Signal transducer and activator of transcription (STAT) proteins are involved in hematopoietic cytokine receptor signaling pathways that regulate cell proliferation, differentiation, and survival. STATs are dysregulated in acute myeloid leukemia (AML); mechanisms of dysregulation include constitutive activation and truncation of the C-terminal transactivation domain; the latter results in a β isoform that has a trans-dominant negative effect on gene induction mediated by the full-length STATα form. It was hypothesized that constitutive STAT activity might correlate with unfavorable treatment outcome in AML. Pretreatment bone marrow samples from 63 adult patients with AML were analyzed by electrophoretic mobility shift assay for the presence of STAT DNA-binding activity. Isoforms and relative levels of STAT proteins were determined by immunoblotting. Constitutive STAT3 activity was detected in samples from 28 (44%) patients. Pretreatment clinical characteristics, expression of STATα/β isoforms, and treatment regimens did not differ significantly between patients with and without constitutive STAT3 activity. Disease-free survival (DFS) was significantly shorter in patients with than in patients without constitutive STAT3 activity (median 8.7 vs 20.6 months;P = .01). Overall survival did not differ significantly. The subgroup of patients with constitutive STAT3 activity and the STAT3β isoform had the shortest DFS (P = .006) and shorter overall survival (P = .049) than all other patients. Whether adverse treatment outcome is attributable to constitutive STAT activity itself or to a process that leads to constitutive STAT activity remains to be determined. This is the first demonstration of a prognostic significance for STAT proteins in a malignancy.
Introduction
Acute myeloid leukemia (AML) is a clonal disease characterized by maturation arrest of a malignant clone of myeloid cells. Because most AML cells are incapable of proliferating ex vivo without growth factor support, growth factors and response to growth factors are likely to determine the growth and differentiation state of leukemic blasts in vivo.1
Signal transducer and activator of transcription (STAT) proteins are latent cytoplasmic transcription factors involved in hematopoietic growth factor signal transduction.2-5 STATs are activated by tyrosine phosphorylation through the action of receptor-associated Janus family tyrosine kinases (JAKs). Receptor-regulated signaling proteins appear to demonstrate specificity: interleukin-6 (IL-6),6 granulocyte (G) colony-stimulating factor (CSF),7 and thrombopoietin (TPO)8 activate STAT3, whereas granulocyte macrophage (GM)–CSF, TPO, and IL-3 primarily activate STAT5.9,10 Receptors for growth factor signaling through STAT3 and STAT5 are present on AML blasts, and these growth factors stimulate AML blast proliferation in vitro.1 Because multipotential, nonleukemic hematopoietic cells undergo differentiation, whereas leukemic cells maintain proliferation rather than differentiation in response to these growth factors, aberration of signaling pathways may contribute to leukemogenesis.
Dysregulation of the growth factor-STAT pathway can occur by 2 mechanisms. One is constitutive activation of the STATs9,10 and the other is the production of C-terminally truncated isoforms called STATβ.11-14 C-terminal transactivation domain truncation does not prevent tyrosine phosphorylation of STATβ, dimerization, or binding to DNA; rather, it either competitively inhibits STATα function or prevents transcription of STAT target genes in a dominant-negative fashion.11-14 Constitutive activation of STAT3 may also result from the introduction of a cysteine residue into the C-terminal domain of the molecule,15 and these C-terminally mutated STAT3 proteins may be oncogenic when constitutively activated. We hypothesized that constitutive STAT3 activity or production of the truncated isoform might be associated with drug resistance and might, therefore, correlate with adverse treatment outcome in AML.
Patients, materials, and methods
Patient population
The patient population consisted of 63 adult patients with AML who had at least 5 × 107 bone marrow cells cryopreserved at the time of diagnosis and were treated in a time frame that allowed for 4 years of follow-up. Pretreatment characteristics are shown in Table 1; the patients were representative of a typical AML population. Thirty-two patients were men and 31 were women. Median age was 64 years (range, 18-85 years). Fifty patients had de novo AML, and 13 had secondary AML; 9 had antecedent hematologic disorders and 4 received chemotherapy for prior malignancy. Peripheral blood mononuclear cells from 3 healthy blood donors and CD34+ cells separated from peripheral stem cell collection samples from 3 patients with breast cancer without bone marrow involvement served as controls. Prior approval was obtained from the Roswell Park Cancer Institute Institutional Review Board. All patients and volunteers signed informed consent.
Pretreatment patient characteristics
| Characteristics . | Total (n = 63) . | STAT3+(n = 28) . | STAT3− (n = 35) . | P . |
|---|---|---|---|---|
| Age, y | .20 | |||
| 60 or younger | 27 | 9 | 18 | |
| Older than 60 | 36 | 19 | 17 | |
| Sex (M/F) | 32/31 | 15/13 | 17/18 | .80 |
| Disease status | 1.00 | |||
| de novo | 50 | 22 | 28 | |
| Secondary | 13 | 6 | 7 | |
| WBC count (median, ×109/L) | 20.25 | 10.31 | 29.86 | .09 |
| FAB | NA | |||
| M0 | 5 | 3 | 2 | |
| M1 | 15 | 6 | 9 | |
| M2 | 25 | 14 | 11 | |
| M3 | 3 | 0 | 3 | |
| M4 | 6 | 3 | 3 | |
| M5 | 6 | 1 | 5 | |
| M6 | 3 | 1 | 2 | |
| Cytogenetics | .73 | |||
| Favorable | 7 | 1 | 6 | |
| Intermediate | 30 | 18 | 12 | |
| Unfavorable* | 26 | 12 | 14 | |
| STAT subtype | .29 | |||
| α | 23 | 9 | 14 | |
| β | 33 | 18 | 15 |
| Characteristics . | Total (n = 63) . | STAT3+(n = 28) . | STAT3− (n = 35) . | P . |
|---|---|---|---|---|
| Age, y | .20 | |||
| 60 or younger | 27 | 9 | 18 | |
| Older than 60 | 36 | 19 | 17 | |
| Sex (M/F) | 32/31 | 15/13 | 17/18 | .80 |
| Disease status | 1.00 | |||
| de novo | 50 | 22 | 28 | |
| Secondary | 13 | 6 | 7 | |
| WBC count (median, ×109/L) | 20.25 | 10.31 | 29.86 | .09 |
| FAB | NA | |||
| M0 | 5 | 3 | 2 | |
| M1 | 15 | 6 | 9 | |
| M2 | 25 | 14 | 11 | |
| M3 | 3 | 0 | 3 | |
| M4 | 6 | 3 | 3 | |
| M5 | 6 | 1 | 5 | |
| M6 | 3 | 1 | 2 | |
| Cytogenetics | .73 | |||
| Favorable | 7 | 1 | 6 | |
| Intermediate | 30 | 18 | 12 | |
| Unfavorable* | 26 | 12 | 14 | |
| STAT subtype | .29 | |||
| α | 23 | 9 | 14 | |
| β | 33 | 18 | 15 |
Twelve patients had one aberration each—r(7), t(6;11), t(5;8), t(9;22), add(1), +8, +mar, +13, add(20), der(13;14), +2mar, or del(X)q23—and 14 had 2 or more cytogenetic aberrations.
FAB indicates French-American-British; NA, not analyzed.
Morphologic studies
Cytogenetic analysis
Cytogenetic analysis was performed on pretreatment bone marrow cells from all patients. Bone marrow samples were processed using short-term unstimulated cultures (24-72 hours). Clonality criteria and descriptions of chromosomal aberrations were according to the International System for Human Cytogenetic Nomenclature.18Patients were divided into 3 prognostic groups based on karyotype, as previously described19; prognostic groups were favorable [t(8;21), inv(16) or t(15;17)], intermediate (normal cytogenetics), and unfavorable (all others).
Treatment
Fifty-one patients received high-dose cytarabine and idarubicin induction therapy20 and 12 received other induction regimens.21-23 Of the 46 patients who achieved complete remission (CR), 44 received postremission therapy as outlined in Table2, and 2 patients did not receive any additional therapy. Of note, postremission therapy included autologous peripheral blood stem cell transplantation in 8 patients and allogeneic transplantation in 2. Additionally, 11 patients, including 5 with and 6 without constitutive STAT3 activity, underwent allogeneic bone marrow transplantation because they had refractory or relapsed disease or they were in second remission.
Treatment outcome
| . | Total (n = 63) . | STAT3+ (n = 28) . | STAT3−(n = 35) . | P . |
|---|---|---|---|---|
| Induction treatment | .75 | |||
| HIDAC/IDA* | 51 | 22 | 29 | |
| Other† | 12 | 6 | 6 | |
| Consolidation | 46 | 21 | 25 | .37 |
| HIDAC/IDA group | 12 | 4 | 8 | |
| HIDAC/IDA alone | 8 | 2 | 6 | |
| HIDAC/IDA + VP/CY‡ | 3 | 1 | 2 | |
| HIDAC/IDA + HIDAC/DNR2-153 | 1 | 1 | 0 | |
| VP/CY | 16 | 10 | 6 | |
| Other | 16 | 6 | 10 | |
| AlloBMT in first remission | 2 | 1 | 1 | |
| AutoPBSCT | 8 | 3 | 5 | |
| ADE2-155 | 2 | 2 | 0 | |
| HIDAC alone | 2 | 0 | 2 | |
| 7 and 32-154 + HIDAC/DNR | 2 | 0 | 2 | |
| None | 2 | 1 | 1 | |
| AlloBMT relapse/refractory AML/2nd remission | 11 | 5 | 6 | NA |
| DFS (median, mo) | 10.6 | 8.7 | 20.6 | .01 |
| Overall survival (median, mo) | 14.7 | 14.0 | 16.8 | .1 |
| Attainment of CR: | ||||
| Yes | 46 | 21 | 25 | .78 |
| No | 17 | 7 | 10 |
| . | Total (n = 63) . | STAT3+ (n = 28) . | STAT3−(n = 35) . | P . |
|---|---|---|---|---|
| Induction treatment | .75 | |||
| HIDAC/IDA* | 51 | 22 | 29 | |
| Other† | 12 | 6 | 6 | |
| Consolidation | 46 | 21 | 25 | .37 |
| HIDAC/IDA group | 12 | 4 | 8 | |
| HIDAC/IDA alone | 8 | 2 | 6 | |
| HIDAC/IDA + VP/CY‡ | 3 | 1 | 2 | |
| HIDAC/IDA + HIDAC/DNR2-153 | 1 | 1 | 0 | |
| VP/CY | 16 | 10 | 6 | |
| Other | 16 | 6 | 10 | |
| AlloBMT in first remission | 2 | 1 | 1 | |
| AutoPBSCT | 8 | 3 | 5 | |
| ADE2-155 | 2 | 2 | 0 | |
| HIDAC alone | 2 | 0 | 2 | |
| 7 and 32-154 + HIDAC/DNR | 2 | 0 | 2 | |
| None | 2 | 1 | 1 | |
| AlloBMT relapse/refractory AML/2nd remission | 11 | 5 | 6 | NA |
| DFS (median, mo) | 10.6 | 8.7 | 20.6 | .01 |
| Overall survival (median, mo) | 14.7 | 14.0 | 16.8 | .1 |
| Attainment of CR: | ||||
| Yes | 46 | 21 | 25 | .78 |
| No | 17 | 7 | 10 |
AlloBMT indicates allogeneic bone marrow transplantation; AutoPBSCT, autologous peripheral blood stem cell transplantation; NA, not analyzed; and STAT, signal transducer and activator of transcription proteins.
High-dose cytarabine 3 g/m2 every 12 hours for a total of 12 doses (1.5 g/m2 for patients older than 50) and idarubicin 12 mg/m2 daily for 3 consecutive days.
Five patients treated with cytarabine 100 mg/m2 continuous infusion over 7 days, daunorubicin 60 mg/m2 daily for 3 days, etoposide 100 mg/m2 for 3 days; three patients treated with cytarabine 100 mg/m2continuous infusion over 7 days, daunorubicin 45 mg/m2daily for 3 days, etoposide 100 mg/m2 for 3 days, and PSC 833 10 mg/m2 for 3 days; one patient treated with cytarabine 200 mg/m2 continuous infusion over 7 days and daunorubicin 45 mg/m2 daily for 3 days; one patient treated with all-trans retinoic acid 40 mg/m2 every 12 hours and idarubicin 12 mg/m2 every other day for 4 doses; one patient treated with cytarabine 1.5 g/m2 every 12 hours for a total of 12 doses; one patient treated with cytarabine 100 mg/m2 continuous infusion over 7 days.
Etoposide 3.6 g/m2 by continuous infusion and cyclophosphamide 50 mg/kg for 4 days.
High-dose cytarabine 2 g/m2 every 12 hours for a total of 8 doses and daunorubicin 45 mg/m2 for 2 days.
Cytarabine 100 mg/m2 continuous infusion over 5 days, daunorubicin 60 mg/m2 daily for 2 days, and etoposide 100 mg/m2 for 2 days.
Cytarabine 100 mg/m2 continuous infusion over 7 days and daunorubicin 45 mg/m2 daily for 3 days.
Response criteria
CR was defined as the normalization of blood counts and bone marrow morphology and the disappearance of all signs of leukemia, lasting for 4 weeks or longer, in accordance with the recommendations of the National Cancer Institute–sponsored workshop.24Relapse was defined as the reappearance and persistence of blasts in the blood or the appearance of 5% or more blasts in the bone marrow not attributable to another cause.
Materials
All chemicals were purchased from Sigma Immunochemicals (St Louis, MO) unless otherwise specified. TPO and G-CSF were kindly provided by Amgen (Thousand Oaks, CA).
Cell collection
Bone marrow samples were collected at diagnosis. Light-density bone marrow cells were isolated by 1.077 g/μL Ficoll-Hypaque density gradient centrifugation and were cryopreserved using standard techniques. Cells were also thawed by standard techniques, and cell viability was verified by the Trypan blue dye exclusion test. Three samples were studied fresh (on the day of collection) and after cryopreservation and thawing to determine whether cryopreservation and thawing affected STAT activity.
Controls consisted of monocytes and granulocytes separated from samples of 3 healthy blood donors. Mononuclear cells were obtained by density centrifugation. Cells were resuspended in RPMI 1640 medium containing 2% fetal calf serum and were incubated in culture dishes for 2 hours at 37°C at 2 × 106 cells/mL. The supernatant containing the nonadherent cells was discarded, and the adherent cells were collected. To obtain granulocytes, the cell pellet was collected after density centrifugation. Red blood cells were lysed with 1.22% ammonium oxalate, allowing the isolation of granulocytes. Additional non-AML controls included CD34+ cells separated from peripheral stem cell collections from 3 patients with breast cancer without bone marrow involvement using the MACS progenitor cell isolation kit (Miltenvy Biotec, Auburn, CA).
Electrophoretic mobility shift assay
The DNA-binding activity of STAT3 and STAT5 in the absence of cytokine or growth factor treatment was assessed by electrophoretic mobility shift assay (EMSA). Whole-cell extracts prepared from AML cells as described13 were incubated with32P-labeled oligomers corresponding to the high-affinity binding element for STAT3, SIE,25 and STAT5 (TB2).26 Complexes were analyzed by 5% polyacrylamide gel electrophoresis and autoradiography. Extracts of TPO-treated MO7E cells displaying activated STAT3α, STAT5α (both A and B27), and CD34+ cells expressing inactive STAT3α, STAT5Aα, and STAT5Bα served as controls for these analyses. Relative DNA-binding activity was determined by densitometry of the autoradiographs; activated MO7E standard was defined as 100%. The detection limit of the EMSA system was set at 2% or less. Binding activity with values greater than 2% was defined as constitutive. Identity of the STAT-containing complexes was determined by antibody supershift with C-terminal–specific anti-STAT3 (C-20) or anti-STAT5 (C-17) monoclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Western blotting
Tyrosine-phosphorylated and unphosphorylated STAT3, STAT5A, and STAT5B proteins were quantitated by Western blot analysis as previously described.13 In brief, whole-cell extracts were separated on 7.5% polyacrylamide sodium dodecyl sulfate gels, and proteins were transferred to nitrocellulose membranes. Membranes were incubated with either antiphospho-STAT3 (Y705) or antiphospho-STAT5A/B (Y694/Y699) antibodies (Upstate Biotechnology, Lake Placid, NY) or with N-terminal–specific anti-STAT3 and anti-STAT5 (Transduction Laboratories, Lexington, KY) to quantitate unphosphorylated proteins. C-terminal–specific anti-STAT3 (C-20) and anti-STAT5 (C17) (Santa Cruz Biotechnology) were used to verify the identity of the C-terminal truncation of the STAT protein forms. Immune complexes were detected by the enhanced chemiluminescence reaction (Amersham Life Science, Arlington Heights, IL).
Statistical methods
The following data were analyzed: constitutive STAT3 activity (present vs absent), patient age (60 years or younger vs older than 60 years), sex, initial white blood cell count (median), karyotype subgroups (favorable vs intermediate vs unfavorable), AML type (de novo vs secondary), STAT isoform (α vs β), treatment (induction treatment with high-dose cytarabine and idarubicin vs other regimens; consolidation treatment with high-dose cytarabine and idarubicin vs high-dose etoposide and cyclophosphamide vs other regimens), attainment of CR (yes vs no), disease-free survival (DFS), and overall survival.
DFS was defined as the time from achievement of CR to relapse, death, or last follow-up visit. Patients alive and still in remission at last follow-up examination were censored in the analysis. Overall survival was calculated from the date of diagnosis to the date of death or the date of last follow-up for living patients. Follow-up time was defined as the time from the date of diagnosis to the date of last contact among patients known to be alive. Time to achieving CR was defined as the interval from date of diagnosis to the date of clinically documented remission. Estimates of DFS and overall survival probabilities were calculated using the Kaplan-Meier method.28 The log-rank statistic29 was used to test for differences in survival times between the 2 STAT3 groups. The proportionality assumption in the Cox proportional hazards regression model was tested for constitutive STAT3 activation for the endpoint of overall survival, and it was found that a Cox model that incorporated a time-dependent covariate30 was a better fit for the data. This model allowed the hazard function for STAT3 expression to vary over time. Patient age, AML type, cytogenetic status, and WBC count were analyzed in univariate Cox proportional hazards models. Variables significant at the 10% level were then analyzed in conjunction with STAT3 and the time-dependent covariate to determine the effect of each of these variables on the prognostic significance of STAT3 expression. Sparseness in the data set did not allow for multivariate analysis of these factors simultaneously or for an appropriate statistical model to be derived for disease-free survival. Associations between both STAT3 groups and categorical variables were evaluated using the Fisher exact test.31The Kruskal-Wallis test32 was used to test for an association between STAT3 and karyotype subgroups. WBC count and time to CR were compared between the 2 STAT3 groups using the Mann-Whitney rank sum test.33 All statistical tests were 2-sided, with statistical significance defined as P < .05.
Results
Constitutive STAT activity
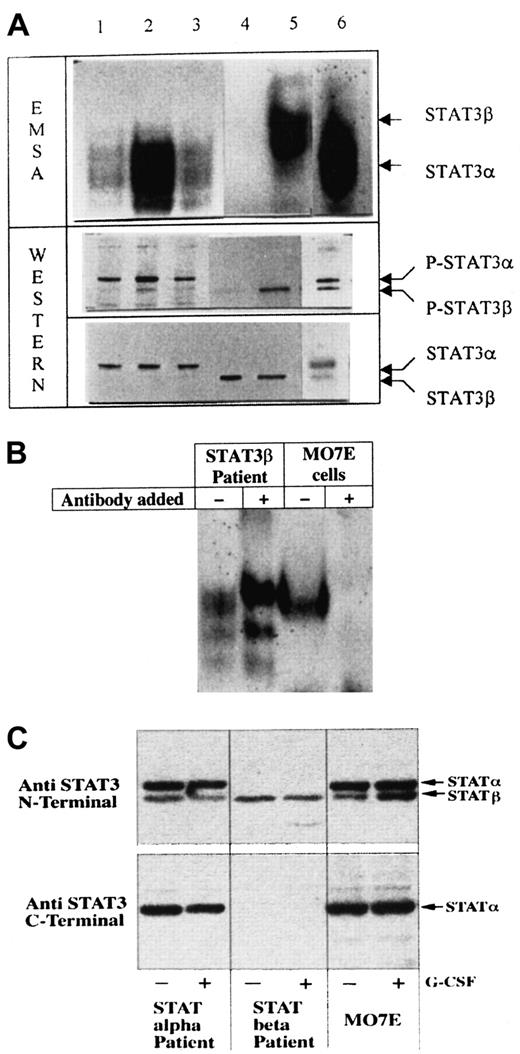
Constitutive STAT3 activity was present in leukemic blasts from 28 of the 63 (44%) patients but in none of the control samples. Constitutive STAT5 activity was detected in blasts from 11 of the 50 (22%) patients. Cryopreservation had no effect on constitutive STAT activity (data not shown), and the results obtained by EMSA and by Western blot analysis were identical. Figure1 depicts EMSA and Western blot analyses for STAT3 from 6 representative patients.
Constitutive STAT3 activity in AML blasts.
(A) The top panel shows EMSA, demonstrating the SIE-binding activity of STAT3, and the lower 2 panels show Western blotting. The middle panel shows hybridization with antibodies against phosphorylated STAT3, and the lower panel shows hybridization with anti-STAT3 antibodies to demonstrate equal loading. The positions of STAT3α and STAT3β are indicated. Lanes 1, 2, 3, 5, and 6 show patient samples with constitutive STAT3 activity. Lane 4 represents a patient sample without constitutive STAT3 activity. Note that the bands in lanes 1, 2, 3, and 6 demonstrate faster migration—that is, STAT3α—whereas the band in lane 5 shows slower migration—that is, STAT3β. (B) Supershift analysis of the SIE band complex formed with C-terminally directed anti-STAT3 antibodies in cells from a patient expressing predominantly STAT3β and MO7E cells expressing predominantly STAT3α. There is no supershift in the patient sample, whereas STAT3α is supershifted in MO7E cells. (C) Western blot analysis of STAT3 hybridized with antibodies directed against the N-terminal (top panel) and the C-terminal (lower panel) domains. When using antibodies directed against the C-terminal domain of STAT3, STAT3β was not detected. To verify that no change occurred after cytokine exposure, samples were exposed to human G-CSF (10 ng/mL) for 10 minutes as previously described.13
Constitutive STAT3 activity in AML blasts.
(A) The top panel shows EMSA, demonstrating the SIE-binding activity of STAT3, and the lower 2 panels show Western blotting. The middle panel shows hybridization with antibodies against phosphorylated STAT3, and the lower panel shows hybridization with anti-STAT3 antibodies to demonstrate equal loading. The positions of STAT3α and STAT3β are indicated. Lanes 1, 2, 3, 5, and 6 show patient samples with constitutive STAT3 activity. Lane 4 represents a patient sample without constitutive STAT3 activity. Note that the bands in lanes 1, 2, 3, and 6 demonstrate faster migration—that is, STAT3α—whereas the band in lane 5 shows slower migration—that is, STAT3β. (B) Supershift analysis of the SIE band complex formed with C-terminally directed anti-STAT3 antibodies in cells from a patient expressing predominantly STAT3β and MO7E cells expressing predominantly STAT3α. There is no supershift in the patient sample, whereas STAT3α is supershifted in MO7E cells. (C) Western blot analysis of STAT3 hybridized with antibodies directed against the N-terminal (top panel) and the C-terminal (lower panel) domains. When using antibodies directed against the C-terminal domain of STAT3, STAT3β was not detected. To verify that no change occurred after cytokine exposure, samples were exposed to human G-CSF (10 ng/mL) for 10 minutes as previously described.13
Constitutive STAT activity and pretreatment clinical characteristics
Differences in patient age, sex, frequency of de novo compared with secondary AML, initial WBC counts, prognostic karyotype subgroups, and expression of STAT3α/β isoforms were not statistically significant between patients with and without constitutive STAT3 activity (Table 1). Patients with and without constitutive STAT5 activity were not compared because of the small proportion of patients with constitutive STAT5 activity.
Constitutive STAT activity and treatment outcome
There was no difference in the distribution of induction (P = .75) and consolidation (P = .37) treatment regimens between the patients with and without constitutive STAT3 activity (Table 2). The CR rate was 75% for patients with constitutive STAT3 activity and 71% for those without. Median follow-up duration was 40 months (range, 19 to 64 months). Fourteen patients are still alive in CR, 4 died in CR of unrelated conditions, and 28 patients have had relapses. The median time to achievement of CR was 42 days (range, 31-78 days) for patients with constitutive STAT3 activity and 39 days (range, 24-92 days) for those without it (P = .37).
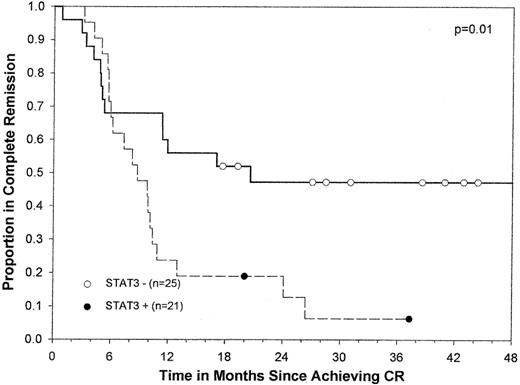
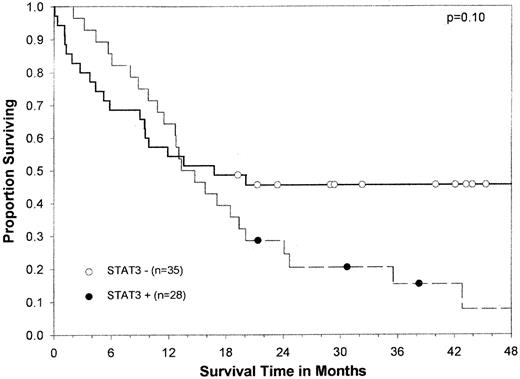
In univariate analysis, DFS was significantly shorter in patients with constitutive STAT3 activity than in those without it (median, 8.7 vs 20.6 months; P = .01) (Figure2). The probability of DFS at 12 months was 0.24 in patients with constitutive STAT3 activity and 0.56 in those without. The difference in overall survival was not statistically significant between the 2 groups (median, 14.0 vs 16.8 months;P = .1) (Figure 3), probably because of the effect of salvage therapy after relapse. However, only 18% of patients with constitutive STAT3 activity were alive at 25 months compared with 37% of those with no constitutive activity.
Kaplan-Meier curve for DFS by constitutive STAT3 activity for 46 patients with AML.
Circles indicate censored patients. (●), patients whose blasts had constitutive STAT3 activity. (○), patients whose blasts did not have constitutive STAT3 activity. Of note, one patient whose blasts did not have constitutive STAT3 activity survived in remission for more than 5 years.
Kaplan-Meier curve for DFS by constitutive STAT3 activity for 46 patients with AML.
Circles indicate censored patients. (●), patients whose blasts had constitutive STAT3 activity. (○), patients whose blasts did not have constitutive STAT3 activity. Of note, one patient whose blasts did not have constitutive STAT3 activity survived in remission for more than 5 years.
Kaplan-Meier overall survival curve by constitutive STAT3 activity for 63 AML patients.
Circles indicate censored patients. (●), patients whose blasts had constitutive STAT3 activity. (○), patients whose blasts did not have constitutive STAT3 activity. Of note, 2 patients whose blasts did not have constitutive STAT3 activity survived for more than 5 years.
Kaplan-Meier overall survival curve by constitutive STAT3 activity for 63 AML patients.
Circles indicate censored patients. (●), patients whose blasts had constitutive STAT3 activity. (○), patients whose blasts did not have constitutive STAT3 activity. Of note, 2 patients whose blasts did not have constitutive STAT3 activity survived for more than 5 years.
Because age, de novo versus secondary AML, and karyotype are known to affect treatment outcome,34 we analyzed the effect of constitutive STAT3 activity on overall survival in association with each of these factors. Table 3 shows the results of the Cox proportional hazards and time-dependent regression analyses. On univariate analysis, age, AML type, and cytogenetic subgroups, but not initial WBC count, were significantly related to overall patient survival. Patients younger than 60 had a 66% reduced risk for death. Patients with de novo AML had a 59% lower risk for death than those with secondary AML. Those with favorable cytogenetics had a 91% lower risk for death than the remaining patients. In the time-dependent model, patients without constitutive STAT3 activity had a 75% lower risk for death (P = .03). However, the differences in risk between patients with constitutive STAT3 activity and those without changed slightly (20%) over time. Constitutive STAT3 activity retained its statistical significance when adjusted for in models with age, AML type, and cytogenetic subgroups in time-dependent covariate analysis. Treatment outcome for patients with and without constitutive STAT5 activity was not analyzed because of the small proportion of patients with constitutive STAT5 activity.
Cox proportional hazard and time-dependent regression analysis for overall survival
| Variable . | n . | Parameter estimate . | Relative risk . | 95% CI . | P . |
|---|---|---|---|---|---|
| Univariate analysis | |||||
| Age, y | |||||
| 60 or younger vs older than 60 | 63 | −1.09 | 0.34 | 0.17-0.66 | .002 |
| AML type | |||||
| de novo vs secondary | 63 | −0.88 | 0.41 | 0.21-0.80 | .009 |
| Cytogenetic status | |||||
| favorable vs intermediate or unfavorable | 62 | −2.41 | 0.09 | 0.01-0.66 | .018 |
| WBC count | 60 | 0.001 | 1.00 | 1.00-1.01 | .615 |
| Time-dependent covariate models | |||||
| STAT3 (no activity vs activity) | 63 | −1.40 | 0.25 | 0.07-0.88 | .031 |
| STAT3 × time-dependent covariate | 0.18 | 1.20 | 1.07-1.34 | .002 | |
| STAT3 | 63 | −1.57 | 0.21 | 0.06-0.75 | .016 |
| STAT3 × time-dependent covariate | 0.17 | 1.19 | 1.06-1.33 | .003 | |
| Age | −0.92 | 0.40 | 0.20-0.79 | .009 | |
| STAT3 | 62 | −1.42 | 0.24 | 0.06-0.75 | .029 |
| STAT3 × time-dependent covariate | 0.17 | 1.19 | 1.06-1.33 | .003 | |
| AML type | −0.61 | 0.55 | 0.28-1.07 | .077 | |
| STAT3 | 62 | −1.56 | 0.21 | 0.06-0.75 | .016 |
| STAT3 × time-dependent covariate | 0.16 | 1.18 | 1.05-1.32 | .005 | |
| Cytogenetic status | −1.97 | 0.14 | 0.02-1.03 | .054 |
| Variable . | n . | Parameter estimate . | Relative risk . | 95% CI . | P . |
|---|---|---|---|---|---|
| Univariate analysis | |||||
| Age, y | |||||
| 60 or younger vs older than 60 | 63 | −1.09 | 0.34 | 0.17-0.66 | .002 |
| AML type | |||||
| de novo vs secondary | 63 | −0.88 | 0.41 | 0.21-0.80 | .009 |
| Cytogenetic status | |||||
| favorable vs intermediate or unfavorable | 62 | −2.41 | 0.09 | 0.01-0.66 | .018 |
| WBC count | 60 | 0.001 | 1.00 | 1.00-1.01 | .615 |
| Time-dependent covariate models | |||||
| STAT3 (no activity vs activity) | 63 | −1.40 | 0.25 | 0.07-0.88 | .031 |
| STAT3 × time-dependent covariate | 0.18 | 1.20 | 1.07-1.34 | .002 | |
| STAT3 | 63 | −1.57 | 0.21 | 0.06-0.75 | .016 |
| STAT3 × time-dependent covariate | 0.17 | 1.19 | 1.06-1.33 | .003 | |
| Age | −0.92 | 0.40 | 0.20-0.79 | .009 | |
| STAT3 | 62 | −1.42 | 0.24 | 0.06-0.75 | .029 |
| STAT3 × time-dependent covariate | 0.17 | 1.19 | 1.06-1.33 | .003 | |
| AML type | −0.61 | 0.55 | 0.28-1.07 | .077 | |
| STAT3 | 62 | −1.56 | 0.21 | 0.06-0.75 | .016 |
| STAT3 × time-dependent covariate | 0.16 | 1.18 | 1.05-1.32 | .005 | |
| Cytogenetic status | −1.97 | 0.14 | 0.02-1.03 | .054 |
Time-dependent covariate shows how the relative risk associated with constitutive STAT3 activity varies over time.
Expression of truncated STAT3β form in leukemic cells with constitutive STAT3 activity identifies a group of patients with short DFS and overall survival
There was a statistically significant difference in DFS when comparing patients with constitutive STAT3 activity and expression of the truncated isoform versus the remaining patients (P = .006). The median DFS for those with constitutive STAT3β expression was 7.4 months, compared with a median time of 18.8 months for the others. The difference in overall patient survival between these 2 groups of patients was also significant (P = .049); patients with constitutive STAT3β expression had a median survival time of 12.9 months compared with 18.9 months for the others.
Discussion
We have demonstrated that constitutive STAT3 activity is associated with short DFS. The difference in DFS persisted when controlling for age, de novo versus secondary AML, and karyotype. Additionally, the subgroup of patients with constitutive STAT3β activity had particularly short DFS and shorter overall survival. To our knowledge, the relationship between constitutive STAT3 activity and short DFS has not been reported. Moreover, this is the first demonstration of a relationship between STAT expression and disease outcome in any malignancy.
Constitutive STAT3 activity has been reported in 30% to 100% of patients with AML.13,35-37 Several mechanisms have been proposed to explain constitutive STAT3 activity in AML. One proposed mechanism is activation of the signaling pathway by an autocrine loop mediated by a hematopoietic growth factor. Schuringa et al38 showed that constitutive STAT3 activity in AML blasts resulted from autocrine secretion of IL-6. However, IL-6 is known to be inhibitory for AML proliferation.39 Therefore, the role of IL-6–induced STAT3 activation in leukemogenesis remains unclear. Other growth factors, TPO, and G-CSF, are also known to activate STAT3.7,8 We and others19,40-42 have previously demonstrated that the TPO receptor is expressed on AML blasts in 45% to 70% of patients with AML. Moreover, we found that DFS was significantly shorter in the subgroup of AML patients whose blasts expressed TPO receptor mRNA.19 Nine of the same patient samples were analyzed in that study and in the current study. Two patient samples that expressed c-mpl mRNA had constitutive STAT3 activity; among the 7 samples that did not express c-mpl mRNA, 6 did not demonstrate constitutive STAT3 activity. Although the numbers are small, the correlation is intriguing in that it suggests that constitutive STAT3 activity might be induced by TPO. Additional work is needed to address this possible mechanism. Similarly, G-CSF has been shown to induce the transformation and proliferation of AML blasts through STAT3α activation.7Finally, cellular transformation triggered by various tyrosine kinase oncoproteins, such as v-Src and Lck, has also been associated with the constitutive activation of STAT3,43-46 but involvement of these oncoproteins has not been demonstrated in AML and cannot explain the high proportion of patients with constitutive STAT3 activity in this disease. These studies strongly suggest that constitutive STAT3 activity plays an important role in malignant transformation. It is unclear whether constitutive STAT3 activity is a cause or a result of the transforming event.
The targets of STAT3 have not yet been identified. One possible target may be the antiapoptotic pathway of Bcl-2. Expression of antiapoptotic proteins Bcl-XL and Mcl-1, members of the Bcl-2 family, was shown to be increased in multiple myeloma cells with IL-6–induced constitutive STAT3 activity.47,48 Abrogation of STAT3 signaling has been demonstrated to block Bcl-XL expression, with subsequent induction of apoptosis.47 These data suggest that constitutive STAT3 activity may confer resistance to apoptosis in multiple myeloma cells. No similar data are available yet for AML.
Introducing a cysteine at the C-terminal loop of the SH2 domain of STAT3 causes the molecule to dimerize, promote transcription, and induce cell transformation,15 suggesting that altering the C-terminal domain of STAT3 induces constitutive activation, resulting in a transformed phenotype. C-terminally truncated STAT3β isoforms were shown to have a trans-dominant negative effect on gene induction in the STAT signal transduction pathway.11-14 In our assays, C-terminal mutation has not been evident; rather, we have found truncation and loss of the transactivating domain with preservation of the tyrosine phosphorylation capability.13 14 In our patient cohort, the expression of C-terminally truncated STAT3β isoform was associated with an even worse outcome when detected in AML blasts with constitutive STAT3 activity.
In summary, we have demonstrated that constitutive STAT3 activity is associated with short DFS in AML. Moreover, a subgroup of patients with constitutive STAT3 activity who displayed STAT3β isoform experienced a particularly poor outcome. The correlation between STAT expression and outcome remains to be explained. The long-term implication of our data is that the development of novel therapies targeting the signal transduction pathways in AML cells may hold promise for improving treatment outcome in this disease.
Supported in part by grants from the Tower Foundation (Buffalo, NY) and by National Cancer Institute grants CA16056 and CA26122. M.B. is a recipient of The Cancer and Leukemia Group B Clinical Research Award supported by Ortho Biotech, Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Meir Wetzler, Leukemia Section, Dept of Medicine, Roswell Park Cancer Institute, Elm and Carlton Sts, Buffalo, NY 14263; e-mail: meir.wetzler@roswellpark.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal