Chronic myeloid leukemia (CML) is caused by a t(9;22)(q34;q11) reciprocal translocation that generates a BCR-ABLfusion gene on the 22q- or Philadelphia (Ph) chromosome and anABL-BCR gene on 9q+.1,2 Large deletions adjacent to the t(9;22) breakpoint on the derivative 9q+ chromosome have now been found, which result in genomic loss at both sides of the translocation breakpoint.3,4,5,6 Recently, Huntly et al7 analyzed a large series of CML patients and confirmed that these deletions identify a subgroup of patients with relatively poor prognosis. However, the molecular basis of this survival disadvantage has not been identified. Due to its unique location spanning the chromosome 9/chromosome 22 junction, ABL-BCRappears as the most likely candidate for this role for the following reasons: (1) it occupies the minimal region of deletion in the reported cases6; (2) it is a “single-allele” gene whose deletion would result directly in total absence of the gene product, without a need to invoke a second event in a “2-hit” model8; and (3) it is known not to be expressed in approximately one third of CML patients,2,9 a frequency similar to that found for the 9q+ deletions.6
To investigate this hypothesis, we performed reverse transcription–polymerase chain reaction (RT-PCR) amplification of the BCR-ABL and ABL/BCR genes in a total of 519 patients (344 from the German CML studies III or IIIA10 and 175 from the Hammersmith Hospital) diagnosed with BCR-ABL–positive CML between December 1981 and December 2000. The entire group comprised 319 males and 200 females with a median age of 50.6 years (range, 12.4-83.2). After diagnosis, patients were treated with either interferon-α and/or other chemotherapy (n = 399), stem cell allograft (n = 100), or autograft (n = 20). RT-PCR amplifications were done as previously described.11,12 Approximately equal numbers expressed b2a2 or b3a2 transcripts, with a minority in whom both types of transcripts were present (Table 1). ABL(1b)-BCR message was detected in 282/520 (54%) patients, encoding the classic 1bb3, 1bb4, or both 1bb3 and 1bb4 junctions in the great majority of cases. ABL(1a)-BCR transcripts were detected in only 8% of the patients. All frequencies are consistent with previous studies.9
BCR-ABL and ABL-BCR expression in CML patients
| Fusion gene . | Transcript expressed . | No. (%) of patients . |
|---|---|---|
| BCR-ABL | b2a2 | 216 (42) |
| b3a2 | 250 (48) | |
| b2a2 & b3a2 | 48 (9) | |
| Other* | 5 (2) | |
| Total | 519 (100) | |
| ABL(1b)-BCR | Positive | 282 (54) |
| 1bb3 | 41 | |
| 1bb4 | 196 | |
| 1bb3 & 1bb4 | 38 | |
| Other† | 7 | |
| Negative | 237 (46) | |
| ABL(1a)-BCR | Positive | 44 (8) |
| 1ab3 | 3 | |
| 1ab4 | 30 | |
| 1ab3 & 1ab4 | 9 | |
| Other‡ | 2 | |
| Negative | 475 (92) |
| Fusion gene . | Transcript expressed . | No. (%) of patients . |
|---|---|---|
| BCR-ABL | b2a2 | 216 (42) |
| b3a2 | 250 (48) | |
| b2a2 & b3a2 | 48 (9) | |
| Other* | 5 (2) | |
| Total | 519 (100) | |
| ABL(1b)-BCR | Positive | 282 (54) |
| 1bb3 | 41 | |
| 1bb4 | 196 | |
| 1bb3 & 1bb4 | 38 | |
| Other† | 7 | |
| Negative | 237 (46) | |
| ABL(1a)-BCR | Positive | 44 (8) |
| 1ab3 | 3 | |
| 1ab4 | 30 | |
| 1ab3 & 1ab4 | 9 | |
| Other‡ | 2 | |
| Negative | 475 (92) |
b2a3 = 3, e1a2 = 1, e19a2 = 1.
1bb6 = 2, 1bb2 = 1, with variable intervening sequence between ABL and BCR exons = 4, as described.15
1aa6 = 2.
We analyzed a representative sample of 24 BCR-ABL–positive cases by fluorescence in situ hybridization (D-FISH) with commercially available BCR and ABL probes (Vysis, Richmond, UK), as described by Huntly et al.7 Eleven of the 24 patients were ABL-BCR positive, exhibited a classical t(9;22) and, as expected, did not have a 9q+ deletion. The remaining 13 patients did not express ABL-BCR and comprised 9 Ph-positive patients, 1 Ph-negative patient, and 3 patients with complex translocations. Only 7 of the 13 (54%) ABL-BCR–negative cases had a deletion: in 3 of these, both chromosome 9– and 22–derived material was missing, 3 had only chromosome 9–derived material missing, and one lacked exclusively chromosome 22–derived material. These results show that the overall correlation betweenABL-BCR expression and deletion at the 9q+ derivative is relatively poor, with 6/24 (25%) cases being discordant.
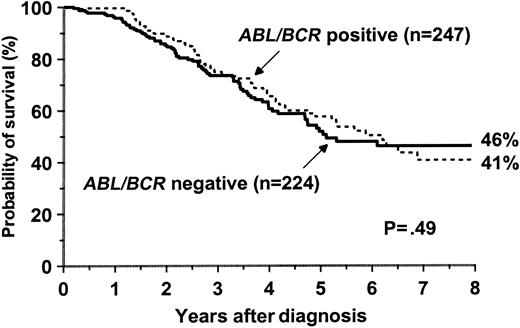
We next assessed whether ABL-BCR expression could influence survival. A total of 471 patients with more than 30 days of follow-up after diagnosis were eligible for analysis. Probability of survival at 5 years was 51.8% (95% confidence interval [CI], 43-60) in theABL-BCR-negative group (n = 224) and 57.6% (95% CI, 49-66) in the positive group (n = 247, P = .49) (Figure 1). There were no differences between theABL-BCR–positive and –negative patients with regard to gender, age, type of BCR-ABL transcript, treatment modality, and Sokal13 or Hasford14 score. Likewise, no significant difference in survival was observed between patients with b3a2 and b2a2 BCR-ABL transcripts or betweenABL-BCR 1bb3 and 1bb4.
These results show that lack of ABL-BCR expression does not correlate with the presence of large deletions on the 9q+ derivative or with a shorter survival in CML patients. Since the powerful negative prognostic value of the 9q+ deletion has been confirmed in Huntly et al's7 large series of patients, it must be assumed that the molecular basis of the survival disadvantage lies in the loss of another gene(s) within the deleted region. However, as the extent and location of the deletion is rather variable, with some patients lacking material derived from either chromosome 9 or 22, it is possible that the group of patients with the 9q+ deletion is still heterogeneous and may encompass subgroups with variable survival disadvantage depending on the specific gene deleted during the translocation. AsABL-BCR has been eliminated as the most likely candidate for the prognostic value of the 9q+ deletion, attention should now be focused on genes surrounding the chromosome 9/chromosome 22 junction.
Supported by grants from the Leukaemia Research Fund, United Kingdom (J.E.S., J.M.G., N.C.P.C., J.V.M.); the German Competence network “Acute and chronic leukemias”; and the Deutsche José-Carreras-Stiftung, Germany (K.M., M.M., O.M., U.B., R.H, A.H.)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal