Recent reports have established the prenatal origin of leukemia translocations and resultant fusion genes in some patients, including MLL-AF4 translocations in infants andTEL-AML1 translocations in children. We now report evidence for the prenatal origin of a translocation in childhood acute myeloid leukemia (AML). The t(8;21) AML1-ETO translocations were sequenced at the genomic level in 10 diagnostic leukemia samples from children with available neonatal Guthrie blood spots. Clonotypic genomic AML1-ETO sequences were detected in the Guthrie spots for 5 individuals, providing unambiguous evidence of prenatal origin in these cases. Two of these patients were older than 10 years of age at diagnosis, indicative of a protracted postnatal latency. Three of the patients were assessed for the persistence of genomic fusion sequences in complete clinical remission samples and were found to be positive. These data indicate that t(8;21) in childhood AML can arise in utero, possibly as an initiating event in childhood AML, and may establish a long-lived or stable parental clone that requires additional secondary genetic alterations to cause leukemia.
Introduction
The etiology of childhood leukemia is currently under scrutiny by means of large-scale epidemiology studies. Despite this intense investigation, a basic understanding of the timing of the origin of the crucial molecular abnormalities, or the “natural history” of leukemic clones, is incomplete. The recognition of crucial temporal and developmental windows for the formation of leukemogenic genetic alterations will help to focus epidemiologic analysis as well as to prompt preventive strategies.
Recent studies using twins with concordant leukemia suggest that a hallmark genetic event in these acute leukemias, ie, chromosomal translocation, can have a prenatal origin.1-5 More explicit evidence is provided by the finding of clonotypic chromosomal fusion sequences in archived neonatal (Guthrie) heel-prick spots matched to children who later contracted leukemia.6-8Additional indirect support for prenatal origin of leukemia clones is derived from the demonstration of the presence of clonotypic rearrangements at the IGH and TCR loci in Guthrie spots.9 10 These studies have concentrated on a limited subset of leukemias primarily of lymphoid phenotype, leaving open the question of whether prenatal origin of childhood leukemia is a possible or common occurrence in other subgroups.
Childhood acute myeloid leukemia (AML) composes about 20% of childhood leukemias overall, and represents a group of diseases with a variety of molecular subtypes. After a peak incidence of leukemias in infants with translocations involving the MLL gene, children with AML exhibit the same range of molecular abnormalities as adults—characteristic translocations, deletions, and aneuploidies. The most frequent translocation in both childhood (1 to 20 years of age) and adult (older than 20 years) AML is the t(8;21) AML1-ETOfusion, resulting in a fusion protein that disrupts the normal function of the transcription factors AML1 andETO.11 Rather than exhibiting an age-associated peak incidence like the MLL translocations (younger than 1 year) and TEL-AML1 translocations (2 to 5 years), the incidence of leukemia with t(8;21) increases slowly during childhood and is constant throughout life.12 Leukemias with t(8;21) translocations also occasionally present following chemotherapy for other cancers or certain occupational exposures (reviewed in Xiao et al13). These facts are compatible with the interpretation of t(8;21) as a postnatal event in temporal proximity to leukemia diagnosis. Alternatively, t(8;21) may occur commonly during early life (during, say, fetal hematopoiesis), yielding long-lived clones that may or may not progress to overt leukemia later in life depending upon the acquisition of appropriate secondary genetic events. To begin to address these questions, we describe a molecular analysis of a series of childhood AML patients with matched Guthrie cards and follow-up remission material.
Patients, materials, and methods
Patients and patient materials
Patients and matched Guthrie cards were from California (n = 7) and the United Kingdom (n = 3). California patients were enrolled in the Northern California Childhood Epidemiology Study or the Children's Oncology Group AML cell bank. Patients in the United Kingdom were enrolled in the United Kingdom Medical Research Council AML 10 trial. High-molecular weight DNA was isolated from patient bone marrow by means of conventional sodium dodecyl sulfate (SDS)/proteinase K methods. Neonatal blood spots, or Guthrie cards, were obtained from a central repository maintained by the Genetic Diseases Branch of the California State Department of Health, for California patients, and from different regional centers for United Kingdom patients. For 3 patients, small portions of leftover bone marrow aspirates obtained for diagnostic purposes following treatment were obtained from the hospitals of treatment. Research ethics committees at participating institutions reviewed and approved all procedures.
Breakpoint sequencing
Patient breakpoints were sequenced by means of long-distance inverse polymerase chain reaction (LDI-PCR) and conventional long distance PCR (LD-PCR), as previously described.13 We tried LD-PCR reactions first when DNA was plentiful (exceeding 10 μg), while we tried LDI-PCR first when DNA was limiting. When one approach failed at amplifying a translocation, we used the other approach.
Guthrie card screening
For nested reactions, we designed specific PCR primers spanning each patient's fusion sequence (Table 1). In most cases, primer design called for a first-round PCR with 60°C annealing temperature followed by a second round with 66°C annealing. Sensitivity of PCR reactions was determined by means of serial dilutions of diagnostic DNA. PCR screening of Guthrie cards was performed with Ampdirect buffers (Nacalai Tesque, Osaka, Japan, or Rockland Immunochemicals, Gilbertsville, PA) exactly as previously described.7Briefly, a first-round amplification in Ampdirect was followed with a subsequent nested amplification with the use of 1 μL the primary reaction as template and conventional Taq polymerase and buffers. Ampdirect buffers allow PCR amplification directly from blood or, in this case, blood-soaked filter papers, by abrogating the effects of PCR inhibitors in blood. One-eighth segments (United Kingdom cards, 1 cm diameter) or one-sixteenth segments (California cards, 1.5 cm diameter) of cards were placed in single tubes. Serial dilutions of diagnostic DNA were amplified in tandem in separate PCR tubes to ensure the sensitivity of each reaction. Preparation of PCR reactions, the cutting of Guthrie cards, and the dilution and allocating of diagnostic DNA were all done in separate rooms.
Polymerase chain reaction primers used for backtracking genomic AML1-ETO translocations in 10 patients
| Patient . | PCR round . | AML1 primers (5′-3′) . | ETO primers (5′-3′) . | Product size . |
|---|---|---|---|---|
| 1 | 1 | CTGCCATTCCGCTGACTAAG | AGATTCTCACCTAAAAGTCTGACT | 126 |
| 2 | CTGCCATTCCGCTGACTAAG | GCAGCGGGGCCTAGCTGAAT | 102 | |
| 2* | 1 | GTCTTCACAAACCCACCGCAAGTCG | CCTCCACATCTAGAAAGCAGGTCTACAGCAC | 165 |
| 2 | CCCGAGAACCTCGAAGTAAGTGCAT | CCTCCACATCTAGAAAGCAGGTCTACAGCAC | 91 | |
| 3 | 1 | GGGAGCAGAAGTGAGATGGT | CTAAATACTGTACCCAAGAAGGG | 541 |
| 2 | GGTTGAGGCAGAATGCAAAAGTTCAAG | GGCTGCTTTATCCAGCGAACCC | 503 | |
| 4 | 1 | TACCGTACATTATGGCTATATTATCA | AATGCCGAATAATTTATTCACCACCACC | 132 |
| 2 | GCCAGTGACATTTAGTTGGATGTGAAAT | CCCATCCTGTTCAAGGTTAACCC | 69 | |
| 5* | 1 | AGGGCCTAATCTTCACTCAGCTAC | AACGCGCATTGATGAATGGTC | 170 |
| 2 | CTTGGTTGAGATCATGTCATATGAG | AACGCGCATTGATGAATGGTC | 125 | |
| 6 | 1 | GGAAGCTCACCAGATAGGCT | GTACTATACTTATGTTAGAAAGAAAACA | 195 |
| 2 | AGCTACCTCTGGGCTCCAGTG | CACAGTGGGAAACCCAACCTTTCAT | 141 | |
| 7 | 1 | ATAATAAAATTCCAAGCAAGTACTTTCA | AAAGCTTAAAGCTACACATGGTC | 447 |
| 2 | AATTTAGGACCTTTTTCAACTCTGCTTAAG | CCACAATTCTCCCCATTCAGTTTTCAG | 320 | |
| 8* | 1 | GAAGCGGCAGTAACCCAGGTG | CCAGGTTCAATGGACTCTTCCC | 249 |
| 2 | GTGCAGCTCTGAGTGAGCTGG | GGCTATTTTCTTCTCAGCAGTCCAG | 136 | |
| 9 | 1 | CTTAGCTTAACGATATGAGAAGAAAT | CTAGCCTTCACCTATGACTTATG | 238 |
| 2 | GTTACGTAGCACTGGACAATATATAAGCA | CTCAGGTCACTTTGATTTTTGCATCTCC | 147 | |
| 10 | 1 | AACCACTGAAATTTGAGTCTTGAG | AGGATATGGAGCCAAGATCTCT | 381 |
| 2 | CTGGCCCTGCTAGCTCAGTCAA | CCAAATTGTATAATAAAAATAGACAATCAATAACATG | 215 |
| Patient . | PCR round . | AML1 primers (5′-3′) . | ETO primers (5′-3′) . | Product size . |
|---|---|---|---|---|
| 1 | 1 | CTGCCATTCCGCTGACTAAG | AGATTCTCACCTAAAAGTCTGACT | 126 |
| 2 | CTGCCATTCCGCTGACTAAG | GCAGCGGGGCCTAGCTGAAT | 102 | |
| 2* | 1 | GTCTTCACAAACCCACCGCAAGTCG | CCTCCACATCTAGAAAGCAGGTCTACAGCAC | 165 |
| 2 | CCCGAGAACCTCGAAGTAAGTGCAT | CCTCCACATCTAGAAAGCAGGTCTACAGCAC | 91 | |
| 3 | 1 | GGGAGCAGAAGTGAGATGGT | CTAAATACTGTACCCAAGAAGGG | 541 |
| 2 | GGTTGAGGCAGAATGCAAAAGTTCAAG | GGCTGCTTTATCCAGCGAACCC | 503 | |
| 4 | 1 | TACCGTACATTATGGCTATATTATCA | AATGCCGAATAATTTATTCACCACCACC | 132 |
| 2 | GCCAGTGACATTTAGTTGGATGTGAAAT | CCCATCCTGTTCAAGGTTAACCC | 69 | |
| 5* | 1 | AGGGCCTAATCTTCACTCAGCTAC | AACGCGCATTGATGAATGGTC | 170 |
| 2 | CTTGGTTGAGATCATGTCATATGAG | AACGCGCATTGATGAATGGTC | 125 | |
| 6 | 1 | GGAAGCTCACCAGATAGGCT | GTACTATACTTATGTTAGAAAGAAAACA | 195 |
| 2 | AGCTACCTCTGGGCTCCAGTG | CACAGTGGGAAACCCAACCTTTCAT | 141 | |
| 7 | 1 | ATAATAAAATTCCAAGCAAGTACTTTCA | AAAGCTTAAAGCTACACATGGTC | 447 |
| 2 | AATTTAGGACCTTTTTCAACTCTGCTTAAG | CCACAATTCTCCCCATTCAGTTTTCAG | 320 | |
| 8* | 1 | GAAGCGGCAGTAACCCAGGTG | CCAGGTTCAATGGACTCTTCCC | 249 |
| 2 | GTGCAGCTCTGAGTGAGCTGG | GGCTATTTTCTTCTCAGCAGTCCAG | 136 | |
| 9 | 1 | CTTAGCTTAACGATATGAGAAGAAAT | CTAGCCTTCACCTATGACTTATG | 238 |
| 2 | GTTACGTAGCACTGGACAATATATAAGCA | CTCAGGTCACTTTGATTTTTGCATCTCC | 147 | |
| 10 | 1 | AACCACTGAAATTTGAGTCTTGAG | AGGATATGGAGCCAAGATCTCT | 381 |
| 2 | CTGGCCCTGCTAGCTCAGTCAA | CCAAATTGTATAATAAAAATAGACAATCAATAACATG | 215 |
PCR indicates polymerase chain reaction.
Patients born and diagnosed in the United Kingdom. The remainder were born and diagnosed in California. Breakpoints for most patients were previously reported in Xiao et al.13 Patient 1 listed in this Table corresponds to patient no. 13 in Xiao, et al. Patient 2 is previous patient no. 1; patient 3, previous no. 15; patient 4, previous no. 9; patient 5, previous no. 6; patient 6, previous no. 19; patient 7, previous no. 14. Patients no. 8 and 10 are the same in both studies.
DNA from archived slides (in the case of available remission material for 3 patients) was purified by conventional SDS/proteinase K methods after scraping the slide contents into a microcentrifuge tube. The DNA was quantified by means of fluorimetry and used directly for PCR in standard buffers with the same primers and assay used for Guthrie analysis, except for the use of conventional PCR buffers for both rounds of PCR. Prior to translocation-specific PCR, the DNA was subjected to PCR of control genes (NQO1 and NRAS)as previously described.5
Results
Matched Guthrie card samples were available for 12 patients. We attempted amplification of clonotypic genomic AML1-ETOfusion in all 12 patient samples and were successful in 10 of them. For the 2 remaining samples, diagnostic karyotypes indicated t(8;21); however, AML1-ETO fusion messenger RNA (mRNA) was not detectable by reverse transcriptase (RT)–PCR, suggesting that the patients might have had a different fusion. The sequences of 9 of these breakpoints are presented elsewhere.13 The one remaining sample (patient no. 9) was a fusion between base 21788663 on Genbank entry NT_011512.4 (AML1 intron 5) and base 242182 on Genbank entry NT_029344.1 (ETO intron 1B) with a single nontemplate“C” at the point of fusion. Short–distance PCR primer sets for all fusions were designed for the genomic fusion of each patient (Table 1) and were checked for sensitivity and specificity by means of serial dilutions of diagnostic patient material and control DNAs (data not shown).
PCR screening of Guthrie cards was performed without prior extraction of DNA. All of the Guthrie material (cut segments of individual blood spots) was placed into the PCR reaction to maximize the opportunity to amplify a rare sequence. Isolation of DNA from a Guthrie spot is variable and depends on age of the card and storage conditions, but cards at least 20 years old have been shown to have equivalent capacity to support PCR amplification in relative equal measure when performed with Ampdirect (Wiemels et al7 and data not shown).
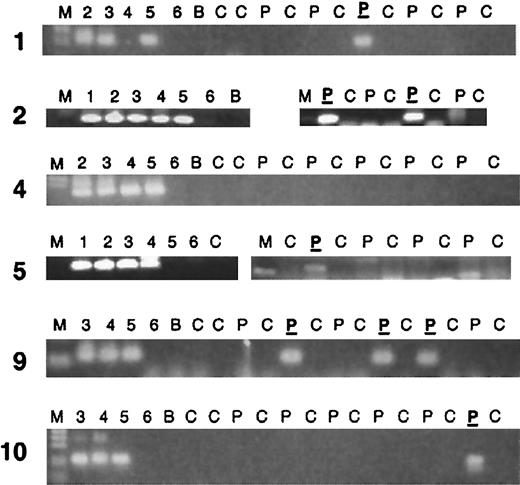
At least 4 segments of a Guthrie card blood spot were available for each patient (Table 2). Of the 10 patients studied, 5 supported amplification of specific clonotypicAML1-ETO sequence on at least one segment (Figure1 and Table 1). PCR products were sequenced with the use of one internal primer. In all 5 cases, the sequence of the Guthrie card–amplified band matched the sequence originally determined from the diagnostic sample. Therefore, in at least 5 of 10 AML cases studied, the clonotypic, genomic fusion sequence and presumptive leukemic or preleukemic cells were present at birth, several years prior to a diagnosis of leukemia.
Patient age at diagnosis and summary of backtrackingAML1-ETO genomic fusions in childhood acute myeloid leukemia
| Study no. . | Age at diagnosis, y, mo . | Guthrie segments tested . | Guthrie segments positive . | Positive or negative* . |
|---|---|---|---|---|
| 1 | 3, 6 | 12 | 2 | + |
| 2 | 3, 11 | 4 | 2 | + |
| 3 | 4, 11 | 12 | 0 | − |
| 4 | 5, 2 | 12 | 0 | − |
| 5 | 6, 3 | 4 | 1 | + |
| 6 | 7, 9 | 12 | 0 | − |
| 7 | 8, 7 | 12 | 0 | − |
| 8 | 10, 0 | 10 | 0 | − |
| 9 | 10, 1 | 12 | 5 | + |
| 10 | 12, 2 | 12 | 1 | + |
| Study no. . | Age at diagnosis, y, mo . | Guthrie segments tested . | Guthrie segments positive . | Positive or negative* . |
|---|---|---|---|---|
| 1 | 3, 6 | 12 | 2 | + |
| 2 | 3, 11 | 4 | 2 | + |
| 3 | 4, 11 | 12 | 0 | − |
| 4 | 5, 2 | 12 | 0 | − |
| 5 | 6, 3 | 4 | 1 | + |
| 6 | 7, 9 | 12 | 0 | − |
| 7 | 8, 7 | 12 | 0 | − |
| 8 | 10, 0 | 10 | 0 | − |
| 9 | 10, 1 | 12 | 5 | + |
| 10 | 12, 2 | 12 | 1 | + |
A plus sign indicates the presence of clonotypicAML1-ETO in the Guthrie card; a minus sign indicates the lack of demonstrable clonotypic translocation sequence, which is indeterminate for prenatal origin.
PCR analysis of clonotypic genomic AML1-ETOfusions in neonatal Guthrie cards of leukemic patients.
Two rounds of PCR (70 total cycles) were performed as described in “Patients, materials, and methods,” and the results are shown only from the secondary round. Partial results are shown; in most cases a second PCR run was performed. Five “positive” PCR patients are shown, as well as one negative. Lane markers: M indicates marker. Lanes 1-6 show 1:10 dilutions of patient DNA, with lane 1 being 100 ng/μL and lane 6, 1 pg/μL DNA. Patient DNAs have varying percentages of leukemic DNA, as some patient samples are from blood rather than bone marrow. C indicates control Guthrie card without patient DNA; P, Guthrie card segment from the patient; P, patient Guthrie card segment that was sequenced and determined to be the same sequence as the diagnostic DNA sample of that patient; B, no DNA sample (blank).
PCR analysis of clonotypic genomic AML1-ETOfusions in neonatal Guthrie cards of leukemic patients.
Two rounds of PCR (70 total cycles) were performed as described in “Patients, materials, and methods,” and the results are shown only from the secondary round. Partial results are shown; in most cases a second PCR run was performed. Five “positive” PCR patients are shown, as well as one negative. Lane markers: M indicates marker. Lanes 1-6 show 1:10 dilutions of patient DNA, with lane 1 being 100 ng/μL and lane 6, 1 pg/μL DNA. Patient DNAs have varying percentages of leukemic DNA, as some patient samples are from blood rather than bone marrow. C indicates control Guthrie card without patient DNA; P, Guthrie card segment from the patient; P, patient Guthrie card segment that was sequenced and determined to be the same sequence as the diagnostic DNA sample of that patient; B, no DNA sample (blank).
Despite the fact that the 2 patients youngest at the time of diagnosis had positive evidence of genomic t(8;21) rearrangement at birth, there was otherwise no clear relationship to age. In fact, the 2 oldest patients (nos. 9 and 10) also demonstrated prenatal origin, which is remarkable in that both of these patients were older than age 10 years at the time of diagnosis. One additional patient (no. 5), aged 6¼ years, was positive.
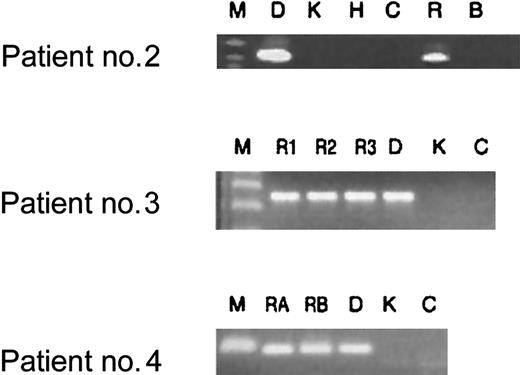
Additional diagnostic bone marrow smears were available for 3 patients enrolled in our study following chemotherapy-induced leukemia remission. These smears were obtained during routine follow-up of patients to monitor treatment success. All 3 patients had cytologically normal bone marrow, ie, lacking the obvious presence of leukemic blasts. As demonstrated in Figure 2, however, all patients had demonstrable clonotypic genomicAML1-ETO fusion sequence in nonquantitative PCR assays in their remission samples. These samples were taken when the patients were either on therapy, had just finished therapy, or had been off all therapy for no longer than 6 months (Figure 2). Longer-term remission samples were not available. Two patients (nos. 3 and 4) continue to be in first remission, while one (no. 2) is now deceased as a result of leukemia relapse. This preliminary assay does not have any prognostic relevance for these patients.
Analysis of patient remission DNA for clonotypicAML1-ETO translocation persistence after treatment.
First, DNA extracted from bone marrow samples was tested for integrity with the use of control gene PCRs (NQO1 and NRASprimers, data not shown). With the same primer sets that were used for screening Guthrie cards, bone marrow samples obtained from treated patients were tested for translocation sequences. All patient samples tested after treatment proved to be positive for clonotypic fusion sequence after 2 rounds of PCR (lanes R). Key to lanes: M indicates marker; D, diagnostic patient–specific DNA; K, Kasumi cell line DNA; H, HL-60 cell line DNA; C, human DNA from a nonpatient control; R, bone marrow sample for patient no. 2 obtained while the patient was on treatment at 5 months after diagnosis; R1, R2, and R3, bone marrow samples from patient no. 3 obtained immediately following induction treatment (2 months after diagnosis), at the end of consolidation (3 months after diagnosis), and 4 months off all therapy (8 months after diagnosis), respectively; RA and RB, DNA from bone marrow from patient no. 4 obtained when the patient was just off all therapy (11 months after diagnosis) and 6 months off all therapy (16 months after diagnosis), respectively.
Analysis of patient remission DNA for clonotypicAML1-ETO translocation persistence after treatment.
First, DNA extracted from bone marrow samples was tested for integrity with the use of control gene PCRs (NQO1 and NRASprimers, data not shown). With the same primer sets that were used for screening Guthrie cards, bone marrow samples obtained from treated patients were tested for translocation sequences. All patient samples tested after treatment proved to be positive for clonotypic fusion sequence after 2 rounds of PCR (lanes R). Key to lanes: M indicates marker; D, diagnostic patient–specific DNA; K, Kasumi cell line DNA; H, HL-60 cell line DNA; C, human DNA from a nonpatient control; R, bone marrow sample for patient no. 2 obtained while the patient was on treatment at 5 months after diagnosis; R1, R2, and R3, bone marrow samples from patient no. 3 obtained immediately following induction treatment (2 months after diagnosis), at the end of consolidation (3 months after diagnosis), and 4 months off all therapy (8 months after diagnosis), respectively; RA and RB, DNA from bone marrow from patient no. 4 obtained when the patient was just off all therapy (11 months after diagnosis) and 6 months off all therapy (16 months after diagnosis), respectively.
Discussion
With this report, we present the first direct evidence for a prenatal origin of t(8;21) AML1-ETO in childhood AML. The assay uses genomic fusion gene sequence, rather than the mRNA, and is therefore specific for the leukemic clone. Of 10 patients assessed, 5 had positive Guthrie cards. This analysis does not, however, rule out a prenatal origin for any or all of the remaining 5 negative Guthries, as negative results are not definitive.7 The limitations of our assay preclude the detection of a clone harbored in a bone marrow compartment and not circulating in the bloodstream at a frequency of at least 1 cell per 30 000 (the approximate number of cells in one 1-cm diameter blood spot6 7), and we have screened, at maximum, the equivalent of 1.5 such blood spots (12 segments) per patient. We therefore cannot definitively say whether the prenatal origin of this leukemia subtype in childhood is typical or usual, although we do conclude that it is possible and common.
Our current report adds one more translocation type, and the first such type for childhood AML in patients older than 2 years, to those previously discovered to have prenatal origin: the MLL andTEL-AML1 translocations. Two of our positiveAML1-ETO patients were older than 10 years of age at diagnosis. These are, to date, the oldest patients to receive an AML diagnosis and to have an AML that was found to have a prenatal origin by means of this assay, although studies with identical twins have also indicated that postnatal latency following initiation in utero can be very protracted.3,5 Taken together, these reports clearly implicate the fetal period as a distinctly susceptible period for chromosomal translocation, perhaps the usual period for such events among pediatric leukemias. It is noteworthy that other childhood cancers, particularly the sarcomas, are also commonly associated with translocations.14 The occasional congenital presentation of such tumors also argues for a prenatal origin and potentially related mechanisms of formation.
A further commonality among the 3 prenatally occurring translocation types (MLL, TEL-AML1, and AML1-ETO) is the structure of the fusion breakpoints. This includes scattered breakpoint distribution with weak clustering, and the evidence of constitutive repair processes for the fusion event.13,15,16 Conspicuously lacking among all 3 types is evidence of formation via a V(D)J recombinase mechanism, a mechanism well-established for the formation of lymphoma translocations17,18 and occasionally hypothesized to be implicated in childhood translocations.19,20 The combined evidence suggests, however, that pediatric translocations that occur in utero form by means of as-yet ill-defined breakage followed by error-prone nonhomologous end joining of broken chromosome ends. These processes could be related to the extensive cell proliferation, oxidative stress, and apoptosis during prenatal hematopoiesis. For infant leukemias with MLL fusions, there is recent epidemiological evidence implicating transplacental chemical exposures.21
Clinical and biologic analysis of t(8;21) leukemia suggests that the translocation may persist in a patient's bone marrow, with evidence ofAML1-ETO RT-PCR cells in patients in long-term remission, up to 12.5 years after treatment.22-24Quantitative assessment by real-time (Taqman) PCR suggests that persistent levels above a threshold may herald relapse but that patients with lower levels of positive cells remain disease free.25-27 Our current result confirms this at the level of genomic sequence, providing direct evidence for the persistence of the same leukemic clone. The longevity of the clone after treatment is paradoxical given that this leukemia subtype has relatively good prognosis in both adults and children, with a cure (greater than 5-year clinical remission) possible in about half of patients.28,29 Persistent AML1-ETO+cells might reflect some control mechanism, immunological or otherwise, restraining the leukemic cells. Alternatively, the persistent cells could be preleukemic cells with AML1-ETO but lack the additional secondary events. From the age of our patients with positive Guthrie spots, we can conclude that such cells both exist and persist for up to 10 years or more. The AML1-ETO persistence phenomenon was studied in detail by Miyamoto and colleagues,30 who sorted and cultured bone marrow cells from patients in remission. AML1-ETO transcripts were present in various stem and myeloid progenitor cell compartments, as well as in B cells and monocytes but not in T cells; the authors interpreted this as reflecting the persistence of a preleukemic multipotential stem cell. An animal model of AML1-ETOleukemia supports a multistep scenario. Mice with anAML1-ETO inserted into one AML1 locus do not develop myeloid leukemia, but will do so at a much higher rate than controls following treatment with the alkylating agent and mutagen ethyl nitrosourea, which presumably induces the necessary secondary event(s).31,32 In these respects, childhood AML parallels ALL with TEL-AML1. In the latter cases, twin concordance data indicate the need for secondary postnatal events4,8coupled with postnatal persistence of the fetal preleukemic clone for up to 14 years.5 Persistent preleukemic clones withTEL-AML1 may, in some cases, also give rise to late relapses of disease following prolonged remission.33
The descriptive epidemiology of t(8;21) leukemia shows a relatively constant age-specific incidence.12 This raises the question of whether translocation-positive preleukemic stem cells formed in utero may persist to adulthood, providing a lifetime reservoir of “initiated” cells that may progress at any time to AML given the appropriate secondary genetic alterations. If correct, then a difference between childhood and adult leukemia might be in the etiologic exposures that induce progression of the t(8;21)-initiated clone to frank leukemia. Future studies investigating such a hypothesis should examine clonotypic AML1-ETO sequences in Guthrie cards of adults with de novo AML. The presence of AML1-ETOin therapy-related adult AML indicates that this fusion is possibly induced in stem cells of adults; however, future studies will have to assess whether or not the fusion gene was pre-existent to the patient's exposure to chemotherapy for the primary cancer.
We thank physicians and hospitals in California and the United Kingdom who contributed diagnostic material to this study; the California Department of Health Services for access to Guthrie cards, including the efforts of Dr Peggy Reynolds, Michael Layefsky, and Dr Fred Lorey; and the Children's Oncology Group and efforts of Dr Anita Khayat and Stacey Dozono for access to patient diagnostic material. We also acknowledge Sandra Hing and John Swansbury for cytogenetic and molecular diagnostic data and samples for United Kingdom patients and Dr Wyn Griffiths for retrieval of Guthrie cards.
Supported by National Institutes of Health grants CA89032 (J.L.W.), ES09137 (P.A.B.), and ES04705 (M.T.S.); a Leukaemia Research Fund Specialist Programme Grant (M.G.); and Portuguese Foundation for Science and Technology grant PRAXIS XXI/BD/19575/99 (A.T.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Joseph L. Wiemels, Laboratory for Molecular Epidemiology, Dept of Epidemiology and Biostatistics, University of California San Francisco, 500 Parnassus Ave MU-420W, San Francisco, CA 94143-0560; e-mail: jwiemels@epi.ucsf.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal