Fusion between the NUP98 and NSD3genes in a patient with acute myeloid leukemia associated with t(8;11)(p11.2;p15), is reported for the first time. The t(8;11)(p11.2;p15) was identified by classical cytogenetics. Fluorescence in situ hybridization (FISH) analysis revealed a split signal with a mix of BAC 118H17 and 290A12, indicating the translocation disrupted NUP98. FISH restriction at 8p11-12 showed a split of BAC 350N15. Molecular investigations into candidate genes in this BAC showed the NUP98 fusion partner at 8p11.2 was the NSD3 gene. To date the NSD3 gene has never been implicated in hematologic malignancies.
Introduction
In myeloid malignancies several chromosomal translocations and corresponding chimeric genes have been identified between NUP98 at 11p15 and PMX1 at 1q23,HOXD13 at 2q31, NSD1 at 5q35, HOXA9 at 7p15, LEDGF at 9p22, DDX10 at 11q22, andTOP1 at 20q11.1-8 In lymphoid malignancies only a subset of T-cell acute lymphoblastic leukemia with a t(4;11)(q21;p15) and a NUP98-RAP1GDS1 fusion gene has been reported to date.9
Chromosomal bands 8p11-p12 are involved in well-characterized leukemic syndromes, such as acute myeloid leukemia (AML) with erythrophagocytosis and t(8;16)(p11;p13.3)10and a chronic myeloproliferative disorder (CMPD) with eosinophilia, concomitant lymphoblastic leukemia–lymphoma, and t(8;13)(p11;q12).11 As a result of translocations,MOZ fuses with the CBP gene at 16p13.3 in AML, and FGFR1 rearranges with ZNF198 at 13q12 in the CMPD–lymphoma syndrome. Alternative recombinations withTIF2/8q13 and p300/22q13 for MOZ gene and with FOP/6q27 and CEP110/9q33 forFGFR1 have also been described.12-16
We report the first patient with AML with recombination between the NUP98 gene and a new partner at 8p11.2, telomeric toFGFR1. NSD3 (nuclear receptor-binding su(var)3-9, enhancer of zeste, trithorax [SET] domain containing gene 3), also named WHSC1L1 (Wolf-Hirschhorn syndrome candidate-1–like-1), belongs to the same family asNSD1, whose mouse homolog is an RARα-interacting protein,17 and NSD2, named WHSC1(Wolf-Hirschhorn syndrome candidate-1) or MMSET(multiple myeloma SET domain).
Study design
Patient history
A 65-year-old man was admitted because of fever and malaise. Clinical examination disclosed slight hepatomegaly. Hematologic data were as follows: hematocrit, 22%; hemoglobin level, 9 g/dL; leukocyte count, 159 × 109/L with 84% blast cells, 5% promyelocytes, 3% myelocytes, 2% neutrophils, 2% basophils, 1% eosinophils, and 3% lymphocytes; platelet count, 81 × 109/L. Bone marrow aspirate was indicative of AML, M1-FAB, with multilineage dysplasia. Blast cells were CD7+, CD13+, CD15+, CD33+, CD34+, HLA-DR+ and CD2−, CD3−, CD19−, CD41−, CD61−. The patient died of septic shock during drug-induced marrow aplasia.
Cytogenetics and fluorescence in situ hybridization
Karyotyping was performed on unstimulated bone marrow cells after short-term culture. Chromosomes were G-banded and classified according to the International System for Human Cytogenetic Nomenclature.18 Fluorescence in situ hybridization (FISH) studies were performed as described elsewhere.19 We used a mixture of 2 BAC clones (118H17 and 290A12) labeled with biotin for theNUP98 gene.19
The 8p11-12 breakpoint was investigated using YAC 176C9 (Dr M. Chaffanet, Marseilles, France) spanning the MOZ gene, BAC 350N15 spanning the FGFR1 gene, and BAC 513D5 telomeric to the FGFR1 locus. Probes were biotin-labeled. A centromeric probe labeled with digoxigenin was added in each experiment for chromosome 8 (D8Z1) or for chromosome 11 (D11Z1) (Oncor, Gaithersburg, MD). At least 8 metaphases were analyzed using an Olympus fluorescence microscope equipped with a cooled CCD camera (Sensys, Photometrics, Tucson, AZ) run by Pathvysion (Vysis, Stuttgart, Germany).
Molecular characterization
Database searches and sequence analyses were performed using BLAST algorithms (http://www.ncbi.nlm.nih.gov/BLAST) and RepeatMasker (http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker). AllNUP98 primers have been described previously19except NUP737F, AGTCACTAGAGGAACTTCG. NSD3 primers were designed on GenBank accession number AK022560 (FLJ536f, CAGAAATTCCAAACACAAGAC; FLJ940f, CCAGTTCAGCCAATACTATC; FLJ1856Af, AGCCAACGCAGAGTGTATCATC; FLJ2907Br, GGTGGGCCTCGTTAGACTGCTC; FLJ3081Cr, TTCCCCCAAAATTTCCATACAA; FLJ1204Df, GAAGAATTACTGGCTGAGGCAAC; FLJ1747Er, ATTTCCCATCCCCTGTAGCATTC) and GenBank accession number AF332469(W1L2863R1, AATTTGCGGACACAGGCTTC; W1L2685R2, TGCAGCACTCTCCCTCAC).
Total RNA was reverse-transcribed using SuperscriptII (Life Technologies, Gaithersburg, MD). Using the Expand Long Template PCR System (Roche, Mannheim, Germany) and the appropriate primers, cDNA aliquots were amplified in 10-μL polymerase chain reaction (PCR) as follows: 35 amplification cycles at 94°C for 5 minutes 10 seconds (94°C for 50 seconds, 58°C for 20 seconds, and 68°C for 4 minutes) and at 68°C for 5 minutes.
Nested PCR was carried out as follows: 35 amplification cycles at 94°C for 5 minutes (94°C for 30 seconds, 58°C for 20 seconds, 68°C for 2 minutes) and at 68°C for 5 minutes. Sequencing was performed with an automated ABI377 sequencing station after purification using Concert Rapid PCR Purification System (Life Technologies).
Results and discussion
The t(8;11)(p11.2;p15) translocation was found as an isolated anomaly in all 20 bone marrow karyotypes from this patient with de novo AML. The mixture of BAC 118H17 and 290A12 was split, thus generating 3 spots of fluorescence: on normal chromosome 11, on der(11), and on der(8) (Figure 1A, left panel). BAC 350N15 spanned the 8p11-12 breakpoint, showing hybridization signals on normal chromosome 8, on der(8), and on der(11) (Figure 1A, right panel).
FISH experiments and molecular characterization of fusion transcripts.
(Ai) FISH with probe D11Z1 (red) for the alpha satellite of centromere 11 and a mixture of BAC 118H17 and BAC 290A12 (green), to encompass theNUP98 gene at 11p15. Red signals are present on normal 11 and der(11), and green signals are present on normal 11, der(11) (arrow), and der(8) (arrowhead). (Aii) FISH with probe D8Z1 (red) for the alpha satellite of centromere 8 and BAC 350N15 (green), to encompass the FGFR1 and NSD3 genes at 8p11.2. Red signals are present on normal 8 and on der(8), and green signals are present on normal 8, der(8) (arrowhead), and der(11) (arrow). (Bi) Nested PCR detects NUP98-NSD3 short fusion transcripts. After primary amplification with primers NUP737F-FLJ3081Cr, diluted aliquots of PCR products were amplified with the following primers: lanes 1-2, NUP737F-FLJ2907Br (2477 base pair [bp]); lanes 3-4, NUP737F-FLJ1747Er (1317 bp); lanes 5-6, NUP1252F-FLJ3081Cr (2136 bp); lanes 7-8, NUP1252F- FLJ2907Br (1962 bp); lanes 9-10, NUP1252F- FLJ1747Er (802 bp). Lane M, molecular weight markers (lambda DNA digested with HindIII). Negative controls without cDNA are marked as (−). (Bii) Nucleotide and deduced amino acid sequences ofNUP98 and NSD3 cDNA at the fusion point. Sequence numbers refer to GenBank accession no. U41815 for NUP98 andAF332469 for NSD3.
FISH experiments and molecular characterization of fusion transcripts.
(Ai) FISH with probe D11Z1 (red) for the alpha satellite of centromere 11 and a mixture of BAC 118H17 and BAC 290A12 (green), to encompass theNUP98 gene at 11p15. Red signals are present on normal 11 and der(11), and green signals are present on normal 11, der(11) (arrow), and der(8) (arrowhead). (Aii) FISH with probe D8Z1 (red) for the alpha satellite of centromere 8 and BAC 350N15 (green), to encompass the FGFR1 and NSD3 genes at 8p11.2. Red signals are present on normal 8 and on der(8), and green signals are present on normal 8, der(8) (arrowhead), and der(11) (arrow). (Bi) Nested PCR detects NUP98-NSD3 short fusion transcripts. After primary amplification with primers NUP737F-FLJ3081Cr, diluted aliquots of PCR products were amplified with the following primers: lanes 1-2, NUP737F-FLJ2907Br (2477 base pair [bp]); lanes 3-4, NUP737F-FLJ1747Er (1317 bp); lanes 5-6, NUP1252F-FLJ3081Cr (2136 bp); lanes 7-8, NUP1252F- FLJ2907Br (1962 bp); lanes 9-10, NUP1252F- FLJ1747Er (802 bp). Lane M, molecular weight markers (lambda DNA digested with HindIII). Negative controls without cDNA are marked as (−). (Bii) Nucleotide and deduced amino acid sequences ofNUP98 and NSD3 cDNA at the fusion point. Sequence numbers refer to GenBank accession no. U41815 for NUP98 andAF332469 for NSD3.
BAC 350N15 was found to contain 2 candidate genes, FGFR1 andNSD3, which were screened for an in-frame fusion transcript with NUP98. PCR experiments excluded FGFR1involvement. The NSD3 gene, like NSD2, is known to encode 2 splicing isoforms, a long form and a short form.20 21 Primers were first designed on the short form (FLJ1856Af, FLJ2907Br, FLJ3081Cr, FLJ1204Df, FLJ1747Er). As reports suggest, the 5′ end of NUP98 bears the oncogenic-related portion. PCR experiments investigated whether (5′)NUP98-NSD3(3′) chimeric transcripts were present in patient-derived cDNA.
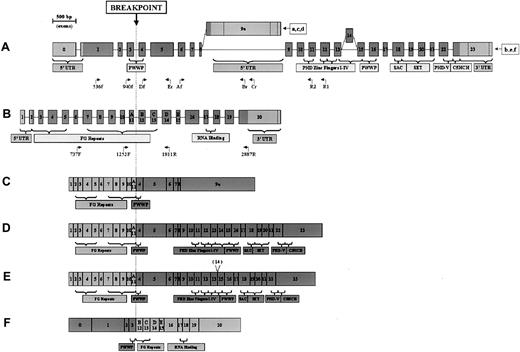
Results showed NUP98-NSD3 fusion transcripts (Figure 1B, upper panel). The NUP1252F-FLJ1747Er amplification product was sequenced, confirming an in-frame fusion between the 2 genes (Figure1B, lower panel). Breakpoints were located between exons 3 and 4 in theNSD3 gene and between exons 11 and 12 in NUP98(Figure 2A-B). Successful amplification from primer FLJ3081Cr implied the expression of short-form chimericNSD3 (Figure 2C).
NSD3 and NUP98 exon structures and chimeric transcripts.
(A, B) NSD3 and NUP98 exon structures showing encoded domains. Coding regions are depicted as dark gray; noncoding regions are light gray. Exon lengths are to scale, as indicated by ruler. Primers used in this study are positioned below the respective gene (only the end parts of the names are indicated). (A)NSD3 exon structure. All transcript sequences start at various points in exon 0. Ending points are indicated for the following GenBank accession numbers: (a) (WHSC1L1s), AF332468; (b) (WHSC1L1l),AF332469; (c) (FLJ12498), AK022560; (d) (NSD3S), AJ295992; (e) (NSD3L),AJ295990; (f) (NSD3L2, lacking exon 14), AJ295991. (B) NUP98exon structure, derived from GenBank accession number U41815. Exons 11 to 15 are also indicated as A-E by Arai et al.7 (C-F) Schematic representation of NSD3-NUP98 fusion transcripts.NUP98-derived exons are depicted as light gray;NSD3-derived exons are dark gray. (C) Short-form transcript, containing NUP98 exons 1 through 11 and NSD3exons 4 through 9a. (D) Long-form transcript, containingNUP98 exons 1 through 11 and NSD3 exons 4 through 23. (E) Alternatively spliced long-form transcript, lackingNSD3 exon 14. (F) Reciprocal transcript, containingNSD3 exons 0 through 3 and NUP98 exons 12 through 20.
NSD3 and NUP98 exon structures and chimeric transcripts.
(A, B) NSD3 and NUP98 exon structures showing encoded domains. Coding regions are depicted as dark gray; noncoding regions are light gray. Exon lengths are to scale, as indicated by ruler. Primers used in this study are positioned below the respective gene (only the end parts of the names are indicated). (A)NSD3 exon structure. All transcript sequences start at various points in exon 0. Ending points are indicated for the following GenBank accession numbers: (a) (WHSC1L1s), AF332468; (b) (WHSC1L1l),AF332469; (c) (FLJ12498), AK022560; (d) (NSD3S), AJ295992; (e) (NSD3L),AJ295990; (f) (NSD3L2, lacking exon 14), AJ295991. (B) NUP98exon structure, derived from GenBank accession number U41815. Exons 11 to 15 are also indicated as A-E by Arai et al.7 (C-F) Schematic representation of NSD3-NUP98 fusion transcripts.NUP98-derived exons are depicted as light gray;NSD3-derived exons are dark gray. (C) Short-form transcript, containing NUP98 exons 1 through 11 and NSD3exons 4 through 9a. (D) Long-form transcript, containingNUP98 exons 1 through 11 and NSD3 exons 4 through 23. (E) Alternatively spliced long-form transcript, lackingNSD3 exon 14. (F) Reciprocal transcript, containingNSD3 exons 0 through 3 and NUP98 exons 12 through 20.
To investigate the expression of long-form mRNA (Figure 2D), nested PCR was performed with primers W1L2685R2 and W1L2863R1 in combination withNUP98 forward primers. Only one band, of the appropriate length, was amplified (data not shown), confirming that the long transcript was present. Because no product was extended after exon 12, the presence of exon 14, which is involved in alternative splicing20 (Figure 2A,E), could not be determined. We also analyzed the expression of the reciprocal transcript (5′)NSD3-NUP98(3′) by matching primers NUP1811R and NUP2887R with 2 NSD3 forward primers FLJ536f and FLJ940f. A specific band of the correct length (data not shown) revealed the patient's cells expressed reciprocal fusion mRNA. The reciprocal transcriptNSD3-NUP98 maintained the 5′ part of NSD3, encoding for a split PWWP domain, and the 3′ part of NUP98, with few FG repeats and the RNA-binding domain (Figure 2F).
As far as we know, NSD3 is the eighth NUP98partner in myeloid malignancies, so NUP98, likeMLL and ETV6, is emerging as a promiscuous gene that recombines and fuses with different genomic sites. In the NSD family, NSD1, located at 5q35, has been shown to rearrange with NUP98 in childhood AML.3NSD2, located at 4p16, is the fusion partner of immunoglobulin heavy- chain gene at 14q32 in a recurrent translocation of multiple myeloma.21,22 Angrand et al20 foundNSD3 as part of an amplicon in breast cancer with 8p karyotypic abnormalities. We also confirmed NSD3amplification in 1 of 30 unselected cases of breast cancer analyzed by semiquantitative PCR (unpublished data, May 2001). This is the first report of NSD3 rearrangements in hematologic malignancies.
These 3 NSD family members have a common region of conserved architecture3,20,21—PHD fingers I-IV, PWWP, SAC/SET, PHD-V, and C5HCH (Figure 2A)—which suggests their involvement in chromatin remodeling and regulation of transcription. The fusion transcripts NUP98-NSD1 in t(5;11)3 andNUP98-NSD3 in t(8;11) both show this region fused to NUP98 FG repeats, which are known to bind transcription factors such as cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB)-binding protein.23 Moreover, transcriptional regulation genes, such as PMX1,1HOXD13,2HOXA9,4,5 andLEDGF,6 are other fusion partners forNUP98 in malignancies. Altogether, these data support the hypothesis that the transcription regulation function is critical forNUP98 fusion gene oncogenicity.
In conclusion, NSD3 has been observed for the first time as a translocation partner of NUP98 in acute myeloid leukemia. Further functional analysis of the chimeric transcripts will elucidate their specific role in leukemogenesis.
Supported by Associazione Italiana per la Ricerca sul Cancro (C.M., M.N.), Associazione Italiana contro le Leucemie-linfomi (C.M.), Ministero dell'Università e della Ricerca Scientifica e Tecnologica (C.M., M.N.), and Associazione Sergio Luciani (R.R.).
R.R. and R.L.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Cristina Mecucci, Hematology and Bone Marrow Transplantation Unit, Policlinico Monteluce, Via Brunamonti, 06123 Perugia, Italy; e-mail: crimecux@unipg.it.

![Fig. 1. FISH experiments and molecular characterization of fusion transcripts. / (Ai) FISH with probe D11Z1 (red) for the alpha satellite of centromere 11 and a mixture of BAC 118H17 and BAC 290A12 (green), to encompass theNUP98 gene at 11p15. Red signals are present on normal 11 and der(11), and green signals are present on normal 11, der(11) (arrow), and der(8) (arrowhead). (Aii) FISH with probe D8Z1 (red) for the alpha satellite of centromere 8 and BAC 350N15 (green), to encompass the FGFR1 and NSD3 genes at 8p11.2. Red signals are present on normal 8 and on der(8), and green signals are present on normal 8, der(8) (arrowhead), and der(11) (arrow). (Bi) Nested PCR detects NUP98-NSD3 short fusion transcripts. After primary amplification with primers NUP737F-FLJ3081Cr, diluted aliquots of PCR products were amplified with the following primers: lanes 1-2, NUP737F-FLJ2907Br (2477 base pair [bp]); lanes 3-4, NUP737F-FLJ1747Er (1317 bp); lanes 5-6, NUP1252F-FLJ3081Cr (2136 bp); lanes 7-8, NUP1252F- FLJ2907Br (1962 bp); lanes 9-10, NUP1252F- FLJ1747Er (802 bp). Lane M, molecular weight markers (lambda DNA digested with HindIII). Negative controls without cDNA are marked as (−). (Bii) Nucleotide and deduced amino acid sequences ofNUP98 and NSD3 cDNA at the fusion point. Sequence numbers refer to GenBank accession no. U41815 for NUP98 andAF332469 for NSD3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/10/10.1182_blood.v99.10.3857/6/m_h81022533001.jpeg?Expires=1769096970&Signature=AUQvoS61Rl1e2m4s1nvLFynNcYtgxS-M3Zcgzu6mQZ-Q10k6P7l9FVdi0rKNdsc~-sbeQr5c0AwmfSdT2wGZr84oXu5xPcdrZwMdVudSeGJ3HxT9xC1i-H3fQ~VqStuuBWuPPEHChvJi7IjM5N2dSgVsujAlw5kNQbOmBqAN4FmXWC5QkdOW2yuRRe9VcdzxwRRPun9DYb5ymykuzcK94O5GHwC3kvkZLDlZSauQaD0gdU5C0y5zhW8y5erb1-QfzNu9k1DoUAhFFW1eHj1ohLHNLA19DKb3uHQN06p4T-v9PHmzwd~PHSxqLq-Y0Gmxob-WKubmNjpNdPOw1BtmPg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal