The acute myelogenous leukemia–1 (AML1)–ETO fusion protein is generated by the t(8;21), which is found in 40% of AMLs of the French-American-British M2 subtype. AML1-ETO interferes with the function of the AML1 (RUNX1, CBFA2) transcription factor in a dominant-negative fashion and represses transcription by binding its consensus DNA–binding site and via protein-protein interactions with other transcription factors. AML1 activity is critical for the development of definitive hematopoiesis, and haploinsufficiency of AML1 has been linked to a propensity to develop AML. Murine experiments suggest that AML1-ETO expression may not be sufficient for leukemogenesis; however, like the BCR-ABL isoforms, the cellular background in which these fusion proteins are expressed may be critical to the phenotype observed. Retroviral gene transfer was used to examine the effect of AML1-ETO on the in vitro behavior of human hematopoietic stem and progenitor cells. Following transduction of CD34+ cells, stem and progenitor cells were quantified in clonogenic assays, cytokine-driven expansion cultures, and long-term stromal cocultures. Expression of AML1-ETO inhibited colony formation by committed progenitors, but enhanced the growth of stem cells (cobblestone area-forming cells), resulting in a profound survival advantage of transduced over nontransduced cells. AML1-ETO–expressing cells retained progenitor activity and continued to express CD34 throughout the 5-week long-term culture. Thus, AML1-ETO enhances the self-renewal of pluripotent stem cells, the physiological target of many acute myeloid leukemias.
Introduction
The recurrent chromosomal translocations found in acute myelogenous leukemia (AML) often involve transcription factors. Translocations may affect the level of gene expression, or more commonly, these genetic abnormalities may generate fusion or chimeric proteins with altered functional properties. Chromosomal translocations often disrupt genes that are required for the normal development of blood cells; one example is the transcription factor core-binding factor (CBF). CBF is a heterodimeric complex that binds to core enhancer elements in viral and cellular genes. Both components of CBF are affected by translocations found in human leukemias, including t(8;21), t(3;21), t(12;21), and inv(16).1,2 The CBFα (AML1/RUNX1) subunit binds DNA directly, whereas the CBFβ subunit enhances binding of CBFα to DNA but does not bind DNA itself. The importance of AML1 in hematopoiesis has been shown by the phenotype of AML1 and CBFβ knockout mice. Both mice lack definitive hematopoiesis and die in utero, and embryonic stem cells from these null mice are unable to contribute to definitive hematopoiesis in chimeric mice.3-7 These findings demonstrate that AML1/CBFβ is required for the development of hematopoietic stem cells.
The (8;21) translocation is associated with approximately 40% of myeloid leukemias of the French-American-British (FAB) M2 subtype and rearranges the AML1 gene on chromosome 21q22 and theETO gene on chromosome 8q22, generating an AML1-ETO fusion protein that contains the first 177 amino acids of AML1 (including the DNA-binding domain but lacking the transcriptional activation domain of AML1) and almost the full length of the ETO protein. The role of AML1-ETO in the pathogenesis of AML has been intensely studied. Thus far, it has been shown that AML1-ETO functions mainly as a transcriptional repressor, while AML1 is mainly a transcriptional activator.8 The importance of the ETO domain to this repression has recently been identified. ETO binds the corepressors N-CoR and mSin3 and recruits histone deacetylase activity. These functions correlate with the ability of AML1-ETO to repress transcription and block myeloid differentiation.9-11 Several model systems have documented the ability of AML1-ETO to inhibit the differentiation of myeloid cell lines in response to chemical signals.12-14 However, the use of cell lines may not accurately model the functional effects of AML1-ETO expression, owing to the different complement of transcription factors found in transformed cells versus hematopoietic progenitor cells (HPCs).
Murine models suggest that AML1-ETO may not be sufficient for leukemogenesis. AML1-ETO “knock-in” mice do not develop leukemia, and factor-dependent cell lines derived from AML1-ETO–expressing murine progenitors are not leukemogenic when transplanted into irradiated syngeneic or severe combined immunodeficient (SCID) mice.15,16 The phenotype of an inducible transgenic mouse model for AML1-ETO is consistent with this hypothesis, as these mice do not develop leukemia upon induction of AML1-ETO expression in hematopoietic cells.17 The AML1-ETO knock-in and transgenic mice have identified several effects of AML1-ETO in hematopoiesis; however, murine HPCs may respond to AML1-ETO expression differently than human HPCs. The importance of expressing these fusion proteins in the “correct” cell type has recently been demonstrated for the t(5;12)–associated protein TEL/PDGFRβ, which is found in chronic myelomonocytic leukemia with eosinophilia. Using a panel of different TEL/PDGFRβ point mutations, Tomasson et al18showed that several mutants could act similarly to wild-type TEL/PDGFRβ and confer factor-independent growth on interleukin (IL)–3–dependent murine cell lines. However, these same mutants do not induce chronic myelomonocytic leukemia when expressed in a murine bone marrow retroviral transduction/transplantation system, whereas the wild-type TEL/PDGFRβ does.
The t(8;21) occurs in a stem cell, as shown by the ability of leukemia cells from patients with AML FAB-M2 to engraft SCID mice.19 Only the CD34+CD38−subset (characteristic of stem cells) contained repopulating ability in the mice. To date there is no accurate model for studying the effects of AML1-ETO on primary human HPCs. We devised a system to express AML1-ETO in human CD34+ cells using retroviral transduction and green fluorescent protein (GFP) as a marker protein. We find differential effects of AML1-ETO expression on human HPCs; AML1-ETO blocks the proliferation of committed progenitors, yet it extends the self-renewal potential of stem cells. The expression of AML1-ETO alone has profound effects on the growth properties of the normal target cell affected by AML, namely, the human hematopoietic stem cell.
Materials and methods
Retroviral vectors and constructs
The MIGRI vector was kindly provided by Dr Warren Pear. The AML1-ETO complementary DNA (cDNA) was tagged with the HA epitope at the aminoterminal end, and a Kozak consensus site was added by polymerase chain reaction (PCR) with the use of the following primers: 5′-CGCATCGATCCATGGCATACCCATACGACGTCCCAGACTACGCTCGTATCCCCGTAGATGCCAG-3′ and 5′-CGTGCCATTAGTTAACGTTGTCGG-3′. This PCR product was substituted into AML1-ETO in pBS-SK with the use of a unique internal HpaI site and a Cla I site in the multiple cloning region. The sequence was verified and the cDNA was subcloned into MIGR1. The pSV-A-MLV-env was obtained from Dr Nathaniel Laudau and Dr Dan Littman through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. The pEQ-PAM3(-E) plasmid was a gift from Dr Elio Vanin at St Jude Children's Research Hospital (Memphis, TN).
Retroviral production and bone marrow transduction
MIGRI, pEQ-PAM3(-E), and pSV-A-MLV-env were transiently transfected into 293T cells in a 1:0.67:0.33 ratio; 12 hours later, the precipitate was washed away, and viral supernatants were collected at 24, 36, and 48 hours. The supernatant was centrifuged at 1800 rpm and filtered through a 0.45-μm filter. Mobilized peripheral blood progenitor cells (PBPCs) were obtained from patients undergoing stem cell transplantation at Memorial Sloan-Kettering Cancer Center (New York, NY) following their informed consent according to the hospital's institutional review board–approved protocol. CD34+ cells were selected from the leukapheresis product by means of the MACS CD34 isolation kit from Miltenyi Biotec (Auburn, CA). Purified CD34+ cells were cycled for 72 hours in Iscoves modified Dulbeccos medium (IMDM); 20% heat-inactivated fetal calf serum (FCS); 2 mM L-glutamine; 100 U/mL penicillin/streptomycin with FLT3 ligand (FLT3L) (100 ng/mL); stem cell factor (SCF) (100 ng/mL); thrombopoietin (TPO) (100 ng/mL); IL-6 (20 ng/mL); granulocyte-macrophage colony-stimulating factor (GM-CSF) (20 ng/mL); and IL-3 (50 ng/mL) (all from Peprotech, Rocky Hill, NJ). We transduced 2 × 105 PBPCs with 4 rounds of retrovirus using retronectin-coated dishes (TaKaRa Shuzo, Shiga, Japan) in IMDM, 20% heat-inactivated FCS, and the same media used for cycling with the addition of 8 μg/mL protamine sulfate (Sigma, St Louis, MO). The cells were expanded in the cycling mix described above for 48 hours and subsequently analyzed on a Becton Dickinson (San Jose, CA) FACScan. Cord blood cells were obtained from the New York Blood Center (kindly provided by Dr Pablo Rubenstein), and CD34+ cells were selected as described for PBPCs. Cells were cycled for 48 hours in IMDM, 20% heat-inactivated FCS, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin with FLT3L (100 ng/mL), SCF (100 ng/mL), TPO (100 ng/mL), and IL-6 (100 ng/mL). CD34+ cord blood cells were transduced as described for CD34+ PBPCs, without the addition of GM-CSF or IL-3.
Hematopoietic progenitor assays
For quantitation of clonogenic progenitors, cells were plated at 2 × 103 cells per milliliter in IMDM with 30% heat-inactivated FCS and 1% methylcellulose (Gibco, Rockville, MD) containing 50 μM β-mercaptoethanol, 2 mM L-glutamine, 0.1 M hemin, 100 U/mL penicillin/streptomycin, 10 ng/mL granulocyte CSF (G-CSF), 20 ng/mL IL-6, 20 ng/mL IL-3, 20 ng/mL SCF, and 600 000 U/L (6 U/mL) erythropoietin (EPO). Granulocyte-macrophage colony-forming unit (CFU-GM), erythroid burst-forming unit (BFU-E), and granulocyte, erythrocyte, megakaryocyte, macrophage CFU (CFU-GEMM) colonies (collectively called CFU cells [CFU-Cs]) consisting of more than 50 cells were scored 14 days later.
Cell culture and cell lines
Transduced CD34+ cells were expanded in IMDM media with 20% heat-inactivated FCS; 2 mM L-glutamine; 100 U/mL penicillin/streptomycin with FLT3L (100 ng/mL); SCF (100 ng/mL); TPO (100 ng/mL); IL-6 (20 ng/mL); GM-CSF (20 ng/mL); EPO (600 000 U/L [6 U/mL]); and IL-3 (50 ng/mL) (Peprotech). Cells were counted weekly and replated at a concentration of 1 × 105/mL. GFP expression was monitored weekly by flow cytometry. To assay for stem cells by the cobblestone area forming cell (CAFC) assay, CD34+ cells were cocultured with the MS-5 monolayer in α–Eagle minimum essential medium (α-MEM) containing 10% heat-inactivated FCS, 10% horse serum, 1 × 10−6 M hydrocortisone, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin. The cultures were counted, demidepopulated, and analyzed by flow cytometry each week. After 5 weeks in culture, total and GFP+ cobblestone areas were counted (a minimum of 8 phase-dark cells were needed to count as a cobblestone area). At week 5 or 6, all nonadherent cells were collected; adherent cells were trypsinized; cells were counted; and progenitors were quantified in methylcellulose assays. Then, 5 × 104 nonadherent cells per milliliter were plated, whereas 2 × 105 adherent cells per milliliter were plated. Secondary CFU-Cs (CFU-GMs/BFU-Es) were counted after 2 weeks. MS-5 stromal cells20 and 293T cells were maintained in α-MEM and Dulbecco modified Eagle medium, respectively, with 10% heat-inactivated FCS containing 2 mM L-glutamine and 100 U/mL penicillin/streptomycin.
Flow cytometry
Cells were washed in 2% FCS/phosphate buffered saline (PBS) and stained with phycoerythrin-conjugated anti-CD34 or anti-CD14 (Becton Dickinson) for 30 minutes at 4°C. Idiotypic controls were used accordingly. Data were analyzed with CellQuest software (Clearwater, FL). For cell sorting, retrovirally transduced cells containing either the control vector or AML1-ETO were expanded for 2 days in expansion mix (as described above) and then sorted for GFP expression by means of a Becton Dickinson FACS Star Plus.
Immunoblotting and histochemistry
Cells were washed with PBS and lysed in 1% TritonX-100, 50 mM NaCl, 50 mM Hepes (pH 7.5), 5 mM EDTA, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, and protease inhibitors (Roche, Indianapolis, IN). Protein lysates were sonicated and centrifuged for 10 minutes, and the protein concentration was determined by means of the Bradford method (Bio-Rad Laboratories, Hercules, CA). Twenty micrograms of protein lysate was mixed with an equal volume of 2 × sodium dodecyl sulfate (SDS) loading buffer (4% SDS, 20% glycerol, 0.72 M 2-mercaptoethanol, 125 mM Tris), boiled for 10 minutes, and run on a 7.5% denaturing SDS–polyacrylamide gel electrophoresis (Bio-Rad). The protein was transferred to nitrocellulose (Amersham, Buckinghamshire, United Kingdom), and the membrane was blocked in 5% bovine serum albumin in Tris-buffered saline with 0.02% Tween-20. The blot was incubated with a primary antibody against ETO (kindly provided by Dr P. Erickson, University of Colorado), washed, and incubated with a secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA). The blot was developed with the use of a chemiluminescent substrate (ECL, Amersham) and exposed on Hyperfilm (Amersham).
Cytospin slides were prepared with the use of 8 × 104cells. Cells were washed with PBS with 2% heat-inactivated FCS. Slides were air-dried, fixed with methanol, and Wright-Giemsa stained. For immunofluorescent staining, cytospins were fixed for 10 minutes in 2% paraformaldehyde and permeabilized for 1 hour by means of 0.1% saponin. An anti-HA antibody (Roche) was added for 1 hour; slides were washed 4 times with PBS containing 0.02% Tween-20; and a secondary antibody conjugated to Cy3 was added for 1 hour (Jackson Immunoresearch Laboratories). After washing, 4′,6-diamidino-2-phenylindole-2HCl (DAPI) was added for 1 minute to stain the nucleus, and cells were visualized by means of an Olympus (Melville, NY) fluorescent microscope.
Southern blot analysis
Genomic DNA was isolated from retrovirally transduced cells by means of the QIAamp DNA Blood mini kit (Qiagen, Valencia, CA). DNA (10 μg) was digested with BamHI overnight, size-separated on a 1% Tris-borate/EDTA-agarose gel, and blotted to a nylon membrane (Amersham) by alkaline transfer. The membranes were then hybridized to [32P]deoxycytidine triphosphate–labeled DNA probes in Rapid-hyb buffer (Amersham) at 65°C.
Results
Expression of AML1-ETO in human HPCs
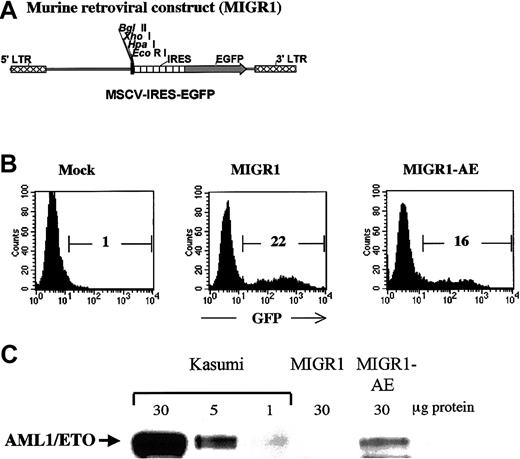
Human CD34+ cells were transduced with either the control MIGR1 virus (shown schematically in Figure1A) or the MIGR1 retrovirus expressing the AML1-ETO fusion protein (MIGR1-AE). The murine stem cell virus long terminal repeat that drives expression in the MIGR1 virus has been shown to be optimally expressed in human CD34+cells.21 The purified CD34+ cells were induced to proliferate by culturing for 3 days in a cytokine cocktail prior to transduction, in order for productive infection to occur.22 Transduction efficiency ranged from 15% to 55% for the control vector and from 10% to 35% for the MIGR1-AE retrovirus, as measured by flow cytometric detection of GFP (Figure1B). Expression of the correct-size AML1-ETO protein was confirmed by Western blotting, with the Kasumi cell line used as a control (Figure1C). The relative level of AML1-ETO expression is comparable to the expression seen in Kasumi, if one considers that only 16% of the PBPCs express the fusion protein. Owing to the IRES element, AML1-ETO and GFP are translated from a single messenger RNA. For this reason and for the sake of clarity, we will refer to “AML1-ETO expression” instead of to “GFP expression” when indicating cells transduced by AML1-ETO, even if the actual readout is for GFP expression.
Transduction of human HPCs and expression of AML1-ETO.
(A) The MIGR1 retroviral vector is shown schematically. AML1-ETO cDNA was subcloned into the multiple cloning site located upstream of the internal ribosome entry site (IRES) element. (B) A representative flow cytometry assay to assess the efficiency of retroviral transduction of human HPCs, showing the percentage of transduced cells calculated on the basis of GFP expression. (C) Western blot analysis demonstrating expression of the correct-size AML1-ETO protein in transduced human HPCs. The Kasumi cell lysate was loaded at 3 different concentrations to allow comparison with the level of AML1-ETO expression in the transduced CD34+cells.
Transduction of human HPCs and expression of AML1-ETO.
(A) The MIGR1 retroviral vector is shown schematically. AML1-ETO cDNA was subcloned into the multiple cloning site located upstream of the internal ribosome entry site (IRES) element. (B) A representative flow cytometry assay to assess the efficiency of retroviral transduction of human HPCs, showing the percentage of transduced cells calculated on the basis of GFP expression. (C) Western blot analysis demonstrating expression of the correct-size AML1-ETO protein in transduced human HPCs. The Kasumi cell lysate was loaded at 3 different concentrations to allow comparison with the level of AML1-ETO expression in the transduced CD34+cells.
AML1-ETO blocks early HPC expansion and inhibits CFU-C activity
Transduced, unsorted HPCs were monitored for expression of GFP over a period of 4 to 5 weeks in cytokine-driven liquid cultures. There was a gradual loss of GFP-expressing cells from the MIGR1-transduced cultures over time (Figure 2A). This reduction in the percentage of transduced cells has been previously described and is probably due to the preferential transduction of committed progenitor cells that mature more rapidly than primitive progenitor cells in liquid culture (Lu et al21 and our unpublished data, March 2001). The loss of GFP does not appear to be due to promoter silencing because the percentage of GFP+ cells detected by fluorescence-activated cell sorting is generally equivalent to the percentage of colonies that express GFP by PCR analysis (data not shown). In contrast to the gradual loss seen in the MIGR1 control culture, cells expressing AML1-ETO were lost more rapidly, with a 70% decrease in AML1-ETO–expressing cells during the first week as compared with a 13% decrease in cells expressing GFP alone (Figure 2A-B). This decrease was due to the diminished expansion of the AML1-ETO–expressing cells during the early period of culture compared with the MIGR1-transduced cells (4-fold versus 12-fold) (Figure 2C, week 1). This effect was specific for cells expressing AML1-ETO, as the overall expansion (of both GFP+ and GFP− cells) was similar in both cultures at various times throughout the culture period (Figure 2D) and the cumulative fold expansion of both cultures was also the same (Figure 2D, inset). The negative effect of AML1-ETO expression was restricted to cells proliferating during the early period of cytokine-driven culture, presumably the more committed progenitors. In contrast, at week 4, the percentage of cells expressing AML1-ETO gradually increased compared with the control transduced cells (Figure 2A), owing to a greater expansion of AML1-ETO–expressing cells at these later time points (3-fold expansion versus no expansion) (Figure 2C, week 4). This positive effect was more apparent when the transduced HPCs were cultured on stroma, as discussed below.
Loss of AML1-ETO–expressing GFP+ cells from culture after transduction of human HPCs.
(A) Transduced HPCs grown in cytokine-driven culture were monitored for expression of GFP protein weekly for 5 weeks. Cells were cultured in complete media with cytokines, counted weekly, and replated at 1 × 105 cells per milliliter. A representative experiment is shown. (B) Decrease in GFP-expressing cells during the first week in cytokine-driven culture, shown as the percentage of loss in GFP-expressing cells (from 3 independent experiments). (C) Fold expansion of GFP-expressing cells over a 4-week period in cytokine-driven culture. (D) Total fold expansion of cells during the same time period shown in panel C. The inset in panel D represents the average total cumulative fold expansion. Cells were counted weekly by trypan blue dye exclusion, and the percentage of GFP expression was determined by flow cytometry. Results are the average of 2 independent experiments. MIGR1 represents cells transduced with the empty retroviral vector. MIGR1-AE represents cells transduced with the AML1-ETO–expressing retrovirus.
Loss of AML1-ETO–expressing GFP+ cells from culture after transduction of human HPCs.
(A) Transduced HPCs grown in cytokine-driven culture were monitored for expression of GFP protein weekly for 5 weeks. Cells were cultured in complete media with cytokines, counted weekly, and replated at 1 × 105 cells per milliliter. A representative experiment is shown. (B) Decrease in GFP-expressing cells during the first week in cytokine-driven culture, shown as the percentage of loss in GFP-expressing cells (from 3 independent experiments). (C) Fold expansion of GFP-expressing cells over a 4-week period in cytokine-driven culture. (D) Total fold expansion of cells during the same time period shown in panel C. The inset in panel D represents the average total cumulative fold expansion. Cells were counted weekly by trypan blue dye exclusion, and the percentage of GFP expression was determined by flow cytometry. Results are the average of 2 independent experiments. MIGR1 represents cells transduced with the empty retroviral vector. MIGR1-AE represents cells transduced with the AML1-ETO–expressing retrovirus.
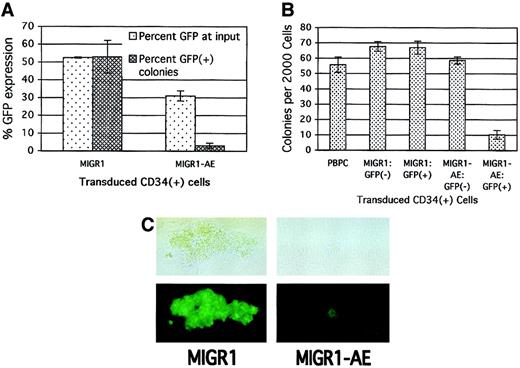
To assay for the more committed progenitors present in the original HPC cultures, we performed colony assays immediately after transduction to further define the effect of AML1-ETO expression on progenitor cell activity. Consistent with its negative effect on cell growth in cytokine-driven culture, the expression of AML1-ETO in committed HPCs decreased the number of colonies that formed in methylcellulose. This negative effect on progenitor activity correlated with the transduction efficiency, with a greater decrease in total colony formation being observed when the transduction efficiency of the MIGR1-AE retrovirus was high (data not shown). The loss in progenitor cell activity was specific for those cells transduced by AML1-ETO, as the percentage of AML1-ETO–expressing colonies was far below the number expected on the basis of the initial transduction rate. A transduction efficiency of 50% for the MIGR1 control virus resulted in approximately 50% GFP+ colonies 2 weeks later (Figure3A). However, with an average transduction efficiency of 30% for the MIGR1-AE retrovirus, fewer than 5% of resulting colonies were GFP+ (Figure 3A).
Decrease in primary CFU-C proliferation due to expression of AML1-ETO.
(A) Human HPCs were transduced with the indicated retrovirus and 2 days later were analyzed for GFP expression and plated in methylcellulose culture. Colonies were counted 2 weeks later, and GFP expression in the colonies was evaluated microscopically. The average of 2 independent experiments is shown. (B) HPCs were transduced as in panel A; the cells were sorted for GFP expression 2 days after transduction and immediately plated in methylcellulose culture. PBPCs represent mock-transduced HPCs. Colonies were counted after 2 weeks, and the average of triplicate plates is indicated. (C) A representative GFP+ colony is shown for both the control and the AML1-ETO–transduced HPCs. The top panels are phase-contrast images, and the bottom panels show fluorescent images. The magnification is 400 ×.
Decrease in primary CFU-C proliferation due to expression of AML1-ETO.
(A) Human HPCs were transduced with the indicated retrovirus and 2 days later were analyzed for GFP expression and plated in methylcellulose culture. Colonies were counted 2 weeks later, and GFP expression in the colonies was evaluated microscopically. The average of 2 independent experiments is shown. (B) HPCs were transduced as in panel A; the cells were sorted for GFP expression 2 days after transduction and immediately plated in methylcellulose culture. PBPCs represent mock-transduced HPCs. Colonies were counted after 2 weeks, and the average of triplicate plates is indicated. (C) A representative GFP+ colony is shown for both the control and the AML1-ETO–transduced HPCs. The top panels are phase-contrast images, and the bottom panels show fluorescent images. The magnification is 400 ×.
To confirm that the negative effect was specific to cells expressing AML1-ETO, we sorted transduced HPCs on the basis of their GFP expression. Only the GFP+ cells expressing AML1-ETO showed a significant decrease in colony formation (Figure 3B, last lane). In addition, the AML1-ETO–expressing colonies (both BFU-Es and CFU-GMs) were significantly smaller than the control colonies (Figure 3C). Thus, several independent assays demonstrate the negative effect of AML1-ETO expression on committed progenitor cell growth.
AML1-ETO expression enhances the self-renewal of human stem cells
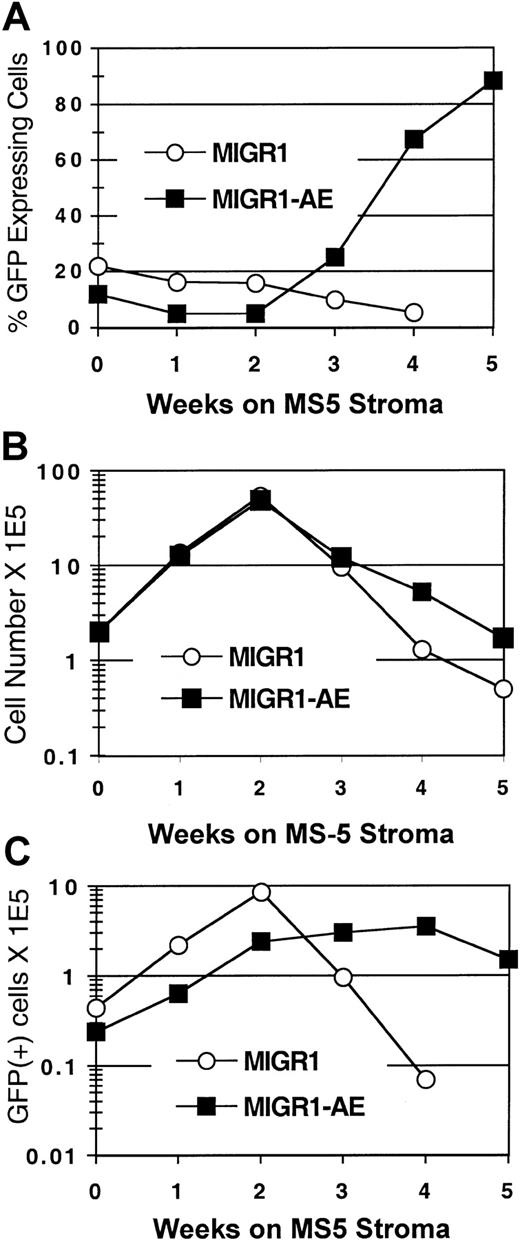
To determine the effect of AML1-ETO expression on human stem cells, we used the CAFC assay.23 The cobblestone areas that form in the MS-5 stroma at week 5 and week 6 are thought to represent the stem cells that were present at the beginning of the assay. Transduced CD34+ cells were seeded onto the MS-5 murine stromal cell line, and GFP expression in the nonadherent cell fraction was monitored over a 5- to 6-week period. In the MIGR1-transduced (control) cultures, the GFP-expressing cells were gradually lost over time (Figure 4A), similar to what was seen in the cytokine-driven stroma-free cultures (Figure 2A). In contrast, the percentage of cells expressing AML1-ETO increased at week 3, and by week 5, 90% of the cells in the MIGR1-AE culture were AML1-ETO positive (Figure 4A). In addition, the total number of cells increased in the MIGR1-AE cultures at the later time points (Figure 4B), and this was due to the increased representation of AML1-ETO–expressing cells (Figure 4C). These AML1-ETO–expressing cells continued to expand during weeks 3 and 4 in CAFC culture, whereas the number of transduced cells in the MIGR1 control culture decreased by more than 2 logs over the same time period (Figure 4C).
AML1-ETO enhances expansion of primitive HPCs.
HPCs were transduced with the indicated virus, expanded for 6 days in cytokine-driven culture, and plated onto the MS-5 stromal cell line in triplicate flasks. Nonadherent cells were counted by trypan blue and analyzed weekly for GFP expression by flow cytometry. (A) Percentage of GFP-expressing cells over time. (B) Total number of nonadherent cells over time. (C) Total number of GFP+ cells during the 5-week stromal cell culture period. Three independent experiments yielded essentially the same results; a representative experiment is shown.
AML1-ETO enhances expansion of primitive HPCs.
HPCs were transduced with the indicated virus, expanded for 6 days in cytokine-driven culture, and plated onto the MS-5 stromal cell line in triplicate flasks. Nonadherent cells were counted by trypan blue and analyzed weekly for GFP expression by flow cytometry. (A) Percentage of GFP-expressing cells over time. (B) Total number of nonadherent cells over time. (C) Total number of GFP+ cells during the 5-week stromal cell culture period. Three independent experiments yielded essentially the same results; a representative experiment is shown.
The number of cobblestone areas present in the MS-5 stroma was quantified, and the percentage that expressed GFP was evaluated by microscopic examination. Transduced stem cells were readily identified in both the control MIGR1 and the MIGR1-AE samples at week 5 (Figure5A-B). The percentage of cobblestone areas expressing AML1-ETO (16%) was significantly increased when compared with the percentage of cobblestone-forming cells transduced by the control MIGR1 (2%), especially when the difference in initial transduction efficiencies for this experiment (12% versus 16%) is taken into account. Overall, this experiment showed a 10-fold enrichment for AML1-ETO–expressing CAFCs. In another experiment, which was evaluated after 6 weeks, 60% of the cobblestone areas expressed AML1-ETO, and the total number of CAFCs was increased 3-fold compared with control MIGR1 (Figure 5C-D). The lack of GFP+ CAFCs for the control MIGR1 sample may relate to the longer time lag between transduction and seeding to CAFC culture in this experiment (6 days), compared with the experiment shown in Figure 5A-B (which was 2 days). This suggests that after 6 days in liquid culture, as compared with 2 days, true stem cells were fewer in number among the cells seeded onto MS-5, and the number of transduced stem cells was even less.
Increased secondary CFU-Cs and cobblestone formation by AML1-ETO–expressing cells.
(A) (B) Transduced CD34+ cells were plated over MS-5 stromal cells 2 days after transduction. Media and cells were demidepopulated weekly. After 5 weeks, cobblestone areas (minimum of 8 phase-dark cells) were counted. (C) (D) Transduced CD34+cells were plated over MS-5 stromal cells 6 days after transduction. Triplicate flasks were examined for the number of cobblestone areas, and these cobblestones were evaluated microscopically for GFP expression. Total cobblestone counts are shown. (E) (F) Nonadherent (suspension) and adherent cells (from the same experiment as panels C and D) were collected from the MS-5 stromal cultures after 6 weeks, counted, and plated in methylcellulose at 5 × 104 cells per dish and 20 × 104 cells per dish, respectively. Two weeks later, colonies were counted and GFP expression of colonies was evaluated microscopically. The average of triplicate plates is indicated. Two independent experiments yielded essentially the same results; a representative experiment is shown.
Increased secondary CFU-Cs and cobblestone formation by AML1-ETO–expressing cells.
(A) (B) Transduced CD34+ cells were plated over MS-5 stromal cells 2 days after transduction. Media and cells were demidepopulated weekly. After 5 weeks, cobblestone areas (minimum of 8 phase-dark cells) were counted. (C) (D) Transduced CD34+cells were plated over MS-5 stromal cells 6 days after transduction. Triplicate flasks were examined for the number of cobblestone areas, and these cobblestones were evaluated microscopically for GFP expression. Total cobblestone counts are shown. (E) (F) Nonadherent (suspension) and adherent cells (from the same experiment as panels C and D) were collected from the MS-5 stromal cultures after 6 weeks, counted, and plated in methylcellulose at 5 × 104 cells per dish and 20 × 104 cells per dish, respectively. Two weeks later, colonies were counted and GFP expression of colonies was evaluated microscopically. The average of triplicate plates is indicated. Two independent experiments yielded essentially the same results; a representative experiment is shown.
Secondary CFU-C formation was analyzed for both the nonadherent and the adherent cells growing in the MS-5 stromal–based cultures. The number of GFP+ CFUs generated from the MIGR1-AE–transduced cells was significantly increased compared with the MIGR1 control, with more than 60% of the CFU-Cs derived from the nonadherent cells continuing to express AML1-ETO and more than 75% of the adherent cell–derived CFU-Cs expressing AML1-ETO (Figure 5E-F). This contrasts with the lack of GFP-expressing colonies in the control transduced cultures (Figure 5F), despite an initially higher transduction rate (22% for MIGR1 versus 16% for MIGR1-AE; Figure 1B). The total number of colonies generated from the MIGR1-AE–adherent fraction was 5-fold higher than that for MIGR1, implying that the number of stem/progenitor cells present in the MS-5 stromal layer was greatly increased by the expression of AML1-ETO. A biologic explanation for the presence of AML1-ETO–expressing CAFCs and CFU-Cs from this experiment is that the expression of AML1-ETO in the stem cells during the cytokine-driven liquid culture prior to seeding on stroma preserved their stem cell phenotype (but did not completely prevent their differentiation in this assay), whereas the MIGR1-transduced cells lost their self-renewal potential. Taken together, these results demonstrate a strong positive effect of AML1-ETO expression on human stem cells, amplifying HPCs capable of generating secondary colonies.
AML1-ETO–expressing HPCs retain an immature phenotype and morphology and show a polyclonal expansion
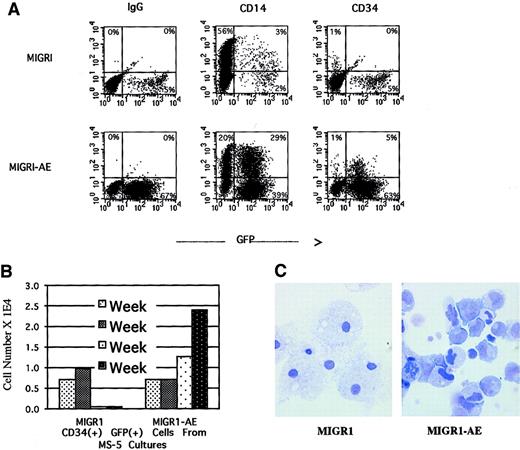
We monitored surface marker expression of the transduced cells from the nonadherent cells in the long-term stromal culture over a period of 4 weeks. During the first week after transduction, there was a small but reproducible decrease in the number of CD14+cells in the AML1-ETO–transduced population, with a slight increase in the number of CD34+ cells (data not shown). By week 4, the percentage of CD34+ cells in the nonadherent fraction was significantly higher in the AML1-ETO–transduced population than in the control population, 5% versus 0% (Figure6A). CD34+ cells are generally not released in any significant numbers from the MS-5 stromal layer and typically represent fewer than 1% of the nonadherent fraction in the long-term stromal culture (the same as for MIGR1-transduced cells, depicted in Figure 6A). By quantifying the number of CD34+ cells generated over the entire experiment, we documented an increase in total CD34+ cell number in the nonadherent fraction, implying a proliferative and/or survival advantage for immature progenitor cells due to expression of AML1-ETO (Figure 6B). This result correlated well with the increase in secondary CFU-Cs for GFP-expressing cells from the AML1-ETO cultures (Figure 5E-F). The morphology of these nonadherent cells was evaluated by Wright-Giemsa staining after 5 weeks in long-term stromal culture, when more than 90% of the cells were expressing AML1-ETO (Figure 4A). Many of the cells had an immature myeloid morphology, whereas the control cells were predominantly terminally differentiated macrophages (Figure6C). The AML1-ETO–expressing cells had more prominent nucleoli and irregular nuclei, with a higher nuclear-to-cytoplasmic ratio, reminiscent of actively proliferating but possibly dysplastic myeloblasts. However, although there is a pronounced bias toward more immature cells due to AML1-ETO expression, these cells do not seem to be blocked at the earliest myeloblast stage, since many of the cells express the CD14 marker indicative of macrophage differentiation (Figure 6A), and the morphology of some cells indicates maturation beyond a blast phenotype (Figure 6C).
AML1-ETO expression in human HPCs preserves an immature myeloid phenotype.
Over a 5-week culture on MS-5 stroma, the nonadherent cells were weekly counted by trypan blue, stained for CD34 and CD14 expression, and analyzed by flow cytometry. (A) CD34 and CD14 expression of cells present at week 4. (B) Quantitation of CD34+ cells present at weekly intervals over a 4-week period. (C) Wright-Giemsa stain of the nonadherent cells after 5 weeks in MS-5 stromal culture, showing macrophage morphology in the MIGR1-transduced cultures and an immature myeloid phenotype in the MIGR1-AE sample. Magnification is 1000 ×.
AML1-ETO expression in human HPCs preserves an immature myeloid phenotype.
Over a 5-week culture on MS-5 stroma, the nonadherent cells were weekly counted by trypan blue, stained for CD34 and CD14 expression, and analyzed by flow cytometry. (A) CD34 and CD14 expression of cells present at week 4. (B) Quantitation of CD34+ cells present at weekly intervals over a 4-week period. (C) Wright-Giemsa stain of the nonadherent cells after 5 weeks in MS-5 stromal culture, showing macrophage morphology in the MIGR1-transduced cultures and an immature myeloid phenotype in the MIGR1-AE sample. Magnification is 1000 ×.
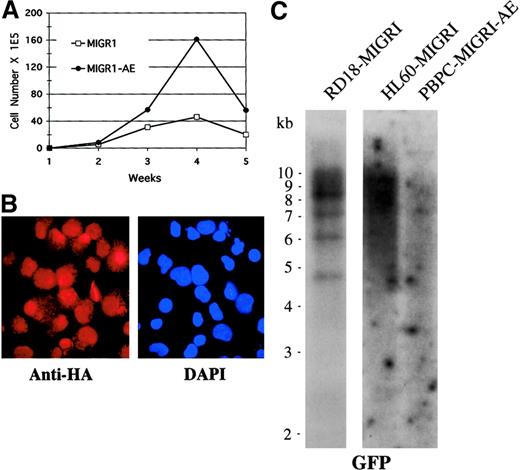
To determine whether other genetic events may be contributing to the phenotype observed over time in AML1-ETO–transduced cells, we analyzed the cells for clonality by Southern blotting and for continued AML1-ETO expression by immunofluorescence, and we also performed conventional cytogenetic analysis to detect any karyotypic abnormalities. We used cells that had been transduced by MIGR1-AE, sorted for GFP expression a few days later, and expanded in cytokine-driven liquid culture over a period of 5 weeks. We confirmed that these AML1-ETO–expressing cells showed a pronounced growth advantage when compared with control cultures (Figure 7A). At week 4, expression of AML1-ETO was analyzed by immunofluorescent staining with the use of an antibody to the HA epitope present at the aminoterminal end of AML1-ETO, and we observed nuclear staining of most of the cells (Figure 7B). This population of cells was subjected to Southern blot analysis with the use of GFP as the radiolabeled probe; the cells expressing AML1-ETO displayed a polyclonal integration pattern, implying no outgrowth of a clonal population over time (Figure 7C). As controls, a known clonal transduced cell population with approximately 4 viral integrants was included (RD18-MIGR1), and a known polyclonal transduced cell population was also hybridized on the same blot (HL60-MIGR1) (Figure 7C). Rehybridizing the blot with a probe to the β-actin gene demonstrated that the DNA was intact (data not shown). Finally, we analyzed cells that had been expanded in cytokines for a number of weeks for cytogenetic abnormalities, and all 20 metaphases displayed a normal karyotype (data not shown). Thus, expression of AML1-ETO itself is probably responsible for the expansion of cells seen in both cytokine-driven liquid culture and long-term stromal coculture assays.
Enhanced expansion of human HPCs by AML1-ETO is polyclonal.
(A) Expansion of transduced, GFP-sorted cells over a 5-week culture period in cytokines. Cell numbers were counted weekly by trypan blue dye exclusion. (B) Immunofluorescent staining of MIGR1-AE–transduced cells for the presence of the fusion protein determined on the basis of detection of the HA epitope. Cells were counterstained with DAPI to identify the nucleus. Magnification is 600×. (C) Southern blot analysis of DNA obtained from MIGR1-AE cells growing in the week-4 culture depicted in panel A. Ten micrograms of genomic DNA was digested with BamHI, which cuts at a single site in the MIGR1-AML1-ETO virus. The full-length GFP cDNA was used as a probe. The membrane was then stripped and rehybridized for the presence of intact DNA with the use of a β-actin probe (data not shown).
Enhanced expansion of human HPCs by AML1-ETO is polyclonal.
(A) Expansion of transduced, GFP-sorted cells over a 5-week culture period in cytokines. Cell numbers were counted weekly by trypan blue dye exclusion. (B) Immunofluorescent staining of MIGR1-AE–transduced cells for the presence of the fusion protein determined on the basis of detection of the HA epitope. Cells were counterstained with DAPI to identify the nucleus. Magnification is 600×. (C) Southern blot analysis of DNA obtained from MIGR1-AE cells growing in the week-4 culture depicted in panel A. Ten micrograms of genomic DNA was digested with BamHI, which cuts at a single site in the MIGR1-AML1-ETO virus. The full-length GFP cDNA was used as a probe. The membrane was then stripped and rehybridized for the presence of intact DNA with the use of a β-actin probe (data not shown).
Discussion
The careful delineation of the functional properties of leukemia-associated fusion proteins depends on the establishment of accurate and reproducible model systems. A number of systems have been developed to study the role of the AML1-ETO fusion protein in leukemogenesis, but all have limitations. To study the activities of AML1-ETO, several laboratories, including our own, have expressed it in cell lines (hematopoietic or nonhematopoietic). Several leukemia cell line–based systems have been established, such as the murine factor–dependent L-G cell line and the human U937 leukemia cell line. AML1-ETO blocks G-CSF–induced granulocytic differentiation of L-G cells12 and inhibits monocytic differentiation of U937 cells following vitamin D and transforming growth factor–β stimulation.10 We have used an NIH3T3 transformation assay to analyze the function of AML1-ETO, which defined its transforming activity and its ability to up-regulate AP-1 activity (with c-jun and ATF-2 as possible downstream effectors in this signaling cascade).24 In addition, AML1-ETO has been shown to repress promoter activation by AML1 in hematopoietic25and nonhematopoietic cell lines.26 Much has been learned from these studies, including the critical role of the ETO portion of the fusion protein in recruiting corepressor molecules, the role of ETO dimerization in the function of AML1-ETO, and the identification of specific target genes that may be involved in AML1-ETO–induced effects.10,12 27
In addition to cell line–based systems, normal murine HPCs have been used to characterize the effects of AML1-ETO expression. The AML1-ETO knock-in mice, which displayed an embryonic lethal phenotype similar to that seen in the AML1 knockout mice,15,16 have detectable HPCs that display a high self-renewal capacity and can be immortalized as factor-dependent myeloid cell lines.16 A recently described transgenic mouse model expresses the AML1-ETO fusion protein from an inducible promoter to bypass the embryonic lethal phenotype. However, no effect on hematopoiesis was seen in these mice upon induction of AML1-ETO.17
We have used a retroviral gene transfer system to express AML1-ETO in normal human hematopoietic stem/progenitor cells, to study its effects in the physiologically relevant cell type, the human hematopoietic stem cell. We show that expression of AML1-ETO in human CD34+ cells produces differential effects, depending on the state of progenitor cell maturation. Expression of AML1-ETO in more mature progenitor cells resulted in growth arrest and abrogated colony formation in primary clonogenic assays. In contrast, AML1-ETO expression resulted in the preferential expansion and/or self-renewal of stem cells. These results suggest that a single oncogenic event, such as a balanced translocation, may have different effects depending on which cell receives the productive translocation; this highlights the importance of targeting the expression of oncogenic fusion proteins to the physiologically relevant cell. Expression of AML1-ETO in primitive human HPCs recapitulates certain aspects of the FAB-M2 subtype of AML in which the translocation is normally found, including the continued expression of CD34 and the propensity to remain in an immature, undifferentiated state with continued self-renewal capacity.
Expression of AML1-ETO in human CD34+ cells led to a rapid decrease in the number of cells positive for AML1-ETO, owing to the decreased expansion of committed HPCs expressing the fusion protein. Similarly, expression of AML1-ETO in 5-fluorouracil–mobilized murine bone marrow cells also led to a negative effect on cell growth in clonogenic assays.16 Despite this apparently inhibitory effect, several laboratories have successfully expressed the AML1-ETO fusion protein in a variety of cellular backgrounds.12,13In addition, expression of AML1-ETO in a tetracycline-inducible transgenic mouse model did not result in the dramatic loss of hematopoietic cells upon induction in vivo,17 further suggesting that the negative effect on cell proliferation may be cell stage–specific. The intensity of expression may also be an important variable, as it has been suggested that a threshold level of AML1-ETO expression is required for its full biologic effects.28For our experiments, we used a constitutively active promoter that is expressed at high levels in a broad range of hematopoietic cells, including primitive hematopoietic progenitors.21Expression from the tetracycline-responsive promoter may not be as strong or as widely expressed.29 However, using this system, Rhoades et al saw a considerable increase in the level of AML1-ETO expression upon continued passage of hematopoietic cells in methylcellulose, which may have been important for the self-renewing effect observed in AML1-ETO–expressing progenitor cells.17 When the regulated expression of AML1-ETO was shut off by the addition of tetracycline, the number of colonies increased significantly, implying the loss of a negative effect when AML1-ETO expression ceased. Similarly, we observed a 5- to 10-fold decrease in the number of AML1-ETO–expressing committed progenitor cell colonies, and the few colonies that did form were significantly smaller than the control colonies (Figure 3C), consistent with a negative effect of AML1-ETO on cell growth.
In contrast to the negative effect seen in committed progenitors, expression of AML1-ETO enhanced the survival and/or proliferative capacity of human stem cells, resulting in their increased representation over time. This effect is seen in both cytokine-driven cultures and, more dramatically, in long-term stromal assays. One possible reason for the more pronounced effect in the MS-5 stromal cultures is that the stem cells reside under the stroma, and weekly demidepopulation of the cultures does not deplete these cells. In contrast, during the weekly reseeding of stroma-free cytokine-driven cultures, the stem cells are discarded along with the other cells. Therefore, the positive effect of AML1-ETO on stem cells, including the increase in cobblestone areas after 5 or 6 weeks and the increased percentage of AML1-ETO–expressing progenitors, is more easily detected in long-term stromal culture.
The morphologic and phenotypic effects of AML1-ETO expression on these stem cells are also consistent with enhanced self-renewal capacity. Many of the cells expressing AML1-ETO retain a more primitive morphology, resembling immature myeloblasts, while the control cells became terminally differentiated. A relatively high percentage of the AML1-ETO–transduced cells continued to express CD34 and retained clonogenic ability for much longer than normal in these assays. These results suggest that AML1-ETO stimulates self-renewal of human stem cells at the expense of differentiation. However, the expression of AML1-ETO does not seem to result in a total block in differentiation, since mature cells are detected by both flow cytometry and histochemistry (Figure 6). Rather, there is an accumulation of immature myeloid cells, which can differentiate in the presence of continued AML1-ETO expression.
Our data allow us to speculate on the contribution of AML1-ETO to leukemogenesis. It is possible that the translocation, when it occurs in a sufficiently primitive hematopoietic cell, slows the differentiation of the cell while simultaneously promoting its self-renewal. The presence of the t(8;21) translocation in a myelodysplastic syndrome patient would be consistent with this interpretation.30 Increased self-renewal expands the pool of candidate cells that are susceptible to second-hit mutations, and when a mutation that confers a strong proliferative signal is acquired, the cell becomes transformed and this results in acute myelogenous leukemia. The sequence of events and the number of hits that are required are likely to vary in different leukemias, but the need for events that promote self-renewal and impair differentiation is probably universal.
The mechanism by which AML1-ETO alters the behavior of human stem cells and HPCs are currently unknown. Biochemical studies support the notion that AML1-ETO dominantly interferes with the function of AML1, presumably by efficiently binding corepressor molecules and targeting histone deacetylase activity to AML1-responsive promoters (reviewed in Downing et al1). AML1 can apparently function as either a transcriptional activator or a transcriptional repressor, and this activity depends on both the cellular and the promoter context.8 From the knockout experiments, it is clear that AML1 is essential for definitive hematopoiesis; however, its exact role in regulating the proliferation, self-renewal, and commitment of hematopoietic cells is only beginning to be understood. The proteins involved in hematopoietic stem cell self-renewal are not well characterized, and the role that AML1 may play in this regard is also unknown. AML1 represses expression of p21waf1/cip1 and shortens the G1 phase of the cell cycle.31,32Also, in studies using a dominant-negative inducible AML1 protein, KRAB-AML1-ER, Lou et al33 observed an inhibition of cell cycle progression with a concomitant decrease in cdk4 levels, again arguing for the involvement of AML1, and by extension AML1-ETO, in the G1-to-S transition. Whether an inhibitory effect of AML1-ETO on cell cycle progression contributes to the negative effects on committed progenitor cells requires further investigation. Studies are underway to identify AML1-ETO target genes in human CD34+ cells that should also shed light on the mechanisms underlying the effects of AML1-ETO on stem cell growth.
In summary, we show that expression of the AML1-ETO fusion protein in human CD34+ cells exerts differential effects on committed progenitors versus stem cells and results in the enhanced self-renewal of stem cells. This is a promising system that will allow us to appreciate the role of AML1-ETO in AML and will prove useful in deciphering the mechanism by which the fusion protein disrupts signaling in human stem cells. The ability to coexpress AML1-ETO with other potentially contributing signaling molecules and to model the effects on human stem cells in immunodeficient mice represent powerful applications of this system.
We thank Geoffrey Jackson and Dianna Ngok for excellent technical assistance, Jae-Hung Shieh for his advice and expertise, Diane Domingo for her help with flow cytometry, and Ellie Park for secretarial and administrative support.
Supported by National Institutes of Health grants R01 DK43025 (S.D.N.) and HL61401 (M.A.S.M), by the Panayiota Makkos Memorial Fund (S.D.N.) and the Gar Reichman Fund of the Cancer Research Institute (M.A.S.M).
Submitted March 26, 2001; accepted August 22, 2001.
J.C.M. and J.C. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
James C. Mulloy, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, Box 575, New York, NY 10021; e-mail:j-mulloy@ski.mskcc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal