Abstract
Interleukin-7 (IL-7) and IL-15 have been recently identified as growth factors for cutaneous T-cell lymphoma (CTCL) cells, and they protect these cells from cell death. Using the CTCL cell line SeAx as a test system now shows that IL-7 and IL-15 are indeed necessary to maintain high levels of bcl-2. The up-regulation of bcl-2 was paralleled by increased DNA-binding activities of the transcription factors STAT2, STAT5, STAT6, and c-Myb to bcl-2gene promoter–enhancer elements. Because STAT5 and c-Myb positively regulate bcl-2, IL-7 and IL-15 may mediate some of their effects on cell death survival gene expression through these 2 factors. Constitutive activities of the 3 STAT factors and c-Myb were found in the IL-7– and IL-15–independent CTCL cell lines HUT78 and MyLa 2059. The c-Myb protein was also present in CTCL cells of the skin lesions of all investigated patients. These results indicate that IL-7 and IL-15 may increase bcl-2 expression in CTCL cells by the activation of c-myb and STAT factors.
Introduction
Cutaneous T-cell lymphoma (CTCL) is a group of lymphoproliferative disorders of the skin.1 The most frequent forms of CTCL are mycosis fungoides (MF) and its leukemic counterpart, the Sézary syndrome (SS). MF proceeds slowly (5-20 years) but leads mostly to death, which is often caused in the late phase of CTCL by rapidly growing and ulcerating tumors and immune disorders. In addition to generalized erythroderma, patients with SS have leukemic T cells in the blood, and life expectancy is generally shorter (3 years) than it is for patients with MF.
Interleukin-7 (IL-7) and IL-15 are growth and survival factors for CTCL cells.2,3 Because CTCL cells may remain restricted to the skin during the longest time in the course of the disease, these 2 cytokines, which are also produced in the skin,4-6 may be responsible for the epidermotropism of this disease. IL-15 has been identified as a T-cell growth-stimulating cytokine produced by many cell types and tissues.7 The IL-15 receptor (R) contains the β and γ chains of the IL-2R and a recently identified specific α chain.7-9 IL-15 can replace IL-2 in several systems,7,8,10 but it proved to be less effective than IL-2. The IL-7 high-affinity receptor also contains the IL-2R γ chain.11
The IL-2Rβ and IL-2Rγ chains interact with the tyrosine kinases Jak1, Jak3, Syk, and Lck, which in turn activate the proto-oncogenes bcl-2, c-myc, c-fos, and c-jun.12 Bcl-2 protects cells from programmed cell death,13 and because IL-7R and IL-15R share one or respectively 2 subunits with IL-2R, one may expect that IL-7 and IL-15 may cause at least partially the same effects as IL-2.
A positive effect of IL-7 on bcl-2 expression has already been shown in murine T-cell lymphoma cells.14 Bcl-2 belongs to a growing gene family whose members either inhibit or promote cell death. Bcl-2 promotes cell survival, whereas the genes bax15and bad promote cell death.16 The survival of a cell depends on the ratio between cell survival–promoting and cell death–promoting molecules.17 Bcl-2 expression has been found in CTCL skin lesions and is supposed to increase the survival and the resistance of CTCL cells against radiotherapy.18 19
Two transcription factors have been found to increase the expression of the bcl-2 gene: c-Myb20-22 and STAT5.23 The c-myb gene was initially identified as an oncogene in acute avian leukemia retrovirus strains.24 Its gene product binds to gene regulatory sequences, and its expression peaks during the transition from G1 to S phase.25 Thus c-myb plays a role in cell cycle progression and cell growth. STAT5 is a member of the signal transducer and activator of transcription factor family, which mediates the effects of growth hormones and cytokines, including IL-2, IL-7, and IL-15, and which have been shown to stimulate the DNA binding of STAT5.26-31
Here we investigated whether IL-7 and IL-15 increase the activities and DNA binding of c-Myb and STAT5. A coregulation of these 2 factors andbcl-2 would make it likely that IL-7 and IL-15 mediate their effects on bcl-2 expression through c-Myb and STAT5. As a test system, we used the IL-7–/IL-15–dependent CTCL cell line SeAx, which can survive 4 to 6 days in the absence of both cytokines. To see the effects of IL-7 and IL-15, SeAx cells were kept for 2 to 4 days in the absence of both interleukins; after this time, the cells either were stimulated with IL-7 or IL-15 or were left unstimulated. This system allowed us to investigate the different effects of IL-7 and IL-15 on bcl-2 and c-myb.
Materials and methods
Cell culture
The cell line HUT78 (Sézary syndrome) was obtained from the European Collection of Animal Cell Cultures. The cell lines MyLa 2059 (mycosis fungoides) and SeAx (Sézary syndrome) were kind gifts of Dr Keld Kaltoft (University of Aarhus, Denmark). HUT 78 and MyLa 2059 and patient Sézary cells were grown in HEPES-buffered RPMI 1640 medium with 2 mM glutamine supplemented with 10% fetal calf serum (FCS), 0.25 mg/mL amphotericin B, 100 U penicillin G, 100 U streptomycin, and 1 mM pyruvate. SeAx cells were grown under the same conditions except that 10% human serum instead of 10% FCS was used. Cytokine concentrations were 5 ng/mL (10 U) IL-7 and 10 ng/mL (10 U) IL-15.
Skin samples
Skin biopsy samples were taken primarily for diagnostic purposes with informed consent of the patients, and the specimens used became surplus material available after all the routine diagnostic procedures. The assignment, to a certain stage, was conducted according to the recommendations of the EORTC Cutaneous Lymphoma Project Group.
Electrophoretic mobility shift assay and supershift experiments
Electrophoretic mobility shift assays were performed according to the method of Barberis et al.32 Double-stranded γ-32P adenosine triphosphate-labeled oligonucleotides (30 000 cpm) containing the binding sites for c-myb (5′ TACAGGCATAACGGTTCCGTAGTG 3′) and the STAT5-binding site of thebcl-2 promoter (5′ AGGACTTCTGCGAATACCGG 3′) were incubated with 3 μg nuclear extract of the investigated cells in binding buffer consisting of 10 mM HEPES, pH 7.9, 60 mM KCl, 4% Ficoll, 1 mM dithiothreitol, and 1 mM EDTA, pH 8. Two micrograms poly IC were used as competitor for unspecific DNA-binding activities. Total volume of the reaction was 30 μL. For supershift experiments, 2 to 4 μg corresponding antibody (TransCruz gel supershift reagent; Santa Cruz Biotechnology, Santa Cruz, CA) was added. Nuclear extracts were prepared according to the method of Gerber et al.33 The reaction mixture was incubated for 30 minutes at 4°C and then was loaded on a 4% native polyacrylamide gel (0.25× TBE). Oligonucleotides were synthesized by Microsynth (Balgach, Switzerland).
Western blot analyses
For Western blotting, 15 to 30 μg protein of nuclear extracts or 30 to 60 μg protein of cytoplasmic extracts were loaded on 7.5% to 9% sodium dodecyl sulfate polyacrylamide gels and separated by polyacrylamide gel electrophoresis. The percentage of polyacrylamide depended on the molecular weight of the protein to be investigated. Proteins were transferred to a nitrocellulose filter using a Mini Trans Blot Cell (Bio-Rad, Glattbrugg, Switzerland) following the instructions of the supplier. Unspecific antibody-binding sites were blocked by incubation of the filter overnight at 4°C in Tris-buffered saline (TBS), 0.3% Tween 20, and 2% milk powder. The filter was incubated with the first antibody (1:1000 dilution; Santa Cruz Biotechnology) for 4 hours at room temperature in TBS, 0.3% Tween 20, and 1% milk powder. Incubation with the second antibody (1:5000 dilution; Roche Diagnostics, Rotkreuz, Switzerland) was conducted in TBS, 0.3% Tween 20, for 4 hours at room temperature. The signal was detected by incubation with BM purple AP substrate (Roche Diagnostics) according to the instructions of the supplier.
Immunohistochemistry
All specimens were snap-frozen in liquid nitrogen, embedded in Tissue-Tek (Gibco, Basel, Switzerland), and stored at −80°C until processing. Cryostat sections of 5 μm were cut onto gelatin-coated microscope slides and air-dried. Alkaline antialkaline phosphatase immunohistochemistry (reagents from Dako, Zug, Switzerland) was performed as published.34Anti–IL-15 antibodies used are commercially available (Peprotech, London, United Kingdom). In addition, all samples were also examined for reactivity with anti-CD3, -CD4, and -CD8 (reagents from Dako).
Results
IL-7 and IL-15 stimulate the expression of cell survival genes
It has been found that malignant T cells of patients with SS and MF express bcl-2.18,19 35 To see whether IL-7 and IL-15 regulate bcl-2, we used the IL-7–/IL-15–dependent SeAx cell line. These cells can survive for 4 to 6 days in the absence of these 2 interleukins, and bcl-2 expression is down-regulated after 2 days (Figure 1). To determine whether IL-7 or IL-15 can re-establish high anti–cell death gene expression after interleukin withdrawal, SeAx cells were kept, in the absence of IL-7 and IL-15, for 48, 54, 60, 72, and 96 hours. Then ten units IL-7 (Figure 1A) or IL-15 (Figure 1C) was added to the cells for another 6 hours. Cells incubated for another 6 hours without IL-7 or IL-15 served as negative controls. Resultant amounts of bcl-2 were determined by Western blot analysis. The β-actin gene served as a control not influenced by interleukins (Figure 1B,D). Figure 1 shows that IL-7 and IL-15 increased the levels of Bcl-2 protein after different times of interleukin starvation. Values after 6 hours of IL-7 and IL-15 addition are even higher than after 48 hours of interleukin withdrawal. Because cell death was beginning, the absolute values for interleukin addition after 72 and 96 hours were lower than those after 54 and 60 hours; however, at this point, the relative bcl-2expression-increasing effects were highest. In contrast tobcl-2, the β-actin gene was only marginally influenced. These effects were attributed solely to the interleukins because the cells were kept under constant serum conditions (10% FCS). The rise in Bcl-2 levels resulted from newly induced bcl-2 gene transcription and protein synthesis—increased Bcl-2 levels were observed after 6 hours but not after 30 minutes. RNA polymerase II inhibitor α amanitin completely abolished the effects of the addition of IL-7 and IL-15 (data not shown).
IL-7 and IL-15 increase Bcl-2 protein levels in interleukin-starved SeAx cells.
(A) Regulation of bcl-2 expression by IL-7 (10 U). (B) Influence of IL-7 on β-actin expression (negative control). (C) Regulation of bcl-2 expression by IL-15. (D) Influence of IL-15 on β-actin expression (negative control). M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 6, 72-h interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15.
IL-7 and IL-15 increase Bcl-2 protein levels in interleukin-starved SeAx cells.
(A) Regulation of bcl-2 expression by IL-7 (10 U). (B) Influence of IL-7 on β-actin expression (negative control). (C) Regulation of bcl-2 expression by IL-15. (D) Influence of IL-15 on β-actin expression (negative control). M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 6, 72-h interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after the 6-hour addition of IL-7 or IL-15.
IL-7 and IL-15 stimulate the DNA binding of c-Myb
Because it has been shown that bcl-2 transcription is up-regulated by the binding of c-Myb to promoter sequences of the bcl-2 promoter, we wanted to know whether IL-7 and IL-15 can increase the binding of c-Myb to an oligonucleotide that contains a c-Myb binding site. Figure 2A shows that IL-7 and IL-15 increased the DNA binding of c-Myb. The increase of the binding of c-Myb occurred within 30 minutes, indicating that de novo protein synthesis is not necessary for c-Myb DNA-binding stimulation. High constitutive c-Myb DNA binding was found in the IL-7– and IL-15–independent CTCL cell lines HUT78 and MyLa 2059. The addition of a c-Myb antibody to the DNA-binding reaction significantly reduced the signal of the largest complex, which probably consists of a double band, indicating that c-Myb is indeed present in this DNA–protein complex (Figure 2B). The addition of a STAT5 antibody had no effect (Figure 2B, lanes 11, 12). Because the signal was not totally repressed by the antibody, it is also possible that related A-Myb and B-Myb proteins bind to this sequence.
Effect of IL-7 and IL-15 on the DNA-binding of c-Myb in SeAx cells.
(A) Nuclear extracts from different cell lines were incubated with a radioactively labeled oligonucleotide containing the c-mybbinding site of the bcl-2 promoter. Lanes 1 to 3, no interleukin added. Nuclear extracts from HUT78 (lane 1), MyLa 2059 (lane 2), and SeAx cells (lane 3). Lane 4, SeAx cells treated with 10 U IL-7. Lane 5, SeAx cells treated with 10 U IL-15. Lane 6, SeAx cells treated with 10 U IL-7 and IL-15. Lane 7, nuclear extract from PBLs of a healthy donor. (B) The binding of c-Myb to its target sequence is inhibited by a c-Myb antibody. Lanes 1 to 6, no antibody added. Lanes 6 to 10, c-Myb antibody added. Lanes 1 and 6, nuclear extract from PBLs. Lanes 2 and 7, nuclear extract from HeLa cells. Lanes 3 and 8, nuclear extract from HUT78 cells. Lanes 4 and 9, nuclear extract from MyLa 2059 cells. Lanes 5 and 10, nuclear extract from SeAx cells. Lanes 11 and 12, STAT5 antibody added as negative control. Lane 11, nuclear extract from HUT78 cells. Lane 12, nuclear extract from MyLa 2059.
Effect of IL-7 and IL-15 on the DNA-binding of c-Myb in SeAx cells.
(A) Nuclear extracts from different cell lines were incubated with a radioactively labeled oligonucleotide containing the c-mybbinding site of the bcl-2 promoter. Lanes 1 to 3, no interleukin added. Nuclear extracts from HUT78 (lane 1), MyLa 2059 (lane 2), and SeAx cells (lane 3). Lane 4, SeAx cells treated with 10 U IL-7. Lane 5, SeAx cells treated with 10 U IL-15. Lane 6, SeAx cells treated with 10 U IL-7 and IL-15. Lane 7, nuclear extract from PBLs of a healthy donor. (B) The binding of c-Myb to its target sequence is inhibited by a c-Myb antibody. Lanes 1 to 6, no antibody added. Lanes 6 to 10, c-Myb antibody added. Lanes 1 and 6, nuclear extract from PBLs. Lanes 2 and 7, nuclear extract from HeLa cells. Lanes 3 and 8, nuclear extract from HUT78 cells. Lanes 4 and 9, nuclear extract from MyLa 2059 cells. Lanes 5 and 10, nuclear extract from SeAx cells. Lanes 11 and 12, STAT5 antibody added as negative control. Lane 11, nuclear extract from HUT78 cells. Lane 12, nuclear extract from MyLa 2059.
IL-7 and IL-15 stimulate the expression of thec-myb gene
It has been reported that c-myb expression is positively regulated by itself.36 If IL-7 and IL-15 should work through c-Myb, one would expect that IL-7 and IL-15 could also stimulate c-myb expression. The stimulation ofc-myb expression by IL-7 and IL-15 would prolong the effect of both interleukins on bcl-2 expression. Figure3A-B shows that IL-7 and IL-15 indeed stimulate c-myb expression and that c-myb is regulated in the same way as bcl-2. There are at least 7 different splice variants of c-myb,36 and they use several alternative exons (eg, 8A, 9A, 13A). IL-7 and IL-15 regulate practically all splice variants. With the polyclonal antibody used, we detected at least one further protein (marked by X) that may be a still unknown c-Myb splice variant given that it was also regulated by both interleukins. The triplet of the 3 splice forms 13A, pMbM-1, and complete coding sequence (CCS) is not always resolved on every gel. Bands above 95 kd were not always reproducible and might have been cross-hybridizing proteins. The strongest IL-7 and IL-15 stimulation after longer times of starvation was seen for splice variants using the alternative exons 8A, 9B, and 13A. The same pattern of splice variants seen in SeAx cells was observed in HUT78 and MyLa 2059 cells (data not shown). Western analysis of peripheral blood leukocytes (PBLs) detected only the splice firms 8A and 9B and the form initially designated as the CCS.37 Gel loading was controlled with an antibody directed against the c-myc gene (Figure 3C-D) that was not regulated by IL-7 or IL-15.38
IL-7 and IL-15 increase c-Myb protein levels in interleukin-starved SeAx cells.
(A) Effects of 10 IL-7. M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 6, 72-hour interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. CCS, splice form initially designated as the complete coding sequence; X, unknown c-Myb splice form. (B) Effects of 10 IL-15. M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 6, 72-hour interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. (C, D) Corresponding experiments with c-myc gene not regulated by IL-7 and IL-15 (negative control).
IL-7 and IL-15 increase c-Myb protein levels in interleukin-starved SeAx cells.
(A) Effects of 10 IL-7. M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 6, 72-hour interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after 6-hour addition of IL-7. CCS, splice form initially designated as the complete coding sequence; X, unknown c-Myb splice form. (B) Effects of 10 IL-15. M, marker. Lane 1, 48-hour interleukin-starved SeAx cells. Lane 2, 54-hour interleukin-starved SeAx cells. Lane 3, 54-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 4, 60-hour interleukin-starved SeAx cells. Lane 5, 60-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 6, 72-hour interleukin-starved SeAx cells. Lane 7, 72-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. Lane 8, 96-hour interleukin-starved SeAx cells. Lane 9, 96-hour interleukin-starved SeAx cells after 6-hour addition of IL-15. (C, D) Corresponding experiments with c-myc gene not regulated by IL-7 and IL-15 (negative control).
IL-7 and IL-15 stimulate the DNA binding of STAT factors to abcl-2 gene promoter element
We recently showed that IL-7 and IL-15 stimulate the binding of the transcription factor STAT5 to the IL-4–responsive element (IL-4RE) in CTCL cell lines.39 Because it has been reported that STAT5 can bind to a promoter element of thebcl-2 gene that has a similar sequence, we tested whether IL-7 and IL-15 can also increase the binding of STAT5 to this promoter element. Using nuclear extracts of IL-7– and IL-15–starved and IL-7– and IL-15–treated SeAx cells and an oligonucleotide representing the STAT5 binding bcl-2 promoter site, we found that both interleukins increased the binding of proteins to this sequence (Figure4A). Electrophoretic mobility shift assay experiments with antibodies against the STAT1 to STAT6 factors and nuclear extracts from CTCL cell lines (Figure 4B-D) showed that antibodies directed against STAT2 and STAT6 partially disrupted the C1 complex and formed a trimeric DNA–protein–antibody complex (supershift). This effect was barely visible with extracts from SeAx cells. The antibody against STAT5 also induced a supershift and caused a total disruption of the C1 complex, indicating that STAT5 is the main component of C1.
Binding of STAT factors to a bcl-2 gene promoter element in CTCL cells.
(A) IL-7 and IL-15 stimulate the binding of STAT factors to abcl-2 gene promoter element. Lane 1, unstimulated PBLs. Lane 2, unstimulated SeAx cells. Lane 3, IL-7–stimulated SeAx cells. Lane 4, IL-15–stimulated SeAx cells. (B-D) Identification of the STAT factors binding to the bcl-2 gene promoter element. (B) IL-7– and IL-15–treated SeAx cells (10 U each). (C) Untreated MyLa 2059. (D) Untreated HUT78 cells. Lane 1, nuclear extract from PBLs. Lanes 2 to 8, nuclear extracts from CTCL cell lines. Lane 2, no antibody added. Lane 3, STAT1 antibody. Lane 4, STAT2 antibody. Lane 5, STAT3 antibody. Lane 6, STAT4 antibody. Lane 7, STAT5 antibody. Lane 8, STAT6 antibody.
Binding of STAT factors to a bcl-2 gene promoter element in CTCL cells.
(A) IL-7 and IL-15 stimulate the binding of STAT factors to abcl-2 gene promoter element. Lane 1, unstimulated PBLs. Lane 2, unstimulated SeAx cells. Lane 3, IL-7–stimulated SeAx cells. Lane 4, IL-15–stimulated SeAx cells. (B-D) Identification of the STAT factors binding to the bcl-2 gene promoter element. (B) IL-7– and IL-15–treated SeAx cells (10 U each). (C) Untreated MyLa 2059. (D) Untreated HUT78 cells. Lane 1, nuclear extract from PBLs. Lanes 2 to 8, nuclear extracts from CTCL cell lines. Lane 2, no antibody added. Lane 3, STAT1 antibody. Lane 4, STAT2 antibody. Lane 5, STAT3 antibody. Lane 6, STAT4 antibody. Lane 7, STAT5 antibody. Lane 8, STAT6 antibody.
The c-myb gene and STAT factors are expressed in skin lesions of patients with CTCL
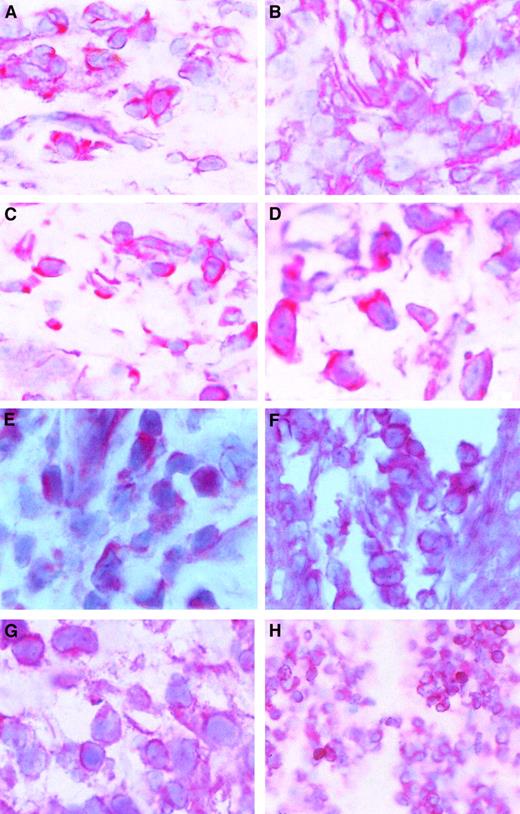
Because c-myb expression is correlated with cell proliferation and because CTCL cells proliferate in the skin but not in the blood, we wondered whether c-myb might be expressed in skin lesions of patients with CTCL. After staining cryostat sections from CTCL lesions with a c-Myb antibody, we found that c-mybwas expressed in all examined skin tumor lesions. Figure5A-D shows that the c-Myb protein is present mainly in the cytoplasm of CTCL cells but is also in the nucleus. Because c-Myb is already present in early stages (early MF; Figure 5A; early SS, Figure 5B), the activation of the c-myboncogene must be an early event in CTCL cancer genesis. Figure 5C shows an advanced MF stage, and Figure 5D shows an advanced SS stage.
c-Myb protein expression and localization in skin lesions.
Immunohistochemical stainings of skin lesions of an early (A) and a late (B) stage of mycosis fungoides and an early (C) and a late (D) stage of Sézary syndrome. Hematoxylin-eosin–stained slices from cryo-conserved biopsy material were tested with a c-Myb–specific antibody. Magnifications are 62.5 × (A-C) and 100 × (D). STAT2 (E), STAT5 (F), STAT6 (G), and bcl-2 (H) expression and localization in CTCL skin lesions. Hematoxylin-eosin–stained slices from cryoconserved biopsy material were tested with a STAT5-specific antibody. Magnifications are 100 × (E, G), 62.5 × (F), and 25 × (H).
c-Myb protein expression and localization in skin lesions.
Immunohistochemical stainings of skin lesions of an early (A) and a late (B) stage of mycosis fungoides and an early (C) and a late (D) stage of Sézary syndrome. Hematoxylin-eosin–stained slices from cryo-conserved biopsy material were tested with a c-Myb–specific antibody. Magnifications are 62.5 × (A-C) and 100 × (D). STAT2 (E), STAT5 (F), STAT6 (G), and bcl-2 (H) expression and localization in CTCL skin lesions. Hematoxylin-eosin–stained slices from cryoconserved biopsy material were tested with a STAT5-specific antibody. Magnifications are 100 × (E, G), 62.5 × (F), and 25 × (H).
The presence of STAT2, STAT5, and STAT6 in the malignant cells of skin lesions has already been shown (Figure 5E-G).39 It has been found in general that STAT5 is the STAT factor with the strongest staining in the nucleus. An example of bcl-2 expression in CTCL skin lesions is given in Figure 5D. In total, 11 patients have been tested.
Discussion
Our results show that IL-7 and IL-15 can stimulate the expression of the cell death survival gene bcl-2 in CTCL cells. DNA binding of the transcription factors c-myb and STAT5 are also stimulated by IL-7 and IL-15 and probably mediate the effects of IL-7 and IL-15 because the promoter of the bcl-2 gene has binding sites for both factors. To a lower extent, STAT2 and STAT6 also bound to the STAT5 binding site. To our knowledge, this is the first time an activation of c-myb by IL-7 and IL-15 has been reported. This result connects the findings that IL-7 and IL-15 can increase mature T-cell survival and bcl-2 expression40,41and that c-myb stimulates bcl-2 expression in lymphocytes.20-22 The stimulation of STAT5 by IL-7 and IL-15 also occurs in normal T cells and other tissues.31However, the stimulation of STAT2 and STAT6 is atypical. A stimulation of STAT6 has until now only been reported for mast cells, which have a cell-specific IL-15 receptor (IL-15RX) (reviewed in42).
The stimulation of c-Myb and of the 3 STAT factors occurs in less than 30 minutes, indicating that, in contrast to bcl-2, no protein synthesis is necessary for the stimulation of both factors by IL-7 and IL-15. Because the c-myb gene promoter also contains a c-Myb binding site, it may also positively regulate its own expression. With respect to bcl-2 and c-myb regulation in SeAx cells, IL-7 and IL-15 are equally effective. Thus, both cytokines may act through the same pathways because IL-7R and IL-15R contain the same γc chain. However, signaling through the γc chain alone is insufficient, because IL-4 and IL-9, whose receptors also contain the γc chain, are inefficient growth factors for CTCL cells.3 The α chains of IL-7R and IL-15R receptors must trigger similar pathways.
Because c-Myb plays a role not only in bcl-2 gene regulation but also in cell cycle progression, it may not only help CTCL cells to survive but also increase the cell division and growth rate of the tumor cells. This assumption is corroborated by the finding that IL-15 is necessary for cell division initiation.43STAT factors mediate the effects of many growth hormones and cytokines and thus not only increase the survival but also the growth of CTCL cells. The c-Myb protein is already present in early CTCL stages. Thus,c-myb activation must be an early step in CTCL cancer genesis.
Like c-Myb, STAT2, STAT5, and STAT6 were found in the 3 cell lines and in skin lesion biopsy specimens from patients with CTCL. As in the CTCL cell lines, the concentration of STAT5 was the highest of all expressed STAT factors. Regarding the presence of constitutive STAT factor activities, CTCL cells resemble leukemic cells from patients with acute myeloid leukemia, Burkitt lymphoma, and adult T-cell leukemia.22,37 44
The constitutive activity of STATs in CTCL cells may caused by deregulated activities of the tyrosine kinases Jak1 and Jak3, which regulate STAT5. However, other STAT factors regulating tyrosine kinases, such as c-Src, Bmx, and c-Yes, may be involved because these 3 tyrosine kinases, which are normally not expressed in T cells, are expressed in CTCL cells (U.D., C.-L.Z, and J.K., unpublished data, May 2000). Another reason for the constitutive activities of the STATs may be the functional loss of STAT inhibitors. Members of the suppressor of cytokine signaling (SOCS) gene family inhibit STAT signaling45-47; indeed, it has recently been found that CTCL cell lines lack functioning SOCS-3 proteins.48
In early stages, CTCL cell survival may depend on IL-7, IL-15, and other growth factors that are delivered by surrounding keratinocytes and dendritic cells. Later, the mutations in tyrosine kinases mentioned above may lead to cytokine-independent transcription factor activation, bcl-2 expression, and cell proliferation. IL-15 produced by the CTCL cells themselves3 may also help the CTCL cells to become independent from the dermal environment.
Our results suggest that the higher amounts of Bcl-2 protein in malignant T cells of patients with CTCL may be a direct consequence of c-Myb and STAT5 activities. Bcl-2 overexpression is of clinical relevance because CTCL tumors with high bcl-2 expression are resistant to radiotherapy.
We suppose that in early CTCL stages c-Myb alone may increase the expression of the bcl-2 gene but that it may not yet be able to increase CTCL cell proliferation because oncogenes such as c-myc,38 with which c-mybcooperates, are not yet expressed.
We thank M. Johnson and M. Bär for the preparation of the photographs.
Supported by the Swiss Cancer League (grant KFS 275-1-1996), the Swiss National Science Foundation (grant 3100-43244.95/1), the United Bank of Switzerland, the Theodor and Ida Herzog-Egli Foundation, and the Kanton of Zürich.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Udo Döbbeling, Department of Dermatology, University Hospital of Zurich, CH-8091 Zurich, Switzerland; e-mail:doebbeli@derm.unizh.ch.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal