensembl v1.2.0 and previous versions). Genes identical to known human genes or protein sequences are referred to as known genes (shaded boxes); genes homologous to, or containing a region of similarity to, gene or protein sequences from human or other species or sequences homologous to only ESTs are referred to as novel genes. Expression profiling was performed on all known and novel genes by means of RT-PCR. Primers were designed to span at least one intron in all cases, and optimal conditions were determined for each primer pair. PBL indicates peripheral blood leukocyte. Expression status is indicated as follows: +, RT-PCR product obtained on at least 2 occasions; −, no RT-PCR product obtained on at least 2 occasions. Note that Ensembl genes 145870 (predicted) and 145906 (predicted) were shown to be positive for expression in human colon and prostate, respectively.
Expression studies in myeloid and other tissues
The expression of all known and novel genes assigned to the CDR was examined in normal human peripheral blood and in normal human bone marrow CD34+ cells (98.5% purity).6 Total RNA was extracted from the selected cell populations by means of the Totally RNA kit (Ambion, Austin, TX). Reverse-transcription polymerase chain reaction (RT-PCR) was performed by means of the Reverse-iT one step RT-PCR kit (ABgene, Epsom, Surrey, United Kingdom) and 1 μg total RNA according to the manufacturer's protocol. Conditions were 47°C for 30 minutes, 94°C for 2 minutes, followed by 40 cycles of 94°C for 20 seconds, an annealing temperature of between 50°C and 65°C for 30 seconds, and 72°C for 1 minute, with a final extension of 72°C for 5 minutes. In each case, a duplicate reaction was set up without reverse transcriptase to act as a control for DNA contamination.
Results and discussion
In this study, we have analyzed 16 cases of 5q− syndrome by FISH or molecular mapping techniques to investigate the deletion or retention of a number of genes and or DNA markers localized to 5q31-5q34 and 5q13 (Figure 1A-B). G-banding analyses, combined with FISH studies, showed that most of the deletions were large, extending from 5q13 to 5q33. The common region of loss in all patients includes the PDEA, CSF1R, CD74, TCOF1, ANX6, SPARC, and MEGF1 genes (Figure 2A). The proximal boundary and the distal boundary of the CDR of the 5q− syndrome are defined in this study by patients 4 and 16, respectively (Figure 1A-B). FISH analysis of the proximal breakpoint in patient 4 has previously been reported as being between D5S207 and ADRβ2 (as patient 3).4 In the present study, further FISH mapping has shown that BAC5483 (mapping approximately 0.5 Mb distal toADRβ2 and containing D5S413) is retained and BAC5464 (proximal and adjacent) is lost in patient 4 (Figure 1A), thus refining the proximal boundary of the critical region. The proximal boundary of the CDR of the 5q− syndrome is defined by patient 4 with a breakpoint between D5S413 and D5S1897.
Patient 16 also had the characteristic clinical and hematological features of the 5q− syndrome.2,8 The karyotype of patient 16 is 46, XX, del(5q)(q13q33), and the percentage of abnormal cells in the bone marrow was high (80%). By means of gene-dosage analysis3,14 the distal breakpoint of the del(5q) of patient 16 was shown to map between the MEGF1 andGRIA1 genes (Figure 1B). The GLRA1gene17 is assigned between the MEGF1gene and the GRIA1 gene (Figure 1B). With the use of Southern analysis and the enzymes EcoRI, PstI,HindIII, and PvuII, additional, non–germ-line sized fragments were observed in the granulocyte DNA of patient 16—but not in the T-lymphocyte DNA of patient 16 or in the DNA of 20 healthy controls—with all enzymes following hybridization with theGLRA1 probe (Figure 1C). These data indicate that the additional bands resulted from rearrangement of theGLRA1 gene, rather than from the presence of DNA polymorphisms. The GLRA1 gene is thus disrupted by the del(5q) in patient 16 and defines the distal boundary of the CDR of the 5q− syndrome.
These data markedly reduce the CDR of the 5q− syndrome, which is flanked by D5S413 and the GLRA1 gene at 5q32. The estimated size of this region is approximately 1.5 Mb, with the use of data from the BAC contig described. This interval contains 3793 single-nucleotide polymorphisms (www.ncbi.nlm.nih.gov/SNP/ [accessed March 17, 2002]), which will be valuable for the further delineation of the CDR.
In virtually all deletion-mapping studies in leukemia, the boundaries of the CDR are defined by only a very small number of cases.4,5 It is also true that the majority of patients, with the 5q− syndrome and AML with the del(5q), possess large deletions in which both the AML CDR at 5q31 and the CDR of the 5q− syndrome at 5q32 will be deleted.5 Indeed, in 15 of the 16 cases reported in this study, both CDRs will be lost. It is important to note, however, that patient 4 of this study, together with 2 other cases with the 5q− syndrome previously reported,3,4 have 5q deletions with a common region of overlap that is molecularly distinct from that described in patients with the del(5q) and AML or some of the more aggressive forms of MDS.4 5
Defining a minimal CDR for a disease allows for the identification of candidate genes within the interval. Sequence data were obtained from a complete BAC tiling path assigned between the DNA marker D5S413 and theGLRA1 gene from the BAC contig mapping to this region of chromosome 5. These sequence data were analyzed by means of the Ensembl gene prediction program.16 The CDR of the 5q− syndrome contains 40 genes: 24 known and 16 novel (predicted) genes (Figure2A-B). All of the known genes previously assigned to this interval were predicted by the Ensembl program. Evidence that each of the novel gene predictions represented bona fide transcribed genes, rather than false positives or processed pseudogenes, was obtained from RT-PCR amplification experiments and from the examination of the predicted exon structure (Figure2B). All RT-PCR products obtained from novel gene predictions were sequenced to confirm their identity.
The annotation of the CDR of the 5q− syndrome in this study confirms and extends data from the initial integrated gene index (IGI) published earlier in the year by the International Human Genome Sequencing Consortium.16 In that earlier publication, 20 genes were assigned to the CDR by means of Ensembl,16 and 18 of these represent known genes.9,11 This compares with a total of 40 genes, both known and novel, assigned to the CDR in this current study. The greater number of novel genes presented here represents a significant difference between the 2 data sets. The IGI data are based upon draft sequence data available in July 2000.16 The annotation of the CDR in this current study is based upon the most recent draft and finished sequence data available and incorporates considerably more sequence data with an improved quality of both sequence assembly and gene prediction.
Given the completeness of the sequence data used in this analysis, together with the high efficiency of the Ensembl program for novel gene prediction, it is reasonable to suggest that the annotation of the CDR of the 5q− syndrome, as presented here, now comprises most if not all of the genes, both known and novel, mapping to this interval.
It has been shown that MDS arises from the transformation of a multipotent hematopoietic stem cell (HSC) or myeloid-committed progenitor cell.18,19 These data suggest that the gene or genes that are inactivated in the 5q− syndrome will be expressed in normal hematopoietic stem and progenitor cells. The expression of all 40 genes assigned to the CDR by means of the Ensembl program was thus examined in normal human bone marrow CD34+ cells by means of RT-PCR. Of the 40 genes in the CDR, 33 were expressed in CD34+ cells and, therefore, represent candidate genes since they are expressed within the HSC/progenitor cell compartment. A number of genes represent good candidate genes for the 5q− syndrome; these include the MEGF1 gene, a member of the cadherin gene family and a human homologue of the Drosophila fat tumor-suppressor gene,12,20,21 and G3BP, the Ras-GTPase–activating protein–binding protein.22
We describe the narrowing of the critical region of the 5q− syndrome to a 1.5-Mb interval and report the complete genomic annotation of this region and the expression status of all known and novel genes in hematopoietic stem cells. It is not known whether a gene (or genes) mapping within the CDR might contribute to tumorigenesis by haploinsufficiency rather than by homozygous inactivation. Nonetheless, whichever mechanism operates in this disease, it is necessary to have a complete genomic annotation of the CDR. The data presented here allow, for the first time, a comprehensive analysis of all candidate genes assigned to this interval and will enable the final elucidation of the genetic nature of this disorder.
We are grateful to the following cytogenetic laboratories for providing fixed cell suspensions for the study: the Wessex Regional Genetics Laboratory (Salisbury, United Kingdom); the ICRF Department of Medical Oncology, St Bartholomew's Hospital (London, United Kingdom); the Division of Human Genetics, University of Newcastle upon Tyne (Newcastle, United Kingdom); the Cytogenetics Laboratory, Department of Haematology, Royal Free Hospital (London, United Kingdom); Oxford Medical Genetics Laboratories, The Churchill Hospital (Oxford, United Kingdom); St Anna Children's Hospital (Vienna, Austria); and the Department of Pathology, Hospital del mar (Barcelona, Spain).
Supported by The Leukaemia Research Fund, United Kingdom (J.B., C.F., A.J.S., F.W., R.J.J., S.T., J.S.W.), and the Medical Research Council (L.K.).
J.B. and C.F. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jacqueline Boultwood, Leukaemia Research Fund Molecular Haematology Unit, Nuffield Department of Clinical Laboratory Sciences, University of Oxford, John Radcliffe Hospital, Headington, Oxford OX3 9DU, United Kingdom; e-mail:jboultwo@enterprise.molbiol.ox.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal