The morphologic, immunophenotypic, genotypic, genomic, and functional features of an undifferentiated acute leukemia with stem cell features are reported. At light and electron microscopy, the leukemic population was represented by primitive progenitor cells with no evidence of differentiation. The blasts were CD34+, AC133+, CD71−, HLA-DR−, CD38−/dim+, CD90+, CD117dim+, flt3+; did not express B, T, or myeloid-associated antigens; and showed a germline configuration of the immunoglobulin and T-cell receptor. Genomic profiling documented the expression of early stem cell and myeloid-associated genes. Receptors for early-acting hemopoietic growth factors (HGFs) were detected, while receptors for unilineage HGF were not expressed. Incubation with the flt3 or Kit ligand induced the expression of unilineage HGF receptors, allowing these cells to respond to their respective ligands. Growth without differentiation was sustained only in the presence of early-acting HGF, namely flt3 ligand, while early and unilineage HGF gave rise to all types of hemopoietic colonies.
Introduction
The use of monoclonal antibodies allows one to identify the lineage affiliation and level of differentiation of virtually all acute leukemias (ALs).1,2 The possibility that AL may display multilineage differentiation potential has been suggested for t(9;22) and t(4;11) acute lymphoblastic leukemia.3-7 The high frequency of CD34 positivity8-10 points to the close relationship of AL with early hemopoietic progenitor cells. Considerable interest has focused on acute myeloid leukemia with minimal signs of differentiation11-13; no study, however, has addressed the multilineage potential of human AL or investigated the genomic profiling and true “stem cell” capacity of such early leukemic cells. Here, we report a very undifferentiated stem cell leukemia in which the lineage affiliation could not be defined by conventional characterization.
Study design
Case report
An 82-year-old woman was referred to the Department of Cellular Biotechnologies and Hematology, University of Rome, Italy, in May 1997 because of mild anemia, progressive asthenia, and malleolar edema. The peripheral blood count showed the following: hemoglobin, 80 g/L [8.07 g/dL]); white blood cell count, 55 × 109/L; polimorphonuclear cells 2.5 × 109/L; undifferentiated lymphoidlike cells, 52.5 × 109/L; platelets, 184 × 109/L. A diagnosis of undifferentiated stem cell leukemia was made. At this stage, the patient gave her informed consent to the storage of viable pathologic cells. In view of the age of the patient, vincristine plus prednisone was administered on an outpatient basis. At 1 month later, a complete recovery of peripheral parameters was achieved, and the patient was put on maintenance treatment. In January 1999, a recurrence was documented. The patient was restarted on vincristine plus prednisone, but died 15 days later.
Electron microscopy, immunophenotype, and hemopoietic growth factor receptor characterization
Ultrastructural examination and immunophenotyping were performed on peripheral blood mononuclear cells. Thin sections were stained with toluidine blue, and ultrathin sections were stained with uranyl acetate and lead citrate; sections were observed under a Philips CM10 transmission electron microscope (FEI, Acht, The Netherlands). Myeloperoxidase (MPO) activity was investigated as described.14
Immunophenotypic characterization was conducted by means of reagents against the following: CD34, surface (s)CD13, cytoplasmic (cy)CD13, CD33, CD11a, CD11b, CD11c, CD14, CD15, CD18, MPO, HLA-DR, terminal deoxynucleotidyl transferase (TdT), CD10, sCD22, cyCD22, CD19, CD23, CD10, CD20, CD79a, immunoglobulin-κ (Igκ), Igλ, sCD3, cyCD3, CD2, CD7, CD5, CD31, CD38, CD117, CD41a, CD61, CD62, CD54, CD56, CD90, glycophorin A, AC133, multidrug resistance (MDR-1) (BD Pharmingen, San Diego, CA). Analyses were performed by flow cytometry with a FACS Scan Flow cytometer (Becton Dickinson, Bedford, MA).
The expression of different hemopoietic growth factor (HGF) receptors was investigated by means of reagents and procedures previously reported.15
Genotypic characterization and genomic profiling
The configuration of the IgH chain gene region was analyzed by single-step polymerase chain reaction,16 while the T-cell receptor–γ (TCR-γ) gene was analyzed through the amplification of the TCR-γ locus V-J junction region by heteroduplex analysis.17
For genomic profiling, total RNA was extracted by means of TRizol reagent (Gibco BRL, Life Technologies, Grand Island, NY) and further purified with the use of SV Total RNA Isolation System (Promega, Madison, WI). The protocol for sample preparation and microarray processing is available from Affymetrix (www.Affymetrix.com; Santa Clara, CA). Genomic profiling was carried out by means of the Affymetrix U95Av2 gene chip, which contains 12 600 gene sequences. To calculate gene expression levels and for normalization with other acute leukemia samples, the d-chip program was used.18 Genes whose expression levels were higher than 100 were considered expressed.
Cell-growth, cell-cycle, apoptosis, and clonogenic activity
Recombinant human interleukin–3 (rhIL-3) (2 × 106 U/mg), rh granulocyte-macrophage colony stimulating factor (rhGM-CSF), and rhIL-6 (2 × 108 U/mg) were supplied by Genetics Institute (Cambridge, MA); rh erythropoietin (rhEpo) (1.2 × 105 U/mg) and bovine basic fibroblast growth factor (rhbFGF; 2 × 107 U/mg) by Amgen (Thousand Oaks, CA); rh flt-3 ligand (FL) (1.9 × 106 U/mg) and rh c-kit ligand (KL) (1 × 105 U/mg) by Immunex (Seattle, WA); rhG-CSF (1 × 108 U/mg) and rhM-CSF (6 × 107 U/mg) by R&D Systems (Minneapolis, MN); rh thrombopoietin (rhTpo) was generously provided by Genentech (San Francisco, CA). Iscoves modified Dulbecco medium (IMDM) (GIBCO, Grand Island, NY) was prepared weekly.
Leukemic cells were grown in liquid suspension in IMDM containing 10% fetal calf serum (GIBCO) and various combinations of HGFs. The cell number was evaluated at each day of culture after trypan blue staining.
Cell-cycle analysis was carried out on nuclei stained with propidium iodide, as described.19
Results and discussion
Morphologic and immunophenotypic characterization
The leukemic population was uniformly composed of highly undifferentiated small- to medium-sized blasts. All cytochemical reactions were negative. Ultrastructural examination showed a large, centrally placed nucleus with prominent nucleoli. The cytoplasm contained small amounts of free ribosomes, a small Golgi apparatus, and endoplasmic reticulum cisternae. No MPO activity was detected.
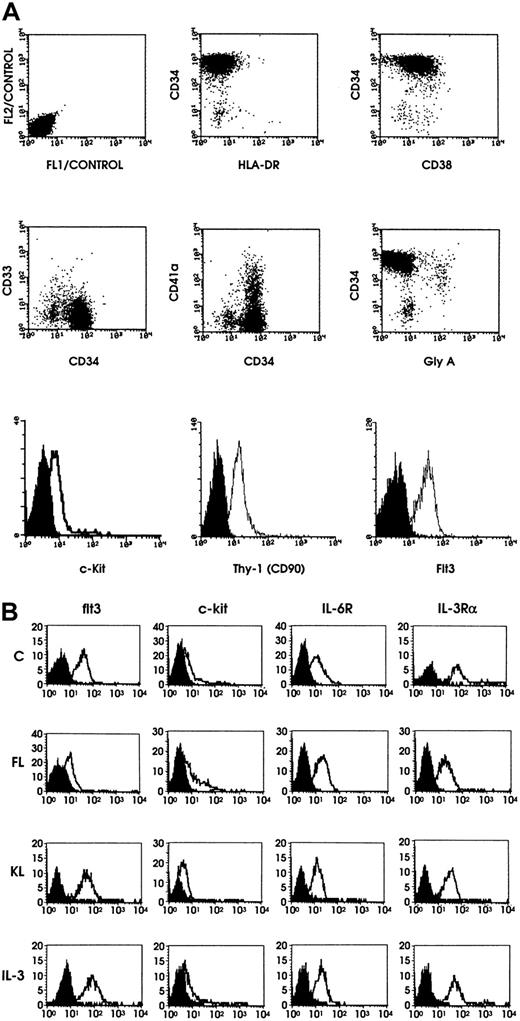
The immunophenotype showed that (1) leukemic cells were CD34+ and did not express MPO; (2) CD34+ cells displayed a phenotype similar to that associated with most normal immature hemopoietic progenitors, CD33−, HLA-DR−, flt3+, CD90+, c-kitdim+, CD38−/dim+, AC133+; and (3) CD34+ cells did not display membrane markers of the erythroid, megakaryocytic, monocytic, granulocytic, B and T lineages.20-22 TdT was expressed on 27% of cells. MDR-1 was negative (Figure 1A).
Flow cytometric analysis of leukemic blasts and results of HGF receptor expression.
(A) Flow cytometric analysis of leukemic blasts. Flow cytometry of leukemic blasts was performed by using a panel of monoclonal antibodies selected to define the stem cell leukemia phenotype. Representative results are shown. (B) Representative results on HGF receptor (HGFR) expression. Expression of flt3, c-kit, IL-6R, and IL-3Rα chain on leukemic blasts grown either in the absence of HGFs (control) or in the presence of FL, KL, or IL-3.
Flow cytometric analysis of leukemic blasts and results of HGF receptor expression.
(A) Flow cytometric analysis of leukemic blasts. Flow cytometry of leukemic blasts was performed by using a panel of monoclonal antibodies selected to define the stem cell leukemia phenotype. Representative results are shown. (B) Representative results on HGF receptor (HGFR) expression. Expression of flt3, c-kit, IL-6R, and IL-3Rα chain on leukemic blasts grown either in the absence of HGFs (control) or in the presence of FL, KL, or IL-3.
The immaturity of the blasts was further supported by the pattern of transcription factor expression: GATA-2+, Tal-1−, GATA-1−.23
Genotypic characterization and genomic profiling
The IgH and TCR-γ gene regions were germline. To more precisely elucidate the nature of the leukemia, a genomic profiling was carried out. While the conventional lymphoid- and myeloid-related genes were not expressed, a set of early stem cell and myeloid-associated genes, particularly of the mixed lineage leukemia (MLL) family, was found (Table 1). CD34 and flt3 were expressed at high levels.
Genomic profile
| Gene description . | Map . | Locus link ID . | Level of expression . |
|---|---|---|---|
| Human myeloid differentiation | 1q21 | 4 170 | 839 |
| Human stem cell progenitor homeobox | 10q26 | 27 287 | 154 |
| Human myeloid elf-1 | Xq26 | 2 000 | 622 |
| Myeloperoxidase | 17q23 | 4 353 | 284 |
| Myelodisplasia/myeloid leukemia factor 2 | 12p13 | 8 079 | 317 |
| CD34 | 1q32 | 947 | 860 |
| Human stem cell leukemia exon 6 | 124 | ||
| Human AML1b protein | 21q22.3 | 861 | 481 |
| Flt3 | 13q12 | 2 322 | 1 417 |
| Breakpoint cluster region | 22q11.23 | 613 | 934 |
| MLL, translocated to 2 | 4q21 | 4 299 | 558 |
| MLL, translocated to 3 | 9p22 | 4 300 | 319 |
| MLL, translocated to 4 | 6q27 | 4 301 | 176 |
| MLL, translocated to 7 | Xq13.1 | 4 303 | 355 |
| Myeloid cell nuclear differentiation antigen | 1q22 | 4 322 | 125 |
| MLL, septinlike fusion | 17q25 | 10 801 | 1 259 |
| Myeloid differentiation primary response gene, 88 | 3p22 | 4 615 | 273 |
| Clathrin-8 assembly lymphoid-myeloid leukemia gene | 11q14 | 8 301 | 656 |
| SET translocation (myeloid leukemia associated) | 9q34 | 6 418 | 1 071 |
| Control genes | |||
| AFFX-HUMGAPDH | 12p13 | 2 597 | 4 511 |
| AFFX-HSACO (β-actin) | 7p15-p12 | 60 | 3 161 |
| Gene description . | Map . | Locus link ID . | Level of expression . |
|---|---|---|---|
| Human myeloid differentiation | 1q21 | 4 170 | 839 |
| Human stem cell progenitor homeobox | 10q26 | 27 287 | 154 |
| Human myeloid elf-1 | Xq26 | 2 000 | 622 |
| Myeloperoxidase | 17q23 | 4 353 | 284 |
| Myelodisplasia/myeloid leukemia factor 2 | 12p13 | 8 079 | 317 |
| CD34 | 1q32 | 947 | 860 |
| Human stem cell leukemia exon 6 | 124 | ||
| Human AML1b protein | 21q22.3 | 861 | 481 |
| Flt3 | 13q12 | 2 322 | 1 417 |
| Breakpoint cluster region | 22q11.23 | 613 | 934 |
| MLL, translocated to 2 | 4q21 | 4 299 | 558 |
| MLL, translocated to 3 | 9p22 | 4 300 | 319 |
| MLL, translocated to 4 | 6q27 | 4 301 | 176 |
| MLL, translocated to 7 | Xq13.1 | 4 303 | 355 |
| Myeloid cell nuclear differentiation antigen | 1q22 | 4 322 | 125 |
| MLL, septinlike fusion | 17q25 | 10 801 | 1 259 |
| Myeloid differentiation primary response gene, 88 | 3p22 | 4 615 | 273 |
| Clathrin-8 assembly lymphoid-myeloid leukemia gene | 11q14 | 8 301 | 656 |
| SET translocation (myeloid leukemia associated) | 9q34 | 6 418 | 1 071 |
| Control genes | |||
| AFFX-HUMGAPDH | 12p13 | 2 597 | 4 511 |
| AFFX-HSACO (β-actin) | 7p15-p12 | 60 | 3 161 |
elf-1 indicates E74-like factor 1; AML, acute myeloid leukemia; MLL, mixed lineage leukemia; AFFX, affimetrix; HUMGAPDH, human GAPDH; HSACO, β-actin.
HGF receptor expression
HGF receptor analysis showed that (1) untreated blasts exhibited a characteristic pattern of HGF receptor expression, in which receptors for early-acting and multilineage HGF were expressed, while receptors for late-acting HGF were virtually absent; (2) blasts grown for 5 days with FL exhibited the expected down-modulation of flt3; no modifications in c-kit, IL-6R, or IL-3Rα; and a significant rise in the expression of all 4 unilineage HGF receptors (Figure 1B); and (3) incubation for 5 days with KL induced a similar phenomenon, with some quantitative differences, consisting of the induction of higher levels of EpoR and lower levels of G-CSFR and c-fms compared with those elicited by FL (Figure 1B).
In vitro growth factor response
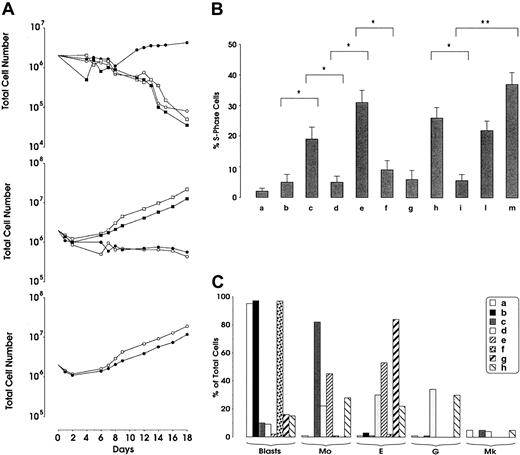
Studies performed with the use of a single HGF showed that only FL, but not other HGFs, had the capacity to sustain the proliferation (Figure 2A) and cell cycling (Figure 2B) of leukemic cells and protect them from apoptosis (data not shown). The combination of different HGFs revealed several peculiar findings: (1) The combination of late-acting HGFs was insufficient to support the growth (Figure 2A-B) and to protect from apoptosis leukemic cells (data not shown). (2) Similarly, multilineage HGF added alone or in combination with late-acting HGF neither stimulated the proliferation of leukemic cells (Figure 2A-B) nor protected them from apoptosis (data not shown). (3) Different combinations of early-acting HGF stimulated the proliferation of leukemic cells only when FL was present in the HGF cocktail (Figure 2A-B). (4) The combination of all early, multilineage, and late-acting HGFs maximally stimulated leukemic blast growth (Figure 2A-B).
Cell-growth and cell-cycle analysis of leukemic blasts and differentiation of leukemic blasts in liquid suspension culture supplemented with different HGFs.
(A) Cell-growth analysis of leukemic blasts. Leukemic blasts were grown in liquid suspension cultures in the presence of different HGFs. Top panel: (○) control; (●) FL; (■) KL, and (▪) IL-3. Middle panel: (○) Epo, Tpo, M-CSF, and G-CSF; (●) IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (■) FL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CF; (▪) KL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, G-CSF. Bottom panel: (○) FL, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (●) KL, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF. Mean values of 3 independent experiments are shown. (B) Cell-cycle analysis of leukemic blasts. Leukemic blasts were grown for 5 days in liquid suspension in the presence of different HGFs. (a) Control; (b) KL; (c) FL; (d) Epo, Tpo, M-CSF, and G-CSF; (e) FL, Epo, Tpo, M-CSF, and G-CSF; (f) KL, Epo, Tpo, M-CSF, and G-CSF; (g) IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (h) FL, KL, IL-6, and bFGF; (i) KL, IL-6, and bFGF; (l) FL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; and (m) KL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF. Mean + SD of 3 independent experiments is presented. *P < .05. **P < .01. (C) Differentiation of leukemic blasts in liquid suspension culture supplemented with different HGFs. Cells were identified on cytospins stained with May-Grünwald Giemsa and were classified as blasts, erythroid (E), monocytic (Mo), granulocytic (G), or megakaryocytic (MK) cells. (a) Control; (b) KL; (c) FL, KL, IL-6, and bFGF; (d) FL, IL-6, bFGF, IL-3, Epo, Tpo, M-CSF, and G-CSF; (e) KL, IL-6, bFGF, IL-3, Epo, Tpo, M-CSF, and G-CSF; (f) Epo, Tpo, M-CSF, and G-CSF; (g) KL, Epo, Tpo, M-CSF, and G-CSF; and (h) FL, Epo, Tpo, M-CSF, and G-CSF. A representative experiment is presented.
Cell-growth and cell-cycle analysis of leukemic blasts and differentiation of leukemic blasts in liquid suspension culture supplemented with different HGFs.
(A) Cell-growth analysis of leukemic blasts. Leukemic blasts were grown in liquid suspension cultures in the presence of different HGFs. Top panel: (○) control; (●) FL; (■) KL, and (▪) IL-3. Middle panel: (○) Epo, Tpo, M-CSF, and G-CSF; (●) IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (■) FL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CF; (▪) KL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, G-CSF. Bottom panel: (○) FL, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (●) KL, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF. Mean values of 3 independent experiments are shown. (B) Cell-cycle analysis of leukemic blasts. Leukemic blasts were grown for 5 days in liquid suspension in the presence of different HGFs. (a) Control; (b) KL; (c) FL; (d) Epo, Tpo, M-CSF, and G-CSF; (e) FL, Epo, Tpo, M-CSF, and G-CSF; (f) KL, Epo, Tpo, M-CSF, and G-CSF; (g) IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; (h) FL, KL, IL-6, and bFGF; (i) KL, IL-6, and bFGF; (l) FL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF; and (m) KL, IL-6, bFGF, IL-3, GM-CSF, Epo, Tpo, M-CSF, and G-CSF. Mean + SD of 3 independent experiments is presented. *P < .05. **P < .01. (C) Differentiation of leukemic blasts in liquid suspension culture supplemented with different HGFs. Cells were identified on cytospins stained with May-Grünwald Giemsa and were classified as blasts, erythroid (E), monocytic (Mo), granulocytic (G), or megakaryocytic (MK) cells. (a) Control; (b) KL; (c) FL, KL, IL-6, and bFGF; (d) FL, IL-6, bFGF, IL-3, Epo, Tpo, M-CSF, and G-CSF; (e) KL, IL-6, bFGF, IL-3, Epo, Tpo, M-CSF, and G-CSF; (f) Epo, Tpo, M-CSF, and G-CSF; (g) KL, Epo, Tpo, M-CSF, and G-CSF; and (h) FL, Epo, Tpo, M-CSF, and G-CSF. A representative experiment is presented.
In parallel, we evaluated the capacity of HGFs to induce the differentiation of leukemic blasts. Single HGF did not allow blasts to differentiate: blasts grown for 1, 2, or 3 weeks in the presence of FL maintained a blast morphology (Figure 2C) and did not display lineage-specific membrane antigens. Interestingly, the addition of early-acting HGFs induced, after 3 weeks of culture, a predominant monocytic differentiation, as shown by morphologic (Figure2C) and immunophenotypic criteria (the cells became CD14+). Addition of KL to multilineage plus unilineage HGFs elicited the differentiation of leukemic blasts into the monocytic and erythroid lineages (Figure 2C), while the addition of KL to unilineage HGFs elicited a selective erythroid differentiation (Figure 2C). The addition of FL to multilineage HGFs in combination with unilineage HGFs or to unilineage HGFs alone induced the differentiation of leukemic cells toward all 4 myeloid hemopoietic lineages (Figure2C).
Taken together, these findings confirm the close stem cell association of this case and further pointed to its likely early myeloid affiliation. It should also be noted that we observed the induction of endothelial cell markers (ie, CD105, CD106, CD62E, and CD62P) after in vitro culture of leukemic cells in the presence of FL and vascular endothelial growth factor.
Further studies on other cases of undifferentiated AL are required to verify whether the atypical pattern of HGF receptor expression and response to HGF and multilineage differentiation potential here reported represents a peculiar property of this leukemia subtype.
Supported by Istituto Superiore di Sanità Italy–USA, project on “Therapy of Tumors” and 1% project; Associazione Italiana per la Ricerca sul Cancro; and Ministero dell'Università e della Ricerca Scientifica.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Robin Foa, Department of Cellular Biotechnologies and Hematology, University of Rome “La Sapienza,” Via Benevento 6, 00161 Rome, Italy; e-mail: rfoa@bce.med.uniroma1.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal