We previously reported that 2 polymorphisms in the 5,10-methylenetetrahydrofolate reductase (MTHFR) gene at positions C677T and A1298C were associated with lower risk of adult acute lymphocytic leukemia (ALL). In the present study, we have examined whether polymorphisms in other folate-metabolizing genes play a role in ALL susceptibility. Polymorphisms in methionine synthase (MS A2756G), cytosolic serine hydroxymethyltransferase (SHMT1 C1420T), and a double (2R2R) or triple (3R3R) 28-bp tandem repeat in the promoter region of thymidylate synthase (TS) were studied and found to modulate ALL risk. In a univariate analysis, SHMT1 1420CT individuals exhibited a 2.1-fold decrease in ALL risk (odds ratio [OR] = 0.48; 95% confidence interval [CI], 0.25-0.91), whereas the 1420TT genotype conferred a 3.3-fold reduction in risk (OR = 0.31; 95% CI, 0.10-0.90). Similarly, TS 2R3R individuals exhibited a 2.8-fold reduction in ALL risk (OR = 0.36; 95% CI: 0.16-0.83), while the TS 3R3R genotype conferred an even greater level of protection (OR = 0.25; 95% CI, 0.08-0.78). However, no significant associations were evident for the MS 2756AG polymorphism (OR = 0.79; 95% CI, 0.38-1.7). In addition, potential interactions between theSHMT1 and TS or MS genes were observed. TS 3R3R individuals who were SHMT1 1420CT/TT had a 13.9-fold decreased ALL risk (OR = 0.072; 95% CI, 0.0067-0.77). Further, MS 2756AG individuals who were SHMT1 1420CT/TT had a 5.6-fold reduction in ALL risk (OR = 0.18; 95% CI, 0.05-0.63). This study suggests an important role for uracil misincorporation and resultant chromosomal damage in the pathogenesis of ALL, and that genetic interactions involving low penetrance polymorphisms in folate-metabolizing genes may increase ALL risk.
Introduction
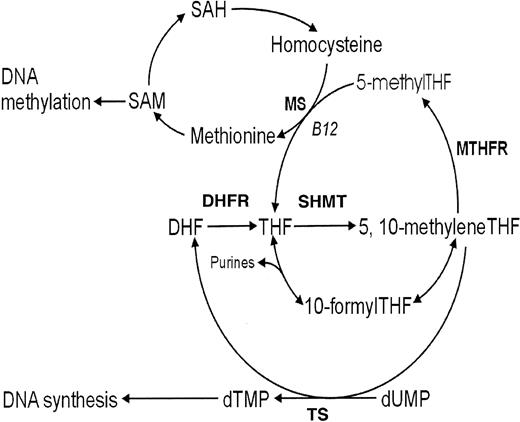
Although the clinical and pathologic aspects of leukemia are well documented, little is known about the genes that influence susceptibility to this disease. While highly penetrant inherited mutations in DNA account for a small number of leukemia cases, the majority of cases likely involve variations in several genes encoding diverse proteins that can interact to define a high-risk phenotype. These gene-gene interactions, as well as their interplay with diet, other environmental exposures, and individual immune function, may be major determinants in leukemia susceptibility. We previously reported an association between polymorphic variants at positions C677T and A1298C in the folate-metabolizing gene 5,10-methylenetetrahydrofolate reductase (MTHFR), and a decreased risk of adult acute lymphocytic leukemia (ALL).1Similar findings in childhood leukemia have recently been reported.2 We propose that this protective effect is due to an increase in the flux of 5,10-methylenetetrahydrofolate (methyleneTHF) available for DNA synthesis and subsequent reductions of uracil in DNA (Figure 1). Accumulation of uracil in DNA and the removal of this abnormal base during excision repair processes could result in DNA double-strand breaks, essential for the formation of chromosomal translocations and deletions. This chromosomal damage may be sufficient to initiate ALL progression through the malignant transformation and clonal expansion of lymphopoeitic progenitor cells.
Overview of the human folate metabolic pathway.
S-adenosylmethionine (SAM); S-adenosylhomocysteine (SAH); dihydrofolate (DHF); dihydrofolate reductase (DHFR); tetrahydrofolate (THF); serine hydroxymethyltransferase (SHMT); 5,10-methylenetetrahydrofolate (5,10-methyleneTHF); 5,10-methylenetetrahydrofolate reductase (MTHFR); 5-methyltetrahydrofolate (5-methylTHF); 10-formyltetrahydrofolate (10-formylTHF); methionine synthase (MS); thymidylate synthase (TS); deoxythymidine monophosphate (dTMP); and deoxyuridine monophosphate (dUMP).
Overview of the human folate metabolic pathway.
S-adenosylmethionine (SAM); S-adenosylhomocysteine (SAH); dihydrofolate (DHF); dihydrofolate reductase (DHFR); tetrahydrofolate (THF); serine hydroxymethyltransferase (SHMT); 5,10-methylenetetrahydrofolate (5,10-methyleneTHF); 5,10-methylenetetrahydrofolate reductase (MTHFR); 5-methyltetrahydrofolate (5-methylTHF); 10-formyltetrahydrofolate (10-formylTHF); methionine synthase (MS); thymidylate synthase (TS); deoxythymidine monophosphate (dTMP); and deoxyuridine monophosphate (dUMP).
To confirm our previous findings of the role of the folate pathway in leukemia susceptibility and to further test the significance of the DNA synthesis pathway in ALL risk, we examined polymorphisms in 3 additional genes involved in various branches of folate metabolism. Thymidylate synthase (TS), located on chromosome 18p11.32, plays a critical role in maintaining a balanced supply of deoxynucleotides required for DNA synthesis (Figure 1). Impairments of the TS enzyme have been associated with chromosome damage and fragile site induction.3,4TS has a unique tandem repeat sequence in the 5′ untranslated region (UTR) immediately upstream of the ATG codon initiation start site that has been shown to be polymorphic, containing either 2 or 3 28-bp repeats.5The presence of the triple versus double 28-bp repeat was shown to enhance gene expression in in vitro and in vivo studies.6 7 This enhanced expression may increase the conversion of deoxyuridine monophosphate (dUMP) to deoxythymidine monophosphate (dTMP), reducing the level of uracil that might otherwise be incorporated into DNA. Theoretically, this could limit DNA damage in rapidly dividing tissues that have the greatest requirement for DNA such as those involved in hematopoeisis, and afford some protection in leukemia risk.
Serine hydroxymethyltransferase (SHMT) encodes a vitamin B6–dependent enzyme8 that catalyzes the reversible conversion of serine and tetrahydrofolate (THF) to glycine and methyleneTHF (Figure 1). There are 2 distinct SHMT isoenzymes: one in the cytosol localized to the SHMT1 gene on chromosome 17p11.2, and the other is in the mitochondrion localized to theSHMT2 gene on chromosome 12q13.2.9 SHMT1 plays a pivotal role in providing one-carbon units for purine, thymidylate, and methionine synthesis, in addition to other metabolic functions.10 Recently, a C1420T polymorphism inSHMT1 numbered from the start ATG codon of the open reading frame or at position C1444T numbered from the start of the 5′-UTR has been described11 that results in reduced plasma and red blood cell folate levels in 1420CC individuals. Based on the role of SHMT in the provision of one-carbon units for multiple folate pathways, disturbances in protein expression or enzyme activity due to this polymorphism could mimic a folate deficiency by reducing the one-carbon moieties available for both remethylation of homocysteine and DNA synthesis, making this an ideal gene candidate to study.
Methionine synthase (MS), located on chromosome 5p15.3-15.2, encodes a vitamin B12–dependent enzyme that catalyzes the remethylation of homocysteine to methionine (Figure 1). An A-to-G transition at position 2756 in the open reading frame of theMS gene that converts an aspartic acid to a glycine residue (D919G) has recently been described.12,13 This polymorphism is predicted to alter enzyme activity since it is located in the domain involved in methylation and reactivation of the B12 cofactor.14 Previous studies have shown that the 2756G allele is associated with decreased plasma homocysteine levels15,16 and a higher proportion of formylated folates,17 the latter of which could result in the increased production of downstream folates favoring purine and pyrimidine synthesis. In addition, lower rates of colorectal cancer have been observed in MS 2756GG versus 2756AA individuals with low alcohol intakes.18 These studies suggest that the MS 2756G allele may be associated with an increased flux of one-carbon moieties available for DNA synthesis and repair. We predicted that if compromised DNA synthesis was a likely mechanism involved in the chromosomal damage associated with susceptibility to ALL, we would expect to observe a higher proportion of ALL cases than controls with the TS 2R2R, SHMT 1420CC, and MS 2756AA genotypes. To test this hypothesis, we examined the same ALL case-control population as in our previous study of MTHFR polymorphisms1 and performed a multivariate analysis of the data obtained in both studies.
Materials and methods
Study population and sample collection
A complete description of the study design has been previously reported.19 Briefly, the study population consisted of 71 individuals diagnosed with ALL that formed part of the British Leukaemia Research Fund Center's case-control study of acute leukemia in adults. Cases comprised of patients 16 to 70 years of age newly diagnosed with acute leukemia between April 1, 1991, and December 31, 1996, while resident in regions of north and southwest England. For each participating case, 2 controls were randomly selected from a list of all persons registered with the same local physician who were of the same sex, year of birth (+/− 2 years), and race as the patient. Cases were considered ineligible if they were diagnosed with acute leukemia within 6 months of being diagnosed with a prior hematologic malignancy or within 2 years of any other cancer. This ineligibility criterion was likewise applied to controls. With their physician's permission, all study subjects were asked to be interviewed and to give a blood sample that was collected by venipuncture. DNA was isolated using a proteinase K treatment, followed by phenol-chloroform extraction and ethanol precipitation. Of the 71 ALL cases, 50 were diagnosed with B-cell ALL, 11 with T-cell ALL, and for 10 cases, the immunophenotype could not be specified. Since the prevalence of the polymorphisms under study may vary with race and the number of nonwhite cases included in the study was small, this analysis was restricted to 71 Caucasian cases and their 114 matched Caucasian controls.
Human subjects concerns
This study was in full compliance with guidelines issued by the Royal College of Pathologists for the investigation of pathologic material from cancer patients. Informed consent was obtained from both case and control subjects. Isolated DNA was sent to the University of Berkeley in a coded, unidentifiable form for genotype analysis.
Analysis of the TS 2R→3R genotypes
Presence of the tandem repeat sequences in the 5′-terminal of the regulatory region of the TS gene were detected using a protocol described by Horie et al.6 Briefly, 0.5–1.0 μg DNA and 0.2 μM each of the forward (5′-GTG GCT CCT GCG TTT CCC CC-3′) and reverse (5′-CCA AGC TTG GCT CCG AGC CGG CCA CAG GCA TGG CGC GG-3′) primers were used. Polymerase chain reaction (PCR) cycling parameters were a 5-minute denaturation cycle at 94°C and 30 cycles of the following: 94°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes. Amplified PCR products were visualized on a 3% agarose gel with ethidium bromide. Homozygotes for the double repeat (2R2R) produced a singlet 220-bp band. Heterozygotes (2R3R) produced 220-bp and 250-bp fragments, and homozygotes for the triple repeat (3R3R) produced a 250-bp fragment.
Analysis of the SHMT1 C1420T genotypes
Genotyping for the SHMT1 variants was first carried out using a standard restriction fragment length polymorphism method and restriction enzyme analysis. The following primers were used to amplify a 292-bp fragment containing the loci of interest: 5′-GTG TGG GGT GAC TTC ATT TGT G-3′ (forward) and 5′-GGA GCA GCT CAT CCA TCT CTC-3′ (reverse). Restriction enzyme digestion was carried out usingEAR1 which cuts the wild-type sequence into 113-bp and 179-bp fragments. However, we found that the EAR1 enzyme was not efficient in cutting at the restriction site, leading to incomplete digestion and inaccurate allele frequencies. We therefore performed allelic discrimination to detect the SHMT1 C1420T polymorphism using fluorogenic 3′-minor groove binding probes in a real-time PCR assay.20 The PCR was conducted in an ABI Prism 7700 thermocycler (Applied Biosystems, Foster City, CA) using fluorescently labeled wild type (5′-FAM-CGC CTC TCT CTT C-MGB-3′) and mutant (5′-VIC-CGC CTC TTT CTT C-MGB-3′) allele probes. Each 15-μL reaction contained 200 nM of each probe, 900 nM each of forward primer 5′-CAG AGC CAC CCT GAA AGA GTT C-3′ and reverse primer 5′-AGT GGG CCC GCT CCT TTA-3′, 1X Taqman Universal PCR Master Mix (Applied Biosystems), and 12 ng DNA. PCR cycling conditions consisted of one 2-minute cycle at 50°C, one 10-minute cycle at 95°C, followed by 38 cycles of 92°C for 15 seconds and 62°C for 1 minute.
Analysis of the MS A2756G genotypes
This A-to-G base pair substitution in the MS gene creates a HaeIII restriction site. Genotyping for the detection of the MS A2756G polymorphism was analyzed by PCR using 0.5 μg to 1.0 μg of human genomic DNA and 0.2 μM each of an exonic forward primer (5′-TGT TCC CAG CTG TTA GAT GAA AAT C-3′) and an intronic reverse primer (5′-GAT CCA AAG CCT TTT ACA CTC CTC-3′). These primers generate a 211-bp fragment spanning the polymorphism. PCR thermal cycling conditions were a 2-minute denaturation period at 94°C, and 40 cycles of the following: 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. This was followed by a 5-minute extension at 72°C. Restriction digestion was carried out using 1.3 μL buffer and 20 units HaeIII restriction enzyme (New England Biolabs, MA) added to 12 μL of PCR product and incubated at 37°C for at least 2 hours. Digestion products were visualized after electrophoresis on a 3% agarose gel with ethidium bromide. Wild types (2756AA) produced a singlet band at 211 bp. Heterozygotes (2756AG) produced 211-bp, 131-bp, and 80-bp fragments. Homozygous mutants (2756GG) produced 2 fragments of 80-bp and 131-bp.
Quality control samples were included in all analyses and laboratory personnel were blinded as to case and control status.
Statistical analysis
Because the data were matched, analyses were carried out using conditional logistic regression to obtain ORs and 95% CIs to estimate relative risks; note that the rate of ALL in the adult population is low (less than one case per 100 000 persons annually), and thus the estimates of the ORs generated by conditional logistic regression are approximations of relative risks. Initially, we examined the possible association of each individual polymorphism and leukemia risk univariately. This was done first categorically and then by treating the number of mutant alleles as a continuous variable (0 = wild type, 1 = heterozygous, or 2 = homozygous mutant) to increase our power to detect trends (referred to as a trend test). For the categorical analyses, we report both the individual ORs and CIs of the non–wild-type (wild type is baseline), as well as the overall likelihood ratio test (χ2) of association. For the trend test, we report only the P value of the trend (association) test. In addition, we also examined potential statistical interactions between genes. Again, both categorical and continuous (number of mutant alleles) analyses were performed, and, as in the univariate analyses, we report the ORs for the categorical analyses (stratified by polymorphisms in each gene) and the likelihood ratio test of interaction, but only the test of interaction for the trend test. The expected frequency of genotypes is a simple application of the Hardy-Weinberg law. Maximum likelihood estimates for the expectation were calculated and tested against the observed counts using a standard likelihood ratio test (against χ2). Maximum likelihoods were also calculated for cases given the Hardy-Weinberg equilibrium of the controls. All analyses were performed using the statistical program Stata (Stata, College Station, TX).
Results
Univariate analyses of the TS 2R→3R, SHMT C1420T, and MS A2756G genotypes
TS 2R→3R.
The TS triple tandem repeat (3R) allele frequency was 50.4% in the controls, within the range of 38% to 54% previously reported in Caucasians.21 22 However, in the cases, we observed a significantly lower 3R allele frequency of 38.0% (P < .01). The TS 2R2R genotype was present in 36.8% of the cases and 20.2% of the controls, the 2R3R variant in 52.2% of the cases and 58.8% of the controls, and the 3R3R tandem repeat in 11.9% of the cases and 21.1% of the controls. DNA from 3 cases did not amplify. We observed that the proportion of patients who were heterozygous for the TS tandem repeat (2R3R) was significantly lower than their respective controls, conferring a 2.8-fold decrease in ALL risk in TS 2R3R individuals when using the TS 2R2R genotype as a reference (OR = 0.36; 95% CI, 0.16-0.83; P = .017). This protective effect was even greater when TS 3R3R individuals were compared with those with the double repeat (2R2R), resulting in a 4.0-fold reduction in ALL risk among TS 3R3R variants (OR = 0.25; 95% CI, 0.08-0.78; P = .017). The trend test yielded a significant P value of .009.
SHMT1 C1420T.
For SHMT1 C1420T, the frequency of the T allele was 23.9% in the cases and 38.2% in the controls. The 1420CC genotype was observed among 57.8% of the cases and 39.5% of the controls, the 1420CT allelic variant in 36.6% of the cases and 44.7% of the controls, and the 1420TT genotype was observed in 5.6% of the cases and 15.8% of the controls. We found that when using SHMT1 1420CC as baseline, 1420CT individuals had a 2.1-fold decrease in ALL risk (OR = 0.48; 95% CI, 0.25-0.91; P = .024), whereas those with the 1420TT genotype exhibited a 3.3-fold decrease in risk of leukemia (OR = 0.31; 95% CI, 0.10-0.90; P = .031). The trend test yielded a significant P value of .008.
MS A2756G.
The overall mutant allele frequency of the MS A2756G polymorphism was 17% in the controls and 15% in the cases, within the range of what has been previously reported.15,18 23-25 The MS A2756G genotype frequencies for AA, AG, and GG were 65.8%, 34.2%, and 0% in the controls, and 71.4%, 27.1%, and 1.4%, in the cases, respectively. One case was not successfully genotyped for MS A2756G. Since there was only one homozygous variant (2756GG) found in our data set, all analyses were performed excluding MS 2756 GG. We did not find that the MS A2756G polymorphism affected ALL risk when comparing heterozygotes (2756AG) to wild types (2756AA) (OR = 0.79; 95% CI, 0.38-1.7). Because essentially only 2 polymorphisms were available in the data, a trend test was irrelevant.
Stratifying the ALL cases according to ALL-B and ALL-T immunophenotypes led to small numbers in each group, resulting in nonsignificant differences in TS 2R→3R, SHMT1 C1420T, and MS A2756G genotype frequencies between groups (data not shown). Therefore, all ALL cases were considered as a single group for this analysis.
Bivariate (interaction) analyses
Following this univariate analysis, we next investigated possible gene-gene interactions using multiplicative interaction terms in a conditional logistic regression model. Here we observed potential statistical interaction between the SHMT1 C1420T and TS 2R→3R and MS A2756G genotypes (Table 2, Table3). To measure effects of interaction between the SHMT1 and TS genes and to gain power in our analyses, we collapsed the SHMT1 1420CT and 1420TT genotype groups since there were no cases within the combined SHMT1 1420TT-TS 3R3R genotype. An interesting observation was that the relative risk of leukemia in SHMT1 1420CT/TT versus 1420CC individuals varied over levels of the TS tandem repeat. Namely, individuals with the TS 3R3R genotype who were SHMT1 1420CT/TT versus 1420CC had a 13.9-fold decrease in ALL risk (OR = 0.072; 95% CI, 0.0067-0.77;P = .03). This estimated reduction in risk was considerably reduced among TS 2R3R variants (OR = 0.39; 95% CI, 0.15-0.99; P = .048), whereas among TS 2R2R genotypes, no significant difference in risk between SHMT1 1420CT/TT and 1420CC individuals was found (OR = 1.1; 95% CI, 0.35-3.7;P = .84). The likelihood ratio test of the existence of multiplicative interaction resulted in a marginal P value of .10.
Odds ratios and 95% confidence intervals for SHMT1 1420CT/TT versus 1420CC genotypes at levels of TS 2R→3R
| SHMT1 C1420T† . | TS 2R → 3R† . | OR* . | 95% CI . | P value . |
|---|---|---|---|---|
| CT/TT v CC | 2R/2R | 1.1 | (0.35, 3.7) | .84 |
| CT/TT v CC | 2R/3R | 0.39 | (0.15, 0.99) | .048 |
| CT/TT v CC | 3R/3R | 0.072 | (0.067, 0.77) | .03 |
| SHMT1 C1420T† . | TS 2R → 3R† . | OR* . | 95% CI . | P value . |
|---|---|---|---|---|
| CT/TT v CC | 2R/2R | 1.1 | (0.35, 3.7) | .84 |
| CT/TT v CC | 2R/3R | 0.39 | (0.15, 0.99) | .048 |
| CT/TT v CC | 3R/3R | 0.072 | (0.067, 0.77) | .03 |
Test for interaction: χ2 = 4.5 (2df) (P = .10).
Odds ratios (ORs) estimated using conditional logistic regression.
Odds ratios and 95% CIs for SHMT1 1420CC/TT versus 1420CC genotypes at levels of MS A2756G
| SHMT1 C1420T3-151 . | MS A2756G3-151 . | OR3-150 . | 95% CI . | P value . |
|---|---|---|---|---|
| CT/TT v CC | AA | 0.59 | (0.28, 1.2) | .15 |
| CT/TT v CC | AG | 0.18 | (0.05, 0.63) | .008 |
| SHMT1 C1420T3-151 . | MS A2756G3-151 . | OR3-150 . | 95% CI . | P value . |
|---|---|---|---|---|
| CT/TT v CC | AA | 0.59 | (0.28, 1.2) | .15 |
| CT/TT v CC | AG | 0.18 | (0.05, 0.63) | .008 |
Test for interaction: χ2 = 2.7 (1df) (P = .10).
Odds ratios (ORs) estimated using conditional logistic regression. CI indicates confidence interval.
Potential interaction was also detected between the MS A2756G and SHMT1 C1420T polymorphisms. Specifically, among individuals with the MS 2756AG variant, the relative risk of SHMT1 CT/TT versus 1420CC was 0.18 (OR = 0.18; 95% CI, 0.05-0.63; P = .008), whereas among MS 2756AA individuals, the same relative risk (1420CT/TT versus 1420CC) was 0.59 (OR = 0.59; 95% CI, 0.28-1.2; P = .15). As above, the P value of the likelihood ratio test of no interaction was marginal at .10. Although the large variation in the estimated ORs among strata implies possible interaction, the study did not have the power to detect this magnitude of interaction statistically; no tests of statistical interaction among the trend tests were close to statistical significance.
Using our previously published data on the MTHFR 677 and 1298 polymorphisms, we next tested whether statistical interaction existed between MTHFR and TS, SHMT1, orMS in our conditional logistic regression model. We found no evidence of statistical interaction in these analyses (data not shown). Due to small sample sizes, the combined effects of more than 2 genotypes (ie, 3-way interactions) could not be examined using conditional logistic regression.
Table 1 shows that for TS andSHMT1, the population-based controls were in Hardy-Weinberg equilibrium. Further, applying the control allele frequency distribution to the cases shows that the case distribution is significantly different to the control distribution. However, the controls are not in Hardy-Weinberg equilibrium for MS (P = .035), although the deficit amounts to only 3 persons, which does not suggest a significant departure from Hardy-Weinberg. Further, there is no significant departure when comparing cases to controls.
Number of acute lymphocytic leukemia cases and controls, odd ratios, and 95% confidence intervals
| Variable . | Case (%) . | Control (%) . | OR* . | 95% CI . | Overall P value† . |
|---|---|---|---|---|---|
| TS 2R → 3R | .01 | ||||
| 2R2R | 25 (36.8) | 23 (20.2) | 1 | — | |
| 2R3R | 35 (52.2) | 67 (58.8) | 0.36 | (0.16, 0.83) | |
| 3R3R | 8 (11.9) | 24 (21.1) | 0.25 | (0.08, 0.78) | |
| SNA‡ | 3 | 0 | |||
| SHMT1 C1420T | .02 | ||||
| CC | 41 (57.8) | 45 (39.5) | 1 | — | |
| CT | 26 (36.6) | 51 (44.7) | 0.48 | (0.25, 0.91) | |
| TT | 4 (5.6) | 18 (15.8) | 0.31 | (0.10, 0.90) | |
| MS A2756G | .5 | ||||
| AA | 50 (71.4) | 75 (65.8) | 1 | ||
| AG | 19 (27.1) | 39 (34.2) | 0.79 | (0.38, 1.66) | |
| GG | 1 (1.4) | 0 (0) | — | — | |
| SNA‡ | 1 | 0 |
| Variable . | Case (%) . | Control (%) . | OR* . | 95% CI . | Overall P value† . |
|---|---|---|---|---|---|
| TS 2R → 3R | .01 | ||||
| 2R2R | 25 (36.8) | 23 (20.2) | 1 | — | |
| 2R3R | 35 (52.2) | 67 (58.8) | 0.36 | (0.16, 0.83) | |
| 3R3R | 8 (11.9) | 24 (21.1) | 0.25 | (0.08, 0.78) | |
| SNA‡ | 3 | 0 | |||
| SHMT1 C1420T | .02 | ||||
| CC | 41 (57.8) | 45 (39.5) | 1 | — | |
| CT | 26 (36.6) | 51 (44.7) | 0.48 | (0.25, 0.91) | |
| TT | 4 (5.6) | 18 (15.8) | 0.31 | (0.10, 0.90) | |
| MS A2756G | .5 | ||||
| AA | 50 (71.4) | 75 (65.8) | 1 | ||
| AG | 19 (27.1) | 39 (34.2) | 0.79 | (0.38, 1.66) | |
| GG | 1 (1.4) | 0 (0) | — | — | |
| SNA‡ | 1 | 0 |
Table depicts number of acute lymphocytic leukemia cases and controls, odd ratios (ORs), and 95% confidence intervals (CIs) by MS A2756G, SHMT1 C1420T, and TS 2R→3R, using MS 2756AA, SHMT1 1420CC, and TS 2R2R as references.
OR estimated using conditional logistic regression.
P value of overall association based upon LR-test of null model.
Samples not amplified.
Discussion
Previously, we have reported that polymorphisms at positions C677T and A1298C of the MTHFR gene are associated with reduced susceptibility to ALL. We postulated that this protective effect was due to increased levels of the methyl group donor methyleneTHF available for DNA synthesis, thereby reducing levels of incorporation of the abnormal base uracil instead of thymidine into DNA. Uracil misincorporation has been shown to cause DNA double-strand breaks,26 presumably through the deficient methylation of dUMP to dTMP. Failure to repair DNA double-strand breaks could result in the formation of chromosome translocations and deletions common in leukemia. This case-control study provides further support of the role of folate deficiency in leukemia risk, and adds greater insight into a proposed biologic mechanism of the relationship between perturbations in one-carbon metabolism and ALL. The results of our current study support the theory of enhanced availability of methyleneTHF for DNA synthesis and repair through the identification of polymorphisms in 2 important DNA-synthesizing genes, TS and SHMT1, and their significant impact on leukemia risk. Specifically, a higher distribution of TS 2R3R or 3R3R versus 2R2R variants was observed among the controls than among the cases, conferring a significant 64% (OR = 0.36; 95% CI, 0.16-0.83) and 75% (OR = 0.25; 95% CI, 0.08-0.78) decrease in risk of ALL among these individuals, respectively. Furthermore, the SHMT1 1420 T allele afforded similar levels of protection. We observed a 52% reduction in ALL risk among individuals with the SHMT1 1420CT genotype (OR = 0.48; 95% CI, 0.25-0.91), and a 69% lower risk in those with the 1420TT variant (OR = 0.31; 95% CI, 0.10-0.90).
However, no significant differences in MS A2756G genotypes among the patients and controls were observed. This may be the result of the small number of MS 2756GG variants in our study population, reducing the power necessary to detect differences. In testing for statistical interaction between the TS 2R→3R, SHMT1 C1420T, and MS A2756G genotypes, we observed interaction approaching significance between theTS and SHMT1 genes. Namely, we observed a 13.9-fold decreased risk of ALL when comparing SHMT1 1420CT/TT versus 1420CC genotypes among TS 3R3R variants (OR = 0.07; 95% CI, 0.007-0.77). However, in comparing SHMT1 1420CT/TT versus 1420CC genotypes among TS 2R2R individuals, no significant difference in risk was found. Although the statistical interactions betweenSHMT1 and TS were not significant at the 0.05 level, the large differences of risk ratios comparing polymorphisms within one position across strata defined by polymorphisms of another, as well as the generally lower power to detect interactions, suggests this statistical interaction might exist and future studies powered to detect these interactions will be needed before stronger conclusions can be made. Interestingly, the statistical interaction observed between TS and SHMT1 may have biologic relevance as outlined below.
TS binds methyleneTHF, which serves as a hydroxymethyl donor in the conversion of dUMP to dTMP in the DNA synthesis pathway.27 Reductions in TS gene expression could likely affect the balanced supply of deoxynucleotides required for normal DNA synthesis, particularly in rapidly proliferating cells such as those found in blood-forming tissues. Horie et al28have recently characterized the human TS promoter region, identifying several potential mechanisms for gene regulation. These studies found that the TS 28-bp triple repeat found in the 5′-UTR near the ATG start site led to gene expression that was 2.6 times greater than that found in the double repeat in a transient expression assay.6 Moreover, at least one set of the repeated sequence and its complementary sequence is necessary for efficient expression of the human TS gene. Possible mechanisms for the enhancing effect of the repeat could be an unknown nuclear factor that binds to the region, or the formation of stem loops in the 5′-terminal region of TS mRNA influencing gene expression.7 These studies may explain the protective effect against ALL that we observed among individuals with at least oneTS triple tandem repeat allele and further substantiates the basis that enhanced flux of methyleneTHF in the DNA synthesis pathway may attenuate cancer risk. During folate stress or as a result of TS inhibition, high levels of uracil accumulate in DNA, which has been previously demonstrated in a number of in vivo studies,26,29,30 resulting in the degradation of newly synthesized DNA due to an active excision repair pathway.31 Early investigations established that high levels of uracil misincorporation followed by extensive repair by uracil DNA glycosylase increase double-strand DNA breaks that may contribute to chromosomal instability,32translocations,33 and chromosome aberrations,34 factors that may contribute to leukemia risk.
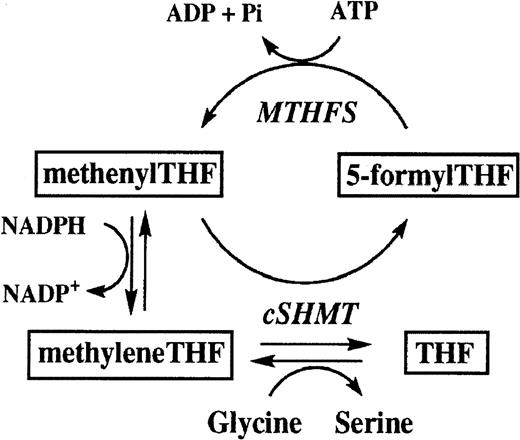
At this time, the functional role of cytosolic SHMT is not fully understood since mitochondrial SHMT can also supply the one-carbon units required for cytosolic folate metabolism.35 In the cytosol, SHMT catalyzes 2 reactions; one involves the reversible interconversion of serine to glycine to form methyleneTHF, the crucial intermediate at the branch point of 3 key pathways involved in thymidylate, purine, and methionine synthesis.36 Cytosolic SHMT also catalyzes the irreversible conversion of methenyltetrahydrofolate (methenylTHF) to 5-formyltetrahydrofolate (5-formylTHF), in what has been termed the “futile cycle”35 (Figure 2). Until recently, the role of 5-formylTHF has been poorly understood, although previous studies identified it as a potent inhibitor of cytosolic SHMT.37 According to recent studies, however, the formation of 5-formylTHF may be critical in maintaining one-carbon homeostasis, particularly during rapid proliferative stages of development.10 Thus, inhibition of cytosolic SHMT may be a mechanism to prevent an unnecessary accumulation of 5-methylTHF (committed to methionine and SAM synthesis) at the expense of depriving one-carbon units for thymidylate and purine formation.10 We speculate that the SHMT1 C1420T polymorphism may influence the flux of one-carbon moieties toward thymidylate synthesis either through increasing formation of 5-formylTHF or by enhancing interactions between SHMT1 and other genes involved in DNA synthesis such as TS or dihydrofolate reductase (DHFR). SHMT1 andTS play crucial roles in the synthesis of DNA that may involve protein-protein interactions in the production of methyleneTHF catalyzed by cytosolic SHMT, the subsequent formation of dihydrofolate (DHF) and thymidylate catalyzed by TS, and the reconversion of DHF to tetrahydrofolate (THF) catalyzed by DHFR (Figure 1).38Furthermore, TS and cytosolic SHMT may share a common translational autoregulatory process that could couple the control of their expression, providing a mechanism to tightly regulate thymidylate and DNA synthesis.39 Our observation of statistical interaction between the TS and SHMT1 genes may relate to some factor that varies between the SHMT1 1420 C and T alleles that affects its biologic interaction with TS and ultimately affects thymidylate synthesis. However, whether the SHMT1 C1420T polymorphism really affects its interaction with theTS gene cannot be concluded from this study, and requires further biochemical analyses.
Schematic of the folate futile cycle (adapted from reference 35).
5-formyltetrahydrofolate (5-formylTHF); methenyltetrahydrofolate synthetase (MTHFS); methenyltetrahydrofolate (methenylTHF); 5,10-methylenetetrahydrofolate (methyleneTHF); cytosolic serine hydroxymethyltransferase (cSHMT); tetrahydrofolate (THF).
Schematic of the folate futile cycle (adapted from reference 35).
5-formyltetrahydrofolate (5-formylTHF); methenyltetrahydrofolate synthetase (MTHFS); methenyltetrahydrofolate (methenylTHF); 5,10-methylenetetrahydrofolate (methyleneTHF); cytosolic serine hydroxymethyltransferase (cSHMT); tetrahydrofolate (THF).
In conclusion, this research reveals an association between aberrations in folate metabolic pathways which affect thymidylate synthesis and lymphocytic leukemia risk that may underscore the widespread importance of compromised DNA fidelity and insufficient folate pools in the pathogenesis of adult ALL. DNA integrity is critically dependent on the bioavailability of deoxynucleotides, particularly in cells with high replication rates such as those found in the hematopoietic system and epithelium. Even minor imbalances in pyrimidine synthesis in these cells can greatly increase the level of DNA double-strand breaks that can lead to point mutations, chromosomal translocations, and other preneoplastic alterations. Moreover, low intakes of folic acid and the folate cofactors, vitamins B2, B6, and B12, may exacerbate ALL risk in individuals with “high risk” genotypes. A combination of unfavorable genotypes, diet, and vitamin B status may conceivably be the key factor in susceptibility to adult ALL that may also hold true for certain subtypes of childhood ALL.40 Continued research in this area may help establish how dietary and nutritional modifications in susceptible individuals may alter individual ALL risk.
Supported by the Leukaemia Research Fund of Great Britain (G.J.M., R.A.C, E.R., G.R.L.), National Institutes of Health (NIH) grant HL58991 (B.S), the National Foundation for Cancer Research (M.T.S.), and NIH grant P30ES01896 from the National Institute of Environmental Health Sciences (M.T.S.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Martyn T. Smith, Division of Environmental Health Sciences, School of Public Health, 216 Earl Warren Hall, University of California, Berkeley, CA 94720-7360; e-mail:martynts@uclink4.berkeley.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal