Abstract
Chronic hepatitis C virus (HCV) infection has been associated with development of mixed cryoglobulinemia type 2 (MC2), a lymphoproliferative disorder characterized by B cell monoclonal expansion and immunoglobulin M/k cryoprecipitable immunoglobulin production. A short sequence (codons 384-410) of the HCV E2 protein, which has the potential to promote B cell proliferation, was investigated in 21 patients with HCV-related MC2 and in a control group of 20 HCV carriers without MC2. In 6 of the 21 (29%) patients with MC2, all the clones isolated from plasma, peripheral blood mononuclear cells, and liver showed sequence length variation compared with the hypervariable region 1 (HVR1) consensus sequence; 5 patients had an insertion at codon 385, and 1 patient had a deletion at codon 384. Inserted residues at position 385 were different within and between patients. No such mutations were observed in any of the HVR1 clones from control patients without MC2, and the difference between the 2 groups was statistically significant (P = .02). Analysis of 1345 HVR1 sequences obtained from GenBank strongly supported the conclusion that the observed insertions and deletion represent a rare event in HCV-infected patients, suggesting that they are significantly associated with MC2. The physical and chemical profiles of the 385 inserted residues detected in the MC2 patients were consistent with the possibility that these mutations, which occurred in a region containing immunodominant epitopes for neutralizing antibodies and binding sites for B lymphocytes, may be selected by functional constraints for interaction with host cells.
Introduction
The hepatitis C virus (HCV), besides being the major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma in many parts of the world, has also been implicated in the pathogenesis of mixed cryoglobulinemia type 2 (MC2).1-3This is a lymphoproliferative disorder characterized by B cell monoclonal expansion, leading to the production of large amounts of circulatory, temperature-sensitive immunocomplexes that accumulate in different tissues and cause the classical syndrome of arthralgia, purpura, weakness, systemic vasculitis, and glomerulonephritis.4-6 In some patients, further expansion of the monoclonal B cell tissue mass into high-grade non-Hodgkin lymphoma has been observed.7 In most published studies, the prevalence of HCV infection in patients with MC2 ranged from 80% to 90%, whereas reported prevalence of MC2 in HCV-infected patients has been variable, with higher rates in Italy and southern Europe than in northern Europe.3,8 How HCV promotes the development of MC2 in a subgroup of chronically infected patients is still unclear, but host and viral factors have been implicated. It has been shown that the HLAB8-DR3 haplotype might confer susceptibility to HCV-related MC2.9 As to the mechanisms promoting B cell proliferation, these might involve direct infection or, more likely, chronic, antigen-driven stimulation of B lymphocytes by HCV. A recent study10 favors the latter hypothesis by suggesting that the E2 envelope protein of HCV could be the chronic antigenic stimulus for clonal selection and expansion of B cells. The E2 protein of HCV contains the hypervariable region 1 (HVR1)–expressing immunodominant epitopes for B cells involved in virus neutralization.11-15 Furthermore, it has been reported that E2 can bind directly to B lymphocytes through interaction with the tetraspanin CD81, and such interaction has been suggested to promote B cell proliferation by lowering their activation threshold.16 Thus, the E2 protein has the potential for multifunctional interaction with B lymphocytes. Still it remains unclear why monoclonal expansion and proliferation leading to MC2 occurs only in a small subset of infected patients. As do other RNA viruses, HCV replicates as a mixture of different, but closely related, genomic variants in the form of virus quasi species.17HVR1 has the highest heterogeneity within the HCV quasi species, and this leads to the possibility that binding to and stimulation of B cells might be favored by specific HVR1 sequences. To assess whether variations in the sequence or conformation of the E2 protein could play a role in the development of HCV-related MC2, we have here compared the HVR1 sequence and quasi species in HCV chronic carriers with or without MC2. In some patients the analysis was extended to HCV derived from plasma, from peripheral blood mononuclear cells, and from liver biopsies to evaluate possible compartmentalization of specific HCV sequences.
Patients, materials, and methods
Patient characteristics
Twenty-one patients with MC2 (purpura, arthralgia, and weakness associated with evidence of circulating monoclonal immunoglobulin (Ig) M/k cryoglobulins) (5 men, 16 women; median age, 66 years; range, 44-67 years) were studied. Serum of all patients was positive for HCV-RNA and was infected with HCV 1b. None of the patients had ever been treated with interferon or with steroid or immunosuppressive treatments. The main clinical and laboratory features in the 21 MC2 patients are described in Table 1. Twenty untreated patients with chronic HCV infection without clinical and laboratory evidence of MC2 (8 men, 12 women; median age, 55 years; range, 38-65 years) were also studied. This control group was a consecutive series of patients infected with HCV 1b and treated in our unit. Mean alanine aminotransferase value at the time of this study was 88 ± 51 IU/L (range, 12-191 IU/L), and liver histology was consistent with mild chronic hepatitis in 10 patients, moderate/severe chronic hepatitis in 8 patients, and liver cirrhosis in 2 patients. The HVR1 sequence was analyzed in all patients and controls using plasma samples. In 4 patients with MC2, plasma, peripheral blood mononuclear cells (PBMC2s), and liver biopsy samples obtained on the same data were analyzed in parallel.
Clinical and laboratory findings of 21 patients with mixed cryoglobulinemia
| Patient no. . | Age (y)/sex . | ALT (IU/L) . | Histology . | Disease duration (mo) . | Cryocrit % . | Monoclonal component . | RF (IU/L) . | Lymphoma . | HCV-HVR1 mutated . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 67/F | 29 | Cirrhosis | 204 | 3 | IgM/k | 429 | NA | + |
| 2 | 61/F | 76 | Mild | 96 | 3 | IgM/k | 165 | NA | + |
| 3 | 71/F | 24 | Severe | 120 | 10 | IgM/k | 57 | NHL | + |
| 4 | 60/M | 14 | Mild | 12 | 3 | IgM/k | 38 | NA | + |
| 5 | 66/F | 162 | Cirrhosis | 68 | 1 | IgM/k | 89 | NO | + |
| 6 | 64/M | 24 | Mild | 108 | 5 | IgM/k | < 30 | NO | + |
| 7 | 73/F | 28 | Mild | 114 | 5 | IgM/k | 109 | NO | − |
| 8 | 67/F | 78 | Cirrhosis | 132 | 2 | IgM/k | < 30 | NA | − |
| 9 | 69/F | 25 | Mild | 66 | 5 | IgM/k | 437 | NA | − |
| 10 | 75/M | 31 | Mild | 60 | 5 | IgM/k | 325 | NA | − |
| 11 | 50/F | 49 | Mild | 156 | 9 | IgM/k | 82 | NHL | − |
| 12 | 59/F | 14 | Mild | 110 | 2 | IgM/k | 55 | NHL | − |
| 13 | 53/M | 33 | Moderate | 54 | 5 | IgM/k | 245 | NA | − |
| 14 | 67/F | 68 | Moderate | 50 | 0.5 | IgM/k | < 30 | NO | − |
| 15 | 53/F | 15 | Mild | 93 | 1 | IgM/λ | < 30 | NA | − |
| 16 | 55/F | 97 | Severe | 72 | 3 | IgM/k | 159 | NO | − |
| 17 | 45/M | 32 | NA | 204 | 3 | IgM/k | 545 | NO | − |
| 18 | 77/F | 104 | NA | 84 | 0.5 | IgM/k | 58 | NA | − |
| 19 | 69/F | 113 | Severe | 90 | 1 | IgM/k | 250 | NO | − |
| 20 | 68/F | 60 | Mild | 48 | 0.5 | IgM/λ | 1450 | NA | − |
| 21 | 44/F | 43 | Cirrhosis | 75 | 1 | IgM/k | 122 | NO | − |
| Patient no. . | Age (y)/sex . | ALT (IU/L) . | Histology . | Disease duration (mo) . | Cryocrit % . | Monoclonal component . | RF (IU/L) . | Lymphoma . | HCV-HVR1 mutated . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 67/F | 29 | Cirrhosis | 204 | 3 | IgM/k | 429 | NA | + |
| 2 | 61/F | 76 | Mild | 96 | 3 | IgM/k | 165 | NA | + |
| 3 | 71/F | 24 | Severe | 120 | 10 | IgM/k | 57 | NHL | + |
| 4 | 60/M | 14 | Mild | 12 | 3 | IgM/k | 38 | NA | + |
| 5 | 66/F | 162 | Cirrhosis | 68 | 1 | IgM/k | 89 | NO | + |
| 6 | 64/M | 24 | Mild | 108 | 5 | IgM/k | < 30 | NO | + |
| 7 | 73/F | 28 | Mild | 114 | 5 | IgM/k | 109 | NO | − |
| 8 | 67/F | 78 | Cirrhosis | 132 | 2 | IgM/k | < 30 | NA | − |
| 9 | 69/F | 25 | Mild | 66 | 5 | IgM/k | 437 | NA | − |
| 10 | 75/M | 31 | Mild | 60 | 5 | IgM/k | 325 | NA | − |
| 11 | 50/F | 49 | Mild | 156 | 9 | IgM/k | 82 | NHL | − |
| 12 | 59/F | 14 | Mild | 110 | 2 | IgM/k | 55 | NHL | − |
| 13 | 53/M | 33 | Moderate | 54 | 5 | IgM/k | 245 | NA | − |
| 14 | 67/F | 68 | Moderate | 50 | 0.5 | IgM/k | < 30 | NO | − |
| 15 | 53/F | 15 | Mild | 93 | 1 | IgM/λ | < 30 | NA | − |
| 16 | 55/F | 97 | Severe | 72 | 3 | IgM/k | 159 | NO | − |
| 17 | 45/M | 32 | NA | 204 | 3 | IgM/k | 545 | NO | − |
| 18 | 77/F | 104 | NA | 84 | 0.5 | IgM/k | 58 | NA | − |
| 19 | 69/F | 113 | Severe | 90 | 1 | IgM/k | 250 | NO | − |
| 20 | 68/F | 60 | Mild | 48 | 0.5 | IgM/λ | 1450 | NA | − |
| 21 | 44/F | 43 | Cirrhosis | 75 | 1 | IgM/k | 122 | NO | − |
ALT indicates serum alanine aminotransferase; NA, not available; NHL, non-Hodgkin lymphoma.
Polymerase chain reaction amplification and cloning of the HVR1
Total RNA was extracted first from samples of plasma and then from PBMC2s and liver biopsy samples by the single-step RNAZol method (Tel-Test, Friendswood, TX) according to the manufacturer's instructions. PBMC2s were isolated from 10 mL heparinized blood by Ficoll-Hypaque (Sigma, St Louis, MO) density gradient centrifugation, resuspended in 10 mL RPMI 1640 (Biowhittaker, Walkersville, MD) and washed thrice in PBS. The third wash of PBMC2s was tested for HCV RNA negativity by polymerase chain reaction (PCR). RNA was extracted from approximately 10 mg homogenized liver biopsy specimen directly into 900 μL RNAZol. In the case of plasma sample, the amount of RNA used corresponded to 100 μL.
Total RNA was resuspended in 10 μL diethylpyrocarbonate-treated distilled water. Reverse transcription (RT) was performed as described previously.18 Briefly, the RNA was incubated at 70°C for 5 minutes and then reverse-transcribed in a 25 μL reaction mixture containing 50 pM outer antisense primer, 3 mM MgCl2, 1 mM deoxynucleoside triphosphate (dNTP), 1 mM dithiothreitol, 75 mM KCl, 5 mM Tris-HCl (pH 8.3), 20 U RNase inhibitor (Pharmacia LKB, Piscataway, NJ), and 130 U Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Gaithersburg, MD). The mixture was incubated at 37°C for 60 minutes and then at 95°C for 5 minutes. E2 HVR1 was amplified after RT by nested PCR. First-round PCR was performed as follows: 10 μL cDNA was added to 40 μL mixture containing 50 pM external sense primer, 1.5 mM MgCl2, 23.5 mM Tris-HCl (pH 8.3), 35.5 mM KCl, and 1.5 U Taq polymerase (Perkin-Elmer, Norwalk, CT). Nested “hot-start” PCR was performed as follows: the bottom mixture contained 2.0 mM MgCl2, 0.2 mM each dNTP, 10 mM Tris-HCl (pH 8.3), 15 mM KCl, and 50 pM each internal primer. A wax layer was used to separate the lower mixture from the top reaction mixture containing 40 mM Tris-HCl (pH 8.3), 60 mM KCl, 1.5 U Taq polymerase, and 1 μL first-round PCR product. For the amplification of a 175 nt fragment (nt 1428-nt 1603) of the E2-HVR1, the following 2 sets of primers were used: outer primers, AS 5′CATTGCAGTTCAGGGCCGTGCTA3′ and S 5′GGTGCTCACTGGGGAGTCCT3′; inner primers, AS 5′TGCCAACTGCCATTGGTGTT3′ and S 5′TCCATGGTGGGGAACTGGGC3′. For first- and second-round PCR, 30 cycles with the following conditions were used: 30 seconds at 94°C, 25 seconds at 55°C, and 30 seconds at 72°C. PCR products were purified (QIAQuik columns; Qiagen, Chatsworth, CA) and ligated into TA cloning vectors (TA cloning kit; Invitrogen, Leek, The Netherlands) according to the manufacturer's instructions.
HVR1 sequencing
For sequencing, 8 clones were isolated from the plasma of 21 patients with MC2 and 20 patients in the chronic hepatitis control group; in total, 328 clones were analyzed. Deduced amino acid (aa) sequences from all the clones were aligned to the HVR1 consensus sequence published by Puntoriero et al.19
Cycle sequencing was performed using an ABI Prism Big Dye terminator ready reaction kit following the manufacturer's instructions (PerkinElmer Cetus, Emeryville, CA). The inner set of primers previously described was used to determine the nucleotide sequence of each strand. Electrophoresis of the sequencing products was performed by an ABI 310 automated DNA sequencer (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Phylogenetic analysis
Sequence alignment was obtained using the Pileup program of the GCG package (Accelcrys, San Diego, CA). Aligned sequences of each patient were compared to the sequence of the clone used as probe and then shaded using the Publish program of the GCG Package.
Phylogenetic trees were derived using the Treecon for Windows package (UIA, Antwerp, Belgium).20 Tree topologies were inferred by the neighbor-joining method with the Kimura 2-parameter distance matrices.
Physical and chemical profiles of HVR1 sequences
Prediction of relative residue accessibility and antigenicity values were calculated over a 7-aa window by the double-prediction method of Antheprot21(http://antheprot-pbil.ibcp.fr/ns_sommaire.html).
Analysis of the HVR1 quasi species and comparison of plasma, peripheral blood mononuclear cells, and liver HVR1
The HVR1 quasi species in plasma, PBMC2s, and liver were studied and compared using the heteroduplex tracking assay (HTA) and the clonal frequency analysis (CFA) techniques. These methods, which have been described in detail elsewhere,18,22 23 allowed us to resolve on nondenaturing gel electrophoresis intra-sample sequence heterogeneity through the mismatches between a radiolabeled probe (from one cDNA clone representing the quasi-species major variant) and the target sequences (the heterogeneous PCR product in the HTA and the PCR product from individual cDNA clones in the CFA). Briefly, to generate a probe, the insert of one clone was amplified by PCR. After column purification, the PCR product was end-radiolabeled with T4 polynucleotide kinase (Gibco BRL) plus γ-32P] adenosine triphosphate. Initially, quasi-species analysis was performed with the HTA technique whereby the probe, which represented the quasi-species major variant of the plasma sample, is hybridized to the heterogeneous HVR1 PCR product from each specimen, and the different viral sequences are then resolved by nondenaturing polyacrylamide gel electrophoresis. Nucleotide changes between the probe and the target DNA led to the formation of heteroduplex bands characterized by retarded mobility during gel electrophoresis. Probe hybridized with its own unlabeled sequence was used to indicate the homoduplex control. Shift control was made by hybridizing a probe with a PCR product obtained from a different patient. As previously demonstrated, the degree of shift between heteroduplex bands and homoduplex bands was directly proportional to the number of nucleotide changes between the probe and the target DNA.
To analyze in detail the quasi-species composition of a patient, 20 independent clones per specimen were then examined by CFA using the same blood-derived, patient-specific probe obtained for the analysis of the heterogeneous PCR product. In 4 selected patients in the MC2 group, the nucleotide sequence was determined from at least 1 clone from each unique variant by CFA in plasma, PBMC2s, and liver biopsy sample.
Statistical analysis
To examine whether the percentages of patients with mutated HVR1 sequence were significantly different in the group with HCV-related MC2 and in the control group with HCV infection without MC2, a statistical test of independence using the Fisher exact test was applied to the data from the 2 groups.
Nucleotide sequence accession number
Results
HVR1 sequences and quasi species in patients with mixed cryoglobulinemia
HCV-RNA from plasma samples of the 21 patients with MC2 and of the 20 HCV carriers without MC2 was used for RT-PCR amplification, cloning, and sequencing of a 175-nt long fragment, including the 384 to 410 HVR1 sequence. For each patient 8 independent clones were studied, and the derived aa sequences were aligned and compared with the consensus HVR1 sequence described by Puntoriero et al.19
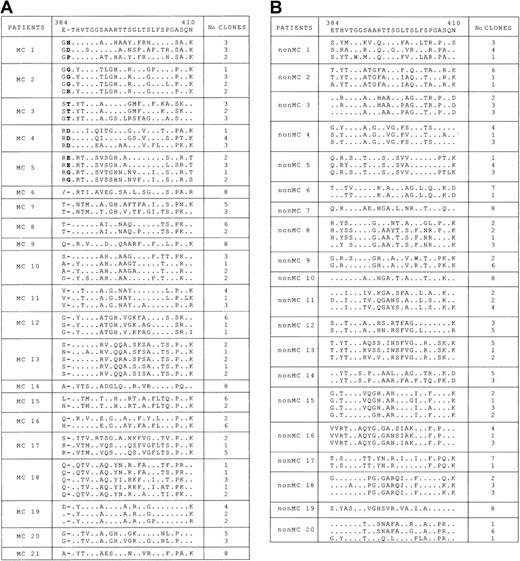
As shown in Figure 1A, different degrees of quasi-species complexity were identified in the patients with MC2. In 4 patients (patients 6, 9, 14, and 21), the quasi species was highly homogeneous, and only 1 major HVR1 variant was detected in all clones; in the remaining 17 patients, 2 to 5 variants were identified. In 6 patients (patients 1 to 6), the HVR1 aa sequences had different lengths than the consensus sequence. In 5 of them (patients 1 to 5), a single aa insertion was detected at position 385, and it was found in all the clones analyzed. Although the insertion site was identical in all 5 patients and in all clones from each patient, the inserted residue often varied among clones. It was the same within the whole quasi species in patients 3 and 4 only, in whom it showed a T385 and a D385 insertion, respectively. In patient 6, a 1-aa deletion at position 384 was detected in all 8 clones analyzed. In the remaining 15 patients with MC2 (patients 7 to 21), no such aa insertion or deletion was detected in any of the clones analyzed. The results obtained in 5 patients with an insertion at aa 385 and in those of patient 6 with a deletion at aa 384 were fully confirmed with a second experiment of PCR amplification, cloning, and sequencing, starting from a second batch of the same plasma sample (data not shown). Phylogenetic analysis of the obtained HVR1 sequences was then performed to exclude the possibility of false results from PCR contamination. By using the neighbor-joining method, a phylogenetic tree was constructed for all 57 unique aa sequences obtained from the 21 patients with MC2. As shown in Figure2, sequences from each clustered together and were well confined within subtrees, as expected from epidemiologically unrelated patients. Furthermore, clusters of sequences from the 6 patients with aa insertions or deletions were intermingled with clusters of nonmutated sequences, thus indicating the lack of a unique lineage for sequences with aa alterations in HVR1, distinct from all the other sequences detected. In agreement with these molecular profiles, there was no evidence of possible epidemiologic relationship among the 6 patients with mutated sequences in whom demographic data, geographic origin, and risk factors were considered.
Analysis of HVR1 aa sequence in clones derived from plasma of patients with and without MC2.
(A) Alignment of HVR1 aa sequences (codons 384-410) of individual clones derived from the plasma of 21 patients with MC2. Dots indicate nucleotide sequence identity with the consensus sequence reported on the top line.19 In patients 1 to 5, dashes indicate the sites of aa insertion as compared to the consensus; in patient 6, a slash denotes deletion. For each patient, the number of clones with identical sequences is indicated on the right. (B) Alignment of HVR1 aa sequences (codons 384-410) of individual clones derived from plasma of a control group of 20 patients with no MC2. Dots indicate nucleotide sequence identity, with the consensus sequence reported on the top line. For each patient, the number of clones with identical sequences is indicated on the right.
Analysis of HVR1 aa sequence in clones derived from plasma of patients with and without MC2.
(A) Alignment of HVR1 aa sequences (codons 384-410) of individual clones derived from the plasma of 21 patients with MC2. Dots indicate nucleotide sequence identity with the consensus sequence reported on the top line.19 In patients 1 to 5, dashes indicate the sites of aa insertion as compared to the consensus; in patient 6, a slash denotes deletion. For each patient, the number of clones with identical sequences is indicated on the right. (B) Alignment of HVR1 aa sequences (codons 384-410) of individual clones derived from plasma of a control group of 20 patients with no MC2. Dots indicate nucleotide sequence identity, with the consensus sequence reported on the top line. For each patient, the number of clones with identical sequences is indicated on the right.
Phylogenetic tree of HVR1 sequences from patients with MC2 obtained by the neighbor-joining method.
Branches are drawn to scale, and the scale bar corresponds to 0.05% nucleotide sequence divergence. Numbers included in the brackets correspond to the number of clones with identical sequences identified in that patient. HVR1 indicates the consensus sequence.
Phylogenetic tree of HVR1 sequences from patients with MC2 obtained by the neighbor-joining method.
Branches are drawn to scale, and the scale bar corresponds to 0.05% nucleotide sequence divergence. Numbers included in the brackets correspond to the number of clones with identical sequences identified in that patient. HVR1 indicates the consensus sequence.
HVR1 sequences and quasi species in patients with no mixed cryoglobulinemia
No aa insertions or deletions within the HVR1 sequence were detected when 160 clones obtained from the 20 patients without MC2 were sequenced (Figure 1B). The frequency of sequence length modification was, therefore, significantly higher in patients with MC2 (6 of 21; 29%) than in patients without MC2 (0 of 20; P = .02). We also investigated the frequency of HVR1 sequence length alterations in a much higher number of sequences obtained from the GenBank database. Among the 1345 HVR1 (codons 384-410) sequences found, only 27 (2%) harbored 1 to 4 insertions, and in all but one the insertion occurred at codon 385. Only 7 sequences (0.5%) had 1 or 2 aa deletions. These results indicate that aa insertions or deletions within the 27 residues of the HVR1 sequence, as detected in our patients with MC2, is a rare finding in HCV-infected patients and is significantly associated with MC2.
Physical and chemical profiles of HVR1 sequences
The insertions at 385 detected within the HVR1 sequences in patients with MC2 were either hydrophilic (H and D in patient 1, R in patient 2, T in patient 3, D in patient 4, and E in patient 5) or were characterized by high turn propensity (P in patient 1, G in patient 2, and G in patient 5) and, therefore, appeared able to confer conformational restriction to this portion of the protein. Furthermore, insertion was located just before T386, a polar aa that might have an important structural and functional role given that it is highly conserved among sequences of HCV. Interestingly, T386 was also maintained in the sequence with the aa deletion found in patient 6. Recently, an HCV envelope protein E2 model was proposed, based on aa similarities with the envelope protein E of tick-borne encephalitis virus.24 According to this model, the insertion 385 we detected in patients with MC2 occurred in a highly exposed portion of the E2 protein that was the target of the immunoresponse and was in close steric contiguity with the CD81 binding site. According to these data, the changes detected in our patients with MC2 could affect antigenicity and binding properties of the HVR1 sequence. Indeed, the prediction of relative residue accessibility and antigenicity values, calculated over a 7-aa window by the double-prediction method of Antheprot, indicated that these values were increased at codon 385 in the HVR1 sequence of patients with MC2 compared with the consensus sequence and with those derived from patients without MC2 (mean relative residue accessibility and antigenicity values, 68.3 and 40.0, respectively).
Heteroduplex gel shift assay and sequencing analysis of HVR1 quasi species in different compartments
To investigate whether the 385 insertion, when detected in plasma, was also present in other compartments, detailed HVR1 quasi-species analysis was performed in plasma, PBMC2s, and liver biopsy specimens from 2 patients with MC2 (patients 3 and 4) using a combination of 2 techniques—heteroduplex tracking assay and clonal frequency analysis. The same analysis was then performed in 2 other patients with MC2, but not in patients with mutations in plasma HVR1 sequences (patients 11 and 12), to verify the presence, if any, of circulating variants with the 385 insertion in PBMC2s or liver biopsy samples.
HVR1 of these 4 patients was amplified by RT-PCR starting from paired samples of plasma, liver biopsy specimens, and PBMC2s. PCR products were then used to characterize the quasi species in the 3 different compartments by heteroduplex gel shift analysis.
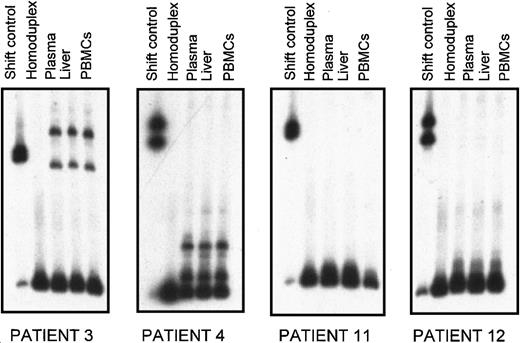
Patient-specific radiolabeled probes from the plasma major variant were generated and initially hybridized with the unlabeled heterogeneous PCR products from plasma, PBMC2s, and liver biopsy samples. HTA allowed rapid determination of the quasi-species composition in the 3 samples from each patient regarding number of variants and their relative genetic distances. Results of HTA are illustrated in Figure3: in each panel, the heteroduplex shift patterns corresponding to the quasi species from plasma, liver, and PBMC2s of each patient are reported. In each patient the migration profile of the HVR1 variants was identical in the different samples, indicating the presence of the same major variants. To more precisely assess quasi-species composition in terms of complexity (number of variants with a distinct shift pattern) and relative abundance of each detected variant, 20 clones from each compartment were then analyzed by CFA. Because it is sensitive enough to discriminate among variants that differ by only a few nucleotide substitutions, CFA allowed the resolution of details in the quasi-species composition. Results of this analysis are reported in Figure 4A for patients 3 and 4 and in Figure 4B for patients 11 and 12.
Heteroduplex tracking assay of HVR1 quasi species from plasma, liver, and PBMC2s from 4 patients with MC2 (patients 3, 4, 11, 12).
For each patient, heterogeneous HVR1 amplification products from different tissues were hybridized with 32P-labeled probe derived from plasma major variant. Lane 1 contains the heteroduplex shift control; lane 2 corresponds to the homoduplex control; lanes 3, 4, and 5 show quasi-species distribution in plasma, liver, and PBMC2s, respectively.
Heteroduplex tracking assay of HVR1 quasi species from plasma, liver, and PBMC2s from 4 patients with MC2 (patients 3, 4, 11, 12).
For each patient, heterogeneous HVR1 amplification products from different tissues were hybridized with 32P-labeled probe derived from plasma major variant. Lane 1 contains the heteroduplex shift control; lane 2 corresponds to the homoduplex control; lanes 3, 4, and 5 show quasi-species distribution in plasma, liver, and PBMC2s, respectively.
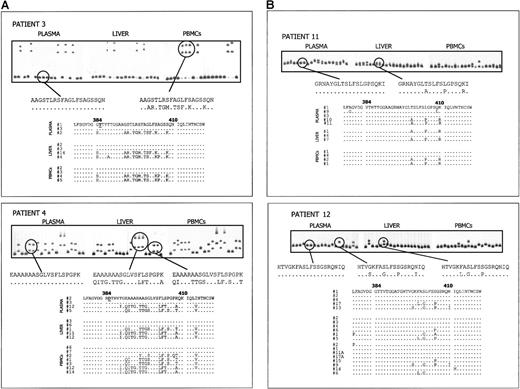
HCV quasi-species analysis in different compartments from patients with and without HVR1 aa insertion.
(A) Analysis of HCV quasi species in plasma, liver, and PBMC2s of 2 patients with MC2 (patients 3 and 4) in whom an HVR1 aa insertion was detected. Upper panels illustrate the CFA profiles of HVR1 quasi-species variants isolated from the 3 different tissues. For each patient, one clone representing a plasma major variant was labeled with 32P and hybridized to 20 HVR1 clones from each of the 3 compartments. In each panel, lane 1 shows the homoduplex control, and lanes 2 to 21 show gel shift patterns of the 20 independent cDNA clones. Gel shift patterns representative of different HVR1 variants and their corresponding aa sequences are indicated. Below, deduced aa sequences of clones from the 3 tissues, selected on the basis of the CFA results, are reported. The 385 insertion, found in all the HCV variants from these patients, is indicated by an underlined letter. Dots indicate aa identity to plasma-derived probe. (B) Analysis of HCV quasi species in plasma, liver, and PBMC2s of 2 patients with MC2 (patients 11 and 12) with no sequence mutations in the HVR1. Upper panels illustrate the CFA profiles of HVR1 quasi-species variants isolated from the 3 different tissues. In each panel, lane 1 shows the homoduplex control, and lanes 2 to 21 show gel shift patterns of the 20 independent cDNA clones. Below, aa sequences of selected clones from the 3 compartments are aligned to the sequence of the CFA probe derived from plasma major variant.
HCV quasi-species analysis in different compartments from patients with and without HVR1 aa insertion.
(A) Analysis of HCV quasi species in plasma, liver, and PBMC2s of 2 patients with MC2 (patients 3 and 4) in whom an HVR1 aa insertion was detected. Upper panels illustrate the CFA profiles of HVR1 quasi-species variants isolated from the 3 different tissues. For each patient, one clone representing a plasma major variant was labeled with 32P and hybridized to 20 HVR1 clones from each of the 3 compartments. In each panel, lane 1 shows the homoduplex control, and lanes 2 to 21 show gel shift patterns of the 20 independent cDNA clones. Gel shift patterns representative of different HVR1 variants and their corresponding aa sequences are indicated. Below, deduced aa sequences of clones from the 3 tissues, selected on the basis of the CFA results, are reported. The 385 insertion, found in all the HCV variants from these patients, is indicated by an underlined letter. Dots indicate aa identity to plasma-derived probe. (B) Analysis of HCV quasi species in plasma, liver, and PBMC2s of 2 patients with MC2 (patients 11 and 12) with no sequence mutations in the HVR1. Upper panels illustrate the CFA profiles of HVR1 quasi-species variants isolated from the 3 different tissues. In each panel, lane 1 shows the homoduplex control, and lanes 2 to 21 show gel shift patterns of the 20 independent cDNA clones. Below, aa sequences of selected clones from the 3 compartments are aligned to the sequence of the CFA probe derived from plasma major variant.
In patient 3, the shift profile of clones derived from plasma, liver, and PBMC2s were identical—2 distinct groups of variants with relative prevalence were conserved in all 3 compartments. In patient 4, 3 major groups of cDNA clones with substantial differences in gel mobility were found in plasma, liver, and PBMC2s, and the prevalence of each type of sequence was again maintained in the 3 compartments. However, although no variants other than the 3 dominants were detected in plasma or liver, 3 additional minor variants with distinct shift patterns were found in PBMC2s (see lanes 2, 8, and 10 on the corresponding CFA panel).
In patient 11, the gel shift patterns revealed the presence of a similar HCV population in plasma, liver, and PBMC2s. In this case, the restricted number of clones with distinct patterns and the slight mobility differences observed between clones was indicative of a tight genetic relation among variants within and between the 3 compartments. Similarly, in patient 12 the quasi species in plasma, liver, and PBMC2 samples was homogeneous and was primarily represented by variants with highly similar migration patterns.
Sequences from clones with unique gel shift profiles on CFA from these 4 patients were determined. In each case, the deduced aa sequences from plasma, PBMCs, and liver were then aligned, and the results were consistent with the presence of identical sequences in the 3 compartments previously observed by CFA.
Discussion
Mixed cryoglobulinemia type 2 has been clearly associated with chronic HCV infection, but how the virus promotes B-cell clonal expansion remains undefined. In this study we have identified a mutation of HVR1 of the E2 protein in a significant proportion of patients with MC2. This mutation consisted of a change in HVR1 sequence length, highly conserved among all HCV genotypes and composed of 27 aa. Five patients with mutated sequence length had 1 residue insertion at codon 385, and one patient had 1 residue deletion at codon 384. Interestingly, 100% of the variants isolated from each of these patients had the mutation, independently on whether HCV had been derived from plasma, PBMC2s, or liver. Thus, though we cannot exclude the presence of nonmutated HVR1 as a minor variant, HVR1 sequence length alterations were predominant in 6 of 21 of our patients with MC2. Whether the mutated HVR1 sequences found in patients with MC2 were the major variant at the time of contamination or emerged and expanded during chronic infection is a matter of speculation. Our findings were consistent with both possibilities. The high prevalence of mutated HCV (each of the 8 clones within the same quasi species) found in patients with MC2—as opposed to the absence of such mutations in patients without MC2, independent of the compartment analyzed, and to the low frequency in the sequence database—seems to favor the hypothesis that these variants could have been selected and expanded to homogeneity by strong replicative advantages over nonmutated sequences. This conclusion is also in agreement with results reported by Casino et al,25 who described an identical insertion at codon 385 in all HCV clones derived from a blood recipient whose donor did not harbor the mutated sequence within the circulating (and infecting) HCV quasi species. Inserted amino acids at position 385 varied among patients and among variants in the same patient, but in all instances they appeared to affect the conformation of the protein, though with different effects. In particular, insertions were represented either by hydrophilic residues often found in protein–protein interaction sites (H, D, R, T, and E) or by aa characterized by high turn propensity (P and G). Because the mutation occurred at a surface-exposed site of the E2 protein and within a viral sequence known to be of highly conserved length among different HCV genotypes, it is reasonable to suggest that the mutation could represent an adaptation of this region in the interaction with other molecules. Although the exact function of the HVR1 remains to be elucidated, previous studies26 have suggested that sequence variations in this portion of the E2 region could affect antigenic specificity and tissue tropism of the virus.
The pathogenic role of HCV in the development of MC2 has been associated with the ability of the virus to induce strong, protracted B-cell clone proliferation. This might be the consequence of chronic antigenic stimulation by HCV antigens acting directly on B cells or of involvement in the immunocomplexes that elicit the IgM monoclonal response. After studying immunoglobulin antigen receptor variable region genes, De Re et al10 suggest that the HCV E2 protein may indeed represent one of the chronic stimuli driving B-cell selection and clonal expansion. On the other hand, Pileri et al16 show that the E2 protein can also bind directly to B lymphocytes through interaction with the cellular receptor CD81 and that such interaction, which might occur through cellular receptors also involved in virus entry, has been suggested to promote B-cell proliferation by lowering the activation threshold. Although in these studies the discrete domains of E2 representing the epitopes for neutralizing antibodies and B-cell binding sites have not been identified, data suggest that the HVR1 could be directly involved in interaction with and in stimulation of B cells. Several studies show that HVR1, including the N-terminus of HVR1, does contain immunodominant epitopes for neutralizing antibodies.11-15According to a recently proposed model of the HCV envelope protein E2, the 385 insertion we detected in patients with MC2 would occur in a highly exposed portion of the E2 protein that is the target of the immunoresponse and is in close steric contiguity with the CD81 binding site.24 We suggest that selection and expression of the mutated HVR1 sequence seen in patients with MC2 could reflect a replicative advantage resulting from optimized fitness with cell receptors or coreceptors involved in virus binding and entry. These receptors or coreceptors could also be responsible for triggering B-cell proliferation by the mutated HCV, thus explaining the association with MC2. Further studies on the binding and stimulatory activity on B lymphocytes of the identified HVR1 sequences are warranted to prove or reject this hypothesis.
Supported by a grant (MM06104479-007) from the 40% MURST, Italy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alfredo Alberti, Department of Clinical and Experimental Medicine, University of Padua, Via Giustiniani, 2 Padua 35128, Italy; e-mail: labepvir@unipd.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal