Juvenile myelomonocytic leukemia (JMML) is an aggressive childhood disorder with few therapeutic options. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and tumor necrosis factor–α (TNF-α) promote JMML cell growth. A hyperactive function of the ras oncogene is a hallmark of JMML. We therefore targeted the protein kinase Raf-1 downstream of Ras using a DNA enzyme that degrades mRNA–Raf-1. Western blots of JMML cell lysates revealed phosphorylated Raf-1 protein, indicating constitutive activation. Addition of GM-CSF, but not TNF-α, increased phosphorylation of both Raf-1 and the mitogen-activated protein kinases (MAPKs) JNK-1 and ERK-1. Depletion of Raf-1 protein markedly impaired activation of MAPKs, induced substantial inhibition of JMML cell colony formation, and virtually abolished GM-CSF hypersensitivity in JMML cells. Exogenous TNF-α, but not GM-CSF, restored colony formation of JMML cells pretreated with the enzyme. We could not detect any effect of the enzyme on the proliferation of normal bone marrow cells, indicating its specificity and potential safety. When immunodeficient mice engrafted with JMML cells were treated continuously with the enzyme via a peritoneal osmotic mini-pump for 4 weeks, a profound reduction in the JMML cell numbers in the recipient murine bone marrows was found. We conclude that GM-CSF is a chief regulator of JMML growth and exerts its proleukemic effects primarily via the Ras/Raf-1 signaling cascade. TNF-α plays a permissive role, being dependent upon GM-CSF to induce JMML cell proliferation. The DNA enzyme efficiently catabolized mRNA–Raf-1 with subsequent inhibition of JMML cell growth, suggesting its potential as a mechanism-based therapy in this fatal leukemia.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare childhood malignancy with a poor prognosis.1Substantial improvement has not been observed in patients offered either conventional chemotherapy or treatment with retinoids.1,2 Only allogeneic bone marrow transplantation can induce durable remissions, but the relapse frequency is high and serious side effects are relatively common.3
A major feature of this disease is that the cytokines granulocyte-macrophage colony-stimulating factor (GM-CSF) and tumor necrosis factor–α (TNF-α) promote proliferation and viability of JMML cells in vitro. JMML cells cocultured with monocytes showed enhanced growth rates compared with monocyte-depleted cultures, and exogenous addition of these cytokines could expand JMML cell colony numbers markedly.4-7 Moreover, neutralizing anti–GM-CSF or anti–TNF-α monoclonal antibodies (MoAbs), diphtheria toxin fused to GM-CSF, or an antagonist directed at the GM-CSF receptor all decreased JMML cell growth, induced apoptosis, or both.6-8Furthermore, reduction of GM-CSF gene expression by DNA triplex formation using an oligonucleotide targeting a part of theGM-CSF gene promoter inhibited both spontaneous JMML cell colony formation as well as growth of JMML cells with added exogenous TNF-α.9 JMML cell growth and viability could also be reduced in vitro with an RNA ribozyme directed against TNF-α mRNA.10 Finally, simultaneous inhibition of GM-CSF and TNF-α in a nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mouse model of JMML abolished extramedullar dissemination and reduced the leukemic cell mass within the recipient murine bone marrow.11 Collectively, these data suggest that the cytokines TNF-α and GM-CSF clearly enhance the aggressiveness of JMML. However, although pathogenic mutations have been identified in the GM-CSF signal transduction pathway in some JMML patients, the full spectrum of aberrancies in signal transduction in JMML is not yet clear.
Cytokines signal via surface receptors and then through multiple intracellular cytoplasmic signaling systems to the nuclear effector mechanisms. The Ras family of proteins plays an important role in relaying these signals through the protein kinase Raf-1 and the mitogen-activated protein kinase (MAPK) cascades.12,13 The neurofibromin protein, encoded by the neurofibromatosis type 1 tumor suppressor gene (NF1), inhibits Ras by hydrolyzing active guanosine triphosphate (GTP)–Ras to the inactive guanosine diphosphate–bound form. Several independent findings link a dysregulated Ras signaling pathway to the pathogenesis of JMML. There are increased frequencies of both NF1 and RASgene mutations among JMML patients,14,15 and these groups appear to be mutually exclusive. Allogeneic transplantation of hematopoietic fetal liver cells from mice with a targeted disruption of the Nf1 gene led to a myeloproliferative disorder resembling JMML.16 Moreover, the loss of the Nf1 gene in these cells rendered them hypersensitive to GM-CSF, similar to JMML cells,16,17 and hematopoietic fetal liver cells lacking both the Nf1 and the GM-CSF genes were dependent on exogenous GM-CSF for efficient homing and proliferation in recipient murine bone marrows.18 Furthermore, inhibition of the prenylation of Ras, mandatory for Ras cell transforming activity, markedly inhibits JMML colony growth.19
These results suggest that GM-CSF is an important regulator of JMML cell growth and indicate that GM-CSF–induced proliferation is mediated via the Ras/Raf-1 signaling system. Specific Raf-1–related inhibition might therefore be a therapeutic option in this fatal childhood leukemia. The advent of catalytic nucleotides, such as ribozymes and DNA enzymes, offers a unique opportunity to selectively inhibit synthesis of individual proteins by degradation of their mRNAs.20 In the present study, we used a DNA enzyme designed to specifically cleave mRNA for Raf-1. The enzyme was tested on JMML cells cultured both in vitro and in a xenograft model of JMML.
Materials and methods
Synthesis of DNA enzymes and in vitro cleavage activity
The Raf-1 enzyme (5′-tatgtgctccaGGCTAGCTACAACGAtgatgca-3′) and its inactive form were chemically synthesized by Eurogentec (Seraing, Belgium). The lower-case letters represent the phosphorothioate sequence (antisense arms), and the upper-case letters denote the phosphodiester sequence (catalytic core). The inactive enzyme has the same sequence as the active enzyme, but with reversed arms. The targeted mRNA–Raf-1 was synthesized by in vitro transcription from a cloned polymerase chain reaction product. In brief, total RNA from the breast cancer cell line T47D was prepared and reversed transcribed with oligo dT as the reverse primer. A part of the mRNA–Raf-1 sequence was amplified using specific primers: forward, 5′-TAAGCTGCATCAATGGAGCA-3′, and reverse, 5′-TGGGAGAGGGAACCTTCAGA-3′. The amplicon was expected to have 770 base pairs. Amplified DNA was agarose gel purified and cloned into the pGEM-T vector (Promega, Madison, WI), and positive clones were sequenced. Plasmid from one of the positive clones was prepared, linearized with SalI restriction enzyme, phenol extracted, precipitated, dissolved in water to a concentration of 5 μg/μL, and then used as template for in vitro transcription using the T7 RNA polymerase.
The DNA enzyme (100 nM) and the in vitro–transcribed RNA (900 nM) were mixed in a reaction buffer containing 50 mM Tris, pH 7.5, and 10 mM MgCl2. The reaction was performed at 37°C for various time periods. Following cleavage, a stop solution was added, and then samples were analyzed by 10% polyacrylamide gels containing 7 M urea.
Sampling of donor cells
The study was approved by the respective Institutional Review Boards, and parental consent was obtained. We studied 8 children, aged 9 months to 3 years, with JMML classified according to established criteria.21 Samples from 6 healthy age-matched children were also included. Bone marrow cells were obtained from aspirates of the iliac crest and processed as described.7 Briefly, mononuclear cells were isolated by sedimentation of erythrocytes, followed by density centrifugation. Further purification of JMML cells was achieved by incubating them with MoAbs directed against the GM-CSF and TNF-α receptors, and magnetic sorting. Before further analyses, both JMML cells and normal bone marrow cells were kept in supplemented liquid RPMI 1640 medium.
Transfection of cells, colony formation, and cytokine and mRNA measurements
We used cationic liposomes (25 μg/mL, DOTAP; Boehringer Mannheim, Mannheim, Germany) alone or complexed with the Raf-1 DNA enzyme or its inactive form. Cells were transfected with the liposomes for 10 to 15 hours in liquid culture before they were plated in semisolid methylcellulose to grow for 2 weeks. Colonies (>40 cells per clone) were then stained supravitally and scored.10
Aliquots were sampled from the liquid medium after a 24-hour culture, and the concentrations of GM-CSF, TNF-α, and granulocyte colony-stimulating factor (G-CSF) were determined using enzyme-linked immunosorbent assay kits, according to the instructions of the manufacturer (sensitivity >0.5 pg/mL; R&D Systems, Minneapolis, MN).
To determine mRNAs, we used RNAse protection assays after extraction of total RNA, as described.9 We used mRNA–reduced glyceraldehyde phosphate dehydrogenase (GAPDH) as an internal standard.
Immunoprecipitation, Western blotting, and kinase assays
Immunoprecipitation of intracellular molecules and Western blotting of phosphorylated molecules were performed according to methods described previously.22 Briefly, 10 million cells per milliliter sample were incubated in liquid medium with or without cytokines for selected time periods before they were harvested and lysed. Total Raf-1, JNK-1, and ERK-1 proteins as well as their respective phosphorylated forms were immunoprecipitated with MoAbs (Pharmingen, San Diego, CA) and resolved with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 7.5%). Phosphorylation of the immunoprecipitated molecules was examined using an antiphosphotyrosine MoAb (Boehringer Mannheim) and an enhanced chemiluminescence detection kit (ECL; Amersham, Little Chalfont, United Kingdom).
The activities of JNK-1 and ERK-1 were measured as described.22 In short, the JMML samples (107cells/mL) were cultured with GM-CSF (10 ng/mL) or TNF-α (10 ng/mL). The cells were lysed, and the supernatant was immunoprecipitated with either an anti–JNK-1 or an anti–ERK-1 MoAb (1 μg/mL; Pharmingen). The immunoprecipitates were then adsorbed to protein A–Sepharose (Pierce, Rockford, IL) before washing in lysis buffer and kinase buffer. Next, the immunocomplexes were resuspended in kinase buffer with 20 μCi (0.74 MBq) [γ-32P]-ATP (adenosine triphosphate; Amersham) and 20 μg of either the fusion protein c-Jun [1-169]–glutathione-S–transferase (GST) (Upstate Biotechnology, Lake Placid, NY) or myelin basic protein (Sigma, St Louis, MO). A 30-minute kinase reaction (30°C) was allowed before termination with an equal volume of Laemmli sample buffer. Phosphorylation of products was examined after SDS-PAGE and quantified with a PhosphoImager (Molecular Dynamics, Sunnyvale, CA).
Transplantation of JMML cells into NOD/SCID mice and phenotyping of engrafted cells
Immunodeficient mice underwent transplantation with primary JMML bone marrow cells, as described.11 Four weeks after transplantation, an osmotic mini-pump (0.25 μL/h; Alzet, Palo Alto, CA) was inserted into the peritoneal cavity. The pump contained 1 of 2 DNA enzymes (active or inactive; 10 μg/d) or saline, and the mice were randomly assigned to 1 of these 3 experimental groups. After death, the femoral bone marrows from the recipient mice were removed, stained with a human-specific anti-CD45 MoAb, and sorted with flow cytometry (FACScan; Becton Dickinson, Mountain View, CA). The extent of engraftment was assessed as the fraction of CD45+cells among the total number of nucleated bone marrow cells. We have previously shown that this measure correlates well with values obtained from either Southern blotting with a human-specific DNA probe or fluorescence in situ hybridization using specific human chromosomal markers.11
Statistics
Each measurement was made in triplicate, and the resulting median value was used to calculate the mean and SEM for the 8 JMML patients and the 6 healthy controls. Differences were evaluated with 2-tailed Wilcoxon rank sum tests and assumed significant for P < .05.
Results
In vitro functional analysis of the DNA enzyme
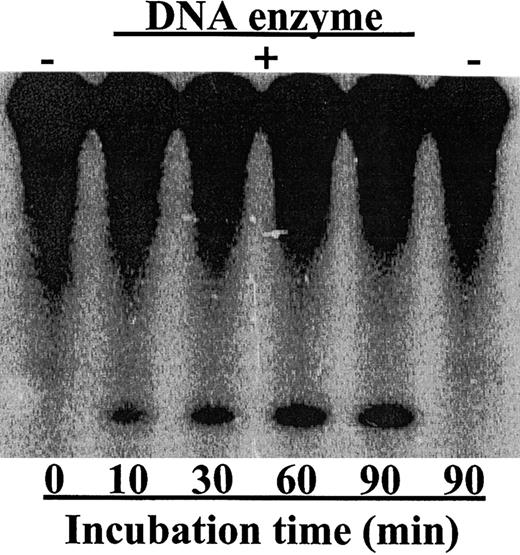
The catalytic core of the DNA enzyme was derived from the in vitro selected 10-23 DNA enzyme.20 This catalytic motif can recognize and cleave mRNA sequences at a phosphodiester bond located between unpaired purine and paired pyrimidines. For the RNA ribozymes,20 the specificity of DNA enzymes was also determined by their antisense arms, which bind to target mRNA through Watson-Crick base pairing. In vitro screening experiments using AU- and GU-cleaving DNA enzymes with random antisense arms identified potential RNA sites that were accessible to DNA enzyme binding and cleavage (data not shown). In this study, we targeted a site located at the translation start AUG. To increase the DNA enzyme stability in biologic fluids, we replaced the hydroxy groups of the phosphate backbone within the DNA antisense arms with sulfur atoms to make it a phosphorothioate-modified DNA enzyme. Incubation of the in vitro–transcribed mRNA–Raf-1 with the DNA enzyme, which was designed to cleave its 5′-end, resulted in significant cleavage activity (Figure1). This result underscores the in vitro accessibility of the targeted site.
The DNA enzyme efficiently cleaved its target mRNA–Raf-1.
The DNA enzyme was incubated with internally labeled 770 nt mRNA–Raf-1. Samples were taken at indicated time points and then analyzed with 10% polyacrylamide gel. The active DNA enzyme degraded 32P internally labeled mRNA–Raf-1 in vitro. No enzyme was added in the left and right lanes. The upper bands represent the Raf-1 mRNA, while the lower bands represent the 5′-cleavage product.
The DNA enzyme efficiently cleaved its target mRNA–Raf-1.
The DNA enzyme was incubated with internally labeled 770 nt mRNA–Raf-1. Samples were taken at indicated time points and then analyzed with 10% polyacrylamide gel. The active DNA enzyme degraded 32P internally labeled mRNA–Raf-1 in vitro. No enzyme was added in the left and right lanes. The upper bands represent the Raf-1 mRNA, while the lower bands represent the 5′-cleavage product.
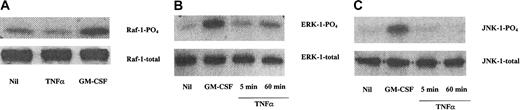
GM-CSF, but not TNF-α, increased Raf-1 activation in JMML cells
The activation status of the Raf-1, ERK-1, and JNK-1 kinases was determined by their phosphorylation patterns in JMML cell lysates. Figure 2A shows that the Raf-1 protein was constitutively phosphorylated in JMML cells, and this activation could be further enhanced upon addition of GM-CSF, but not TNF-α. Similarly, the MAPKs ERK-1 and JNK-1 were also present within the JMML cells, and GM-CSF, but not TNF-α, increased the phosphorylation of both molecules (Figure 2B,C).
GM-CSF, but not TNF-α, increased Raf-1 activation in JMML cells.
(A) Raf-1 was constitutively phosphorylated in JMML cells, and this activation was further enhanced upon adding GM-CSF (5 minutes, 10 ng/mL), but not TNF-α (5 minutes, 10 ng/mL). Similarly, the weak but constitutive activation of the MAPKs ERK-1 (B) and JNK-1 (C) was increased upon addition of GM-CSF (10 ng/mL, 5 minutes), but not TNF-α (10 ng/mL). The data are from immunoprecipitated proteins of JMML cell lysates and combined with Western blotting of their phosphorylated forms (denoted PO4). The total amounts of the proteins were equal (bottom lanes). The data are from one experiment on one JMML case and are representative of 7 other JMML cases.
GM-CSF, but not TNF-α, increased Raf-1 activation in JMML cells.
(A) Raf-1 was constitutively phosphorylated in JMML cells, and this activation was further enhanced upon adding GM-CSF (5 minutes, 10 ng/mL), but not TNF-α (5 minutes, 10 ng/mL). Similarly, the weak but constitutive activation of the MAPKs ERK-1 (B) and JNK-1 (C) was increased upon addition of GM-CSF (10 ng/mL, 5 minutes), but not TNF-α (10 ng/mL). The data are from immunoprecipitated proteins of JMML cell lysates and combined with Western blotting of their phosphorylated forms (denoted PO4). The total amounts of the proteins were equal (bottom lanes). The data are from one experiment on one JMML case and are representative of 7 other JMML cases.
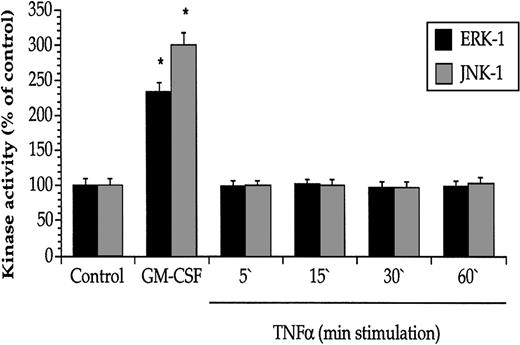
The lack of effect on Raf-1/MAPK activation by TNF-α was further explored with specific kinase activity assays. Whereas GM-CSF exerted a marked stimulatory effect on both ERK-1 and JNK-1 activity, TNF-α had no effect (Figure 3), even if a 10-fold amount of TNF-α was added (data not shown).
GM-CSF, but not TNF-α, enhanced the activities of the MAPKs ERK-1 and JNK-1 in JMML cells.
GM-CSF (5 minutes, 10 ng/mL), but not TNF-α (10 ng/mL), increased the kinase activity of ERK-1 and JNK-1 derived from JMML cells. Values are the means + SEM, n = 8. *P < .05 compared with control.
GM-CSF, but not TNF-α, enhanced the activities of the MAPKs ERK-1 and JNK-1 in JMML cells.
GM-CSF (5 minutes, 10 ng/mL), but not TNF-α (10 ng/mL), increased the kinase activity of ERK-1 and JNK-1 derived from JMML cells. Values are the means + SEM, n = 8. *P < .05 compared with control.
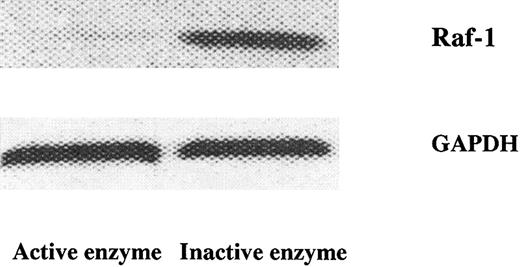
The DNA enzyme reduced Raf-1 gene expression and colony formation of JMML cells, but not of normal bone marrow progenitor cells
Figure 4 shows that the active DNA enzyme markedly impaired the expression of the Raf-1 gene in the JMML cells, whereas the inactive form was without any apparent effect.
DNA enzyme–mediated catabolism of mRNA–Raf-1 in JMML cells.
The active, but not the inactive, DNA enzyme (1 μg/mL) reduced mRNA–Raf-1 in JMML cells. The control mRNA-GAPDH did not change appreciably upon enzyme treatment. The results were obtained from RNAse protection assays and are from one experiment on one JMML case and representative of 7 other JMML cases.
DNA enzyme–mediated catabolism of mRNA–Raf-1 in JMML cells.
The active, but not the inactive, DNA enzyme (1 μg/mL) reduced mRNA–Raf-1 in JMML cells. The control mRNA-GAPDH did not change appreciably upon enzyme treatment. The results were obtained from RNAse protection assays and are from one experiment on one JMML case and representative of 7 other JMML cases.
We next tested the DNA enzyme on the proliferative capacity of JMML cells in colony assays. As expected, the JMML cells spontaneously formed a large number of colonies, and this was further enhanced upon addition of GM-CSF and TNF-α, but not G-CSF (Table1). Importantly, the active DNA enzyme markedly reduced the colony scores, whereas its inactive form was without effect, thus confirming the specificity of the treatment.
The DNA enzyme inhibited spontaneous JMML cell colony formation
| Added compound . | Colony numbers . |
|---|---|
| Nil | 94 ± 6 |
| DOTAP | 92 ± 8 |
| Active DNA enzyme | 33 ± 4* |
| Inactive DNA enzyme | 93 ± 8 |
| GM-CSF | 297 ± 34† |
| TNF-α | 185 ± 19† |
| G-CSF | 90 ± 8 |
| Active DNA enzyme + GM-CSF | 55 ± 6* |
| Active DNA enzyme + TNF-α | 63 ± 8* |
| Active DNA enzyme + G-CSF | 39 ± 6* |
| Added compound . | Colony numbers . |
|---|---|
| Nil | 94 ± 6 |
| DOTAP | 92 ± 8 |
| Active DNA enzyme | 33 ± 4* |
| Inactive DNA enzyme | 93 ± 8 |
| GM-CSF | 297 ± 34† |
| TNF-α | 185 ± 19† |
| G-CSF | 90 ± 8 |
| Active DNA enzyme + GM-CSF | 55 ± 6* |
| Active DNA enzyme + TNF-α | 63 ± 8* |
| Active DNA enzyme + G-CSF | 39 ± 6* |
Values are means ± SEM (n = 8) of the numbers of colonies per 75 000 plated JMML cells. The cytokines GM-CSF, TNF-α, and G-CSF were all added at concentrations of 10 ng/mL, whereas the DNA enzymes were added at concentrations of 1 μg/mL.
GM-CSF indicates granulocyte-macrophage colony-stimulating factor; TNF-α, tumor necrosis factor-α; G-CSF, granulocyte colony-stimulating factor.
Denotes a value significantly less than for the Nil group (P < .05).
Denotes a value significantly larger than for the Nil group (P < .05).
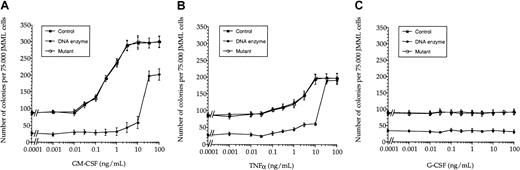
Given the known hypersensitivity of JMML cells to GM-CSF,5 we examined this in JMML cells cocultured with cytokines and the active and inactive DNA enzymes. Figure5A indicates that this GM-CSF hypersensitivity was virtually lost upon treatment with the active DNA enzyme, being only partially restored with high concentrations of exogenous GM-CSF. JMML cells did not show any apparent hypersensitivity to TNF-α (Figure 5B). However, in contrast to the GM-CSF cultures, high doses of TNF-α apparently restored colony formation of JMML cells pretreated with the active DNA enzyme. G-CSF had no effect on JMML cell proliferation (Figure 5C). A 5-fold higher dose of the active DNA enzyme did not change these findings appreciably (data not shown).
DNA enzyme treatment abolished GM-CSF hypersensitivity in JMML cells.
Whereas JMML cells showed hypersensitivity to GM-CSF (A), this could not be detected for either TNF-α (B) or G-CSF (C). The values (mean ± SEM, n = 3) are shown as dose-response curves based on colony scores from one JMML case and are representative of 7 other JMML cases.
DNA enzyme treatment abolished GM-CSF hypersensitivity in JMML cells.
Whereas JMML cells showed hypersensitivity to GM-CSF (A), this could not be detected for either TNF-α (B) or G-CSF (C). The values (mean ± SEM, n = 3) are shown as dose-response curves based on colony scores from one JMML case and are representative of 7 other JMML cases.
The lack of effect of the DNA enzymes on the formation of colony subsets by normal bone marrow progenitor cells is shown in Table2. This suggests the specificity and possible safety of this DNA enzyme–based treatment of JMML.
The DNA enzyme had no effect on colony formation of normal bone marrow cells
| Compound . | Colony type . | |||
|---|---|---|---|---|
| G . | GM . | M . | Total . | |
| Nil | 10 ± 3 | 34 ± 7 | 40 ± 8 | 88 ± 6 |
| DOTAP | 11 ± 4 | 41 ± 8 | 37 ± 9 | 90 ± 7 |
| Active DNA enzyme | 13 ± 4 | 41 ± 8 | 39 ± 8 | 91 ± 8 |
| Inactive DNA enzyme | 8 ± 2 | 38 ± 9 | 37 ± 7 | 88 ± 6 |
| Compound . | Colony type . | |||
|---|---|---|---|---|
| G . | GM . | M . | Total . | |
| Nil | 10 ± 3 | 34 ± 7 | 40 ± 8 | 88 ± 6 |
| DOTAP | 11 ± 4 | 41 ± 8 | 37 ± 9 | 90 ± 7 |
| Active DNA enzyme | 13 ± 4 | 41 ± 8 | 39 ± 8 | 91 ± 8 |
| Inactive DNA enzyme | 8 ± 2 | 38 ± 9 | 37 ± 7 | 88 ± 6 |
Values are colony numbers and given as the means ± SEM (n = 6). We cultured 100 000 bone marrow cells supplemented with G-CSF (10 ng/mL) and stem cell factor (10 ng/mL).
G indicates granulocyte; GM, granulocyte/macrophage; M, macrophage.
The DNA enzyme inhibited GM-CSF and TNF-α cytokine expression and production in JMML cells
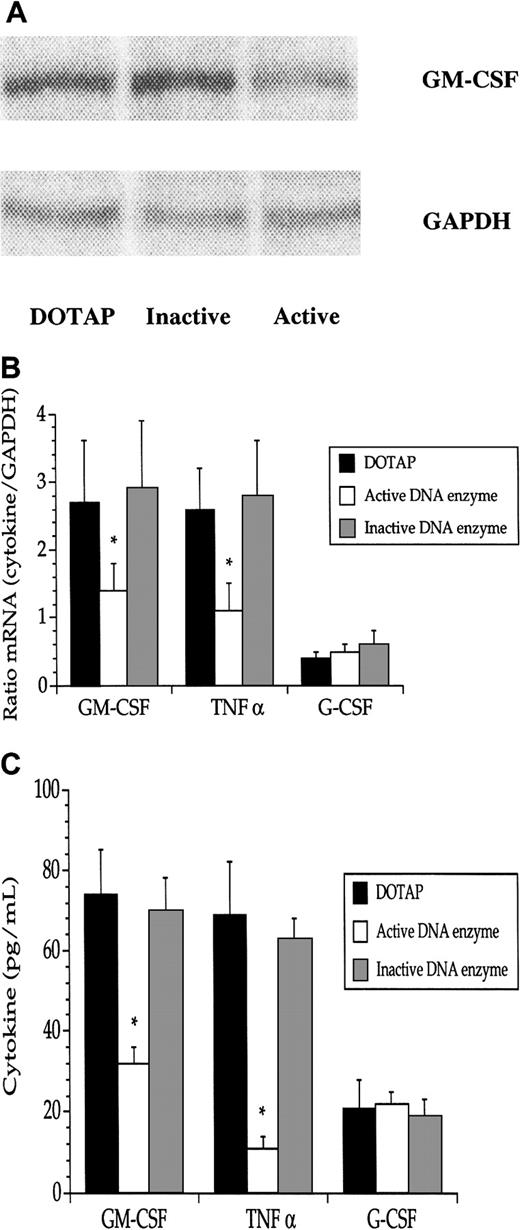
A hitherto unexplained link between GM-CSF and TNF-α has been suggested as a pathogenic mechanism in JMML.10 The activation of Raf-1 by GM-CSF, but not TNF-α (Figures 2 and 3), prompted us to investigate the levels of GM-CSF and TNF-α in JMML cells following treatment with the Raf-1 DNA enzyme. The active DNA enzyme exerted a marked repression of the mRNAs for both GM-CSF and TNF-α, but not for mRNA–G-CSF (Figure6A,B). JMML cell production of both GM-CSF and TNF-α protein, but not of G-CSF protein, was accordingly diminished (Figure 6C). There were no significant differences between GM-CSF and TNF-α in the relative reductions of either the transcripts or proteins.
The DNA enzyme reduced both GM-CSF mRNA and protein in JMML cells.
(A) A decrease of mRNA–GM-CSF was noted after addition of the active, but not the inactive, DNA enzyme (1 μg/mL). Data are from one experiment of one JMML case. Pooled data (means + SEM) from the 8 JMML cases show significant (*) reductions in mRNA (B) and protein (C) for GM-CSF and TNF-α, but not for G-CSF. The data in A and B were obtained from RNAse protection assays.
The DNA enzyme reduced both GM-CSF mRNA and protein in JMML cells.
(A) A decrease of mRNA–GM-CSF was noted after addition of the active, but not the inactive, DNA enzyme (1 μg/mL). Data are from one experiment of one JMML case. Pooled data (means + SEM) from the 8 JMML cases show significant (*) reductions in mRNA (B) and protein (C) for GM-CSF and TNF-α, but not for G-CSF. The data in A and B were obtained from RNAse protection assays.
Antileukemic effect of the DNA enzyme given to immunodeficient mice engrafted with JMML cells
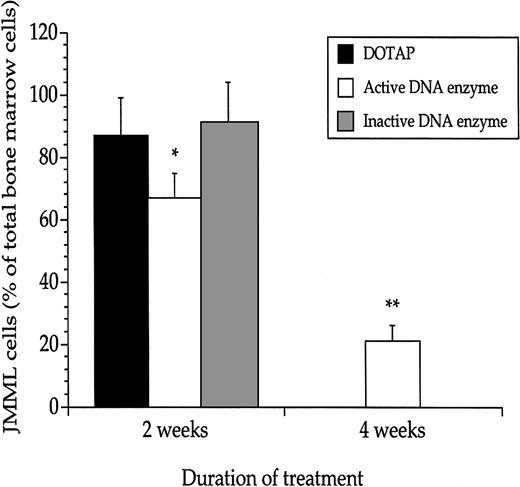
We employed the previously described xenograft model of JMML to test the efficacy of the active DNA enzyme in inhibiting leukemogenesis.11 Four weeks after transplantation, the mice developed overt leukemia (data not shown). At that time, a 2-week continuous treatment with the active DNA enzyme commenced, which led to a reduction in the JMML cell mass, as evidenced by a reduction from about 90% to 70% of the total number of nucleated femoral bone marrow cells (Figure 7). Importantly, mice receiving treatment with active DNA enzyme for a total of 4 weeks had a further substantial reduction in the leukemic cell mass to about 20%. All mice treated for 4 weeks with the active DNA enzyme survived; however, untreated mice or those treated with the inactive DNA enzyme all died of leukemia between 2 to 3 weeks after implantation of the peritoneal pump. Thus, none of the mice in these 2 latter groups were alive at the 4-week analysis (Figure 7).
The DNA enzyme reduced the JMML cell mass in vivo.
Enumeration of JMML cells in recipient mouse femoral bone marrows revealed that continuous treatment with the active DNA enzyme decreased the leukemic cell mass and reduced the mortality. Values are means and SEM based on mice undergoing transplantation with JMML cells from 8 patients (3 mice per JMML patient). *P < .05; **P < .01 when compared with mice treated with DOTAP only.
The DNA enzyme reduced the JMML cell mass in vivo.
Enumeration of JMML cells in recipient mouse femoral bone marrows revealed that continuous treatment with the active DNA enzyme decreased the leukemic cell mass and reduced the mortality. Values are means and SEM based on mice undergoing transplantation with JMML cells from 8 patients (3 mice per JMML patient). *P < .05; **P < .01 when compared with mice treated with DOTAP only.
Discussion
JMML is a rare hematologic malignancy with few therapeutic options and a high mortality rate.1 There is considerable evidence linking GM-CSF stimulation of a dysregulated Ras pathway to the pathogenesis of JMML.4-8,14-19 Recent studies have revealed a crucial role of the MAPKs, downstream of Ras, in the proliferation and survival of many cell types.23 Hence, there is a rationale for a mechanism-based therapy targeting the Ras/Raf-1/MAPK pathway in this disease.24
For many chemical drugs, such as those directed against Ras, the mechanism of action is not well defined. By contrast, the specificity of Watson-Crick hybridization is the basis for rational drug design of nucleic acid enzymes, including ribozymes.25,26 Because of Watson-Crick base pairing, it is possible to specifically inhibit the expression of related proteins such as isoenzymes.27Specific targeting of Raf-1 gene expression by a DNA enzyme inhibited JMML cell growth both in vitro and in vivo. Thus, the Raf-1 signaling pathway seems to be important in JMML.
GM-CSF exerts its biologic effects by first binding to its cognate receptor, resulting in activation of intracellular signaling pathways. Although it has been shown that mutated GM-CSF receptor subunits can induce autonomous proliferation of various cell lines, pathogenic mutations in the genes coding for the α and the β common subunits of the GM-CSF receptor in primary JMML cells have not yet been identified.28 Moreover, in addition to the inducible assembly of the 2 subunits of the GM-CSF receptor upon ligand binding, a pool of preformed receptor complexes is also present and imparts constitutive signaling.29 Whether this pool is enhanced in malignancies such as JMML is not known.
GM-CSF ligand-receptor binding stimulates the Ras pathway at least partly via the Shc adaptor protein, so that the active Ras-GTP–bound form can associate with the inner surface of the cell membrane.30,31 This reaction is necessary for activation of molecules downstream of Ras, such as Raf-1 and the MAPKs, and it requires addition of a farnesyl group to Ras through a prenyl reaction. We recently showed that addition of specific farnesyl transferase inhibitors reduces JMML cell proliferation in vitro.19Whether farnesyl transferase inhibitors are specific forRAS-mutated cells and whether they can be effective therapies for JMML patients are the subjects of an active JMML clinical protocol (no. AAML0122) in the Children's Oncology Group in North America.
It has been shown that GM-CSF can phosphorylate Raf-1 in hematopoietic cells and thereby activate the MAPK signaling cascade, leading to increased proliferation and cell survival.12,13 In contrast, TNF-α reportedly activates MAPK molecules in a Raf-1–independent way in monocytic cells.32 33 Hence, targeting the Raf-1 gene could be beneficial in reducing JMML cell proliferation and provide us with clues about the aberrant GM-CSF/TNF-α signaling pathways in JMML. The finding that Raf-1 was phosphorylated in unstimulated JMML cells indicated constitutive activation. This activation was enhanced upon addition of GM-CSF, but not TNF-α. We also demonstrated increased phosphorylation of both ERK-1 and JNK-1 in JMML cells treated with GM-CSF, but not with TNF-α. Moreover, a stimulating activity of GM-CSF, but not of TNF-α, was noted in specific kinase assays. Therefore, it seems that the activation of Raf-1 in these cells is predominantly mediated by GM-CSF.
To further explore the role of GM-CSF in stimulating Raf-1 in JMML, we treated the JMML cells with a DNA enzyme designed to selectively cleave mRNA–Raf-1 and thereby block protein synthesis. This active DNA enzyme efficiently reduced the Raf-1 gene expression in JMML cells, leading to a reduced proliferation in these cells. In contrast, growth of normal bone marrow progenitor cells was not affected, and addition of an inactive DNA enzyme had no effect on either JMML cells or normal bone marrow progenitor cells in any assay. These data indicate that this DNA enzyme may eventually prove to be a specific, and potentially safe, therapeutic agent. Using growth-kinetic studies and colony assays of JMML cells treated with the active DNA enzyme, we demonstrated an apparent loss of GM-CSF hypersensitivity, the most consistent hallmark of this disease.5 In contrast to these findings, JMML cells showed no hypersensitivity to TNF-α, and high amounts of exogenous TNF-α, but not GM-CSF, could restore proliferation in JMML cells pretreated with the active DNA enzyme. Addition of the active DNA enzyme also reduced both the GM-CSF and TNF-α transcripts as well as the endogenous production of these proteins in JMML cells. In a final set of experiments, we tested the efficacy of the active DNA enzyme in vivo using immunodeficient mice engrafted with primary JMML cells. We could show that the continuous treatment of the leukemic mice with the active DNA enzyme markedly reduced the leukemic cell mass in the bone marrow, and it apparently reduced mortality due to the leukemia.
Collectively, these findings strongly suggest that GM-CSF is a chief regulator in JMML cell proliferation. The data also argue that the stimulatory action of GM-CSF is at least partly conveyed via the Ras/Raf-1/MAPK signaling cascade. In the present study, we included JMML patients who had either RAS or NF1 gene mutations, as well as some who had neither mutation. The inhibitory effect of the DNA enzyme was similar regardless of specific mutational status. This indicates that targeted therapeutic inhibition of Raf-1 can potentially be accomplished regardless of the mutation that activates the Ras pathway upstream of Raf-1. Whether there is a possible additional dysregulated activation of the JAK/STAT pathway,34 or aberrant cross-talk between the Ras/Raf-1 and JAK/STAT pathways upon GM-CSF stimulation in JMML cells, is not known. Although the action of TNF-α in JMML is still not exactly defined, our present findings, as well as the results of previous studies,6,9 10 indicate that TNF-α plays a more permissive role in this disease, being dependent on GM-CSF to stimulate JMML cell growth.
In conclusion, the active DNA enzyme effectively led to the degradation of mRNA–Raf-1 with disruption of the downstream MAPK signaling cascade and the subsequent inhibition of JMML cell growth. Whether this mechanism-based approach holds promise as a clinical therapeutic in this fatal childhood leukemia warrants further examination.
Supported by the Norwegian Cancer Society, the Norwegian Research Council, and the Throne Holst Foundation, and by National Institutes of Health grant CA80916.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Per Ole Iversen, Institute for Nutrition Research, University of Oslo, PO Box 1046 Blindern, 0316 Oslo, Norway; e-mail:poiversen@hotmail.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal