Abstract
Severe congenital neutropenia (SCN) was originally described as an autosomal recessive disorder. Subsequently, autosomal dominant and sporadic forms of the disease have been recognized. All forms are manifest by persistent severe neutropenia and recurrent bacterial infection. In contrast, cyclical hematopoiesis is characterized by periodic neutropenia inter-spaced with (near) normal neutrophil counts. Recently, linkage analysis on 13 affected pedigrees identified chromosome 19p13.3 as the likely position for mutations in cyclical hematopoiesis. Heterozygous mutations in the ELA2 gene encoding neutrophil elastase were detected in all families studied. Further work also demonstrated mutations in ELA2 in sporadic and autosomal dominant SCN. However, all mutations described to date are heterozygous and thus appear to act in a dominant fashion, which is inconsistent with an autosomal recessive disease. Therefore, the current study investigated whether mutations in ELA2could account for the disease phenotype in classical autosomal recessive SCN and in the sporadic and autosomal dominant types. All 5 exons of ELA2 and their flanking introns were studied in 18 patients (3 autosomal recessive, 5 autosomal dominant [from 3 kindreds], and 10 sporadic) using direct automated sequencing. No mutations were found in the autosomal recessive families. A point mutation was identified in 1 of 3 autosomal dominant families, and a base substitution was identified in 8 of 10 patients with the sporadic form, though 1 was subsequently shown to be a low-frequency polymorphism. These results suggest that mutations in ELA2are not responsible for classical autosomal recessive Kostmann syndrome but provide further evidence for the role of ELA2 in SCN.
Introduction
Kostmann1 first described severe congenital neutropenia (SCN) as an autosomal recessive disorder in a large, intermarried Swedish family in 1956. Subsequently, autosomal dominant and sporadic forms of the disease have been recognized. Typically, children are noted in early infancy to have persistent severe neutropenia of less than 0.2 × 109/L, recurrent bacterial infections, and maturation arrest at the promyelocyte-myelocyte stage in the bone marrow. Mutations in the cytoplasmic region of the granulocyte colony-stimulating factor (G-CSF) receptor have been reported in a proportion of the 10% of children with SCN in whom acute myeloid leukemia (AML) develops.2However, such mutations in the cytoplasmic region are an acquired event and are not the cause of the underlying disease.
Cyclical hematopoiesis is characterized by periodic neutropenia inter-spaced with normal or near normal neutrophil counts with a remarkably regular 21-day periodicity.3 In contrast to SCN, autosomal dominant patterns of inheritance predominate. Recently, linkage analysis on 13 affected pedigrees identified chromosome 19p13.3 as the likely position for mutations in cyclical hematopoiesis.4 This area contains a family of genes encoding azurocidin, neutrophil elastase, and proteinase 3. Heterozygous mutations in the ELA2 gene-encoding neutrophil elastase were detected in all families. Additional work by this group also demonstrated mutations in ELA2 in patients with sporadic and autosomal dominant SCN.5 Structural modeling has suggested that in cyclical hematopoiesis the mutations appear to cluster around the active site of the enzyme, whereas in congenital neutropenia the opposite face of the enzyme seems to be predominantly affected. Neutrophil elastase is synthesized in promyelocytes and promonocytes and is packaged into the azurophilic cytoplasmic granules.6 The mature 218-amino acid glycoprotein is released at sites of inflammation and appears to have a critical pathophysiological role in a variety of pulmonary diseases.7 To date, neutrophil elastase has no known function in myelopoiesis.
In the current study, autosomal recessive and autosomal dominant familial cases of SCN, in addition to sporadic cases, were screened to investigate whether mutations in the same gene could account for the pathogenesis of classic, apparently autosomal recessive Kostmann disease and also to confirm the published results in sporadic SCN.
Patients, materials, and methods
Patients and clinical details
Children were recruited directly from the neutropenia clinic at Great Ormond Street Hospital for Children. Additional patients were referred directly to the laboratory from other centers in the United Kingdom. Local Regional Ethical Committee approval was received for the study. Diagnoses of all patients were based on the conventional criteria of persistent severe neutropenia, recurrent bacterial infection, and bone marrow aspirate showing maturation arrest at the myelocyte-promyelocyte stage. Cytogenetic analyses at diagnosis showed normal findings. Children with anti-neutrophil antibodies or myelodysplastic features were excluded. Clinical details are summarized in Table 1.
Clinical details
| Patient . | Inheritance . | Date of birth . | Baseline neutrophil count × 109/L . | G-CSF administration . | Date of transformation to AML . | Comment . |
|---|---|---|---|---|---|---|
| 1 | AR | Dec 1993 | 0.15 | Never | ||
| 2 | AR | Apr 1988 | 0.0 | Regular | Dec 1998 | Relapsed 7 months after UD BMT |
| 3 | AR | May 1999 | 0.05 | Regular | ||
| 4 | AD | Sep 1975 | 0.2 | Regular | ||
| 5* | AD | May 1989 | 0.1 | Regular | ||
| 6* | AD | Dec 1966 | 0.1 | (Regular) | Poor patient compliance | |
| 7† | AD | Dec 1988 | 0.0 | Regular | ||
| 8† | AD | Dec 1988 | 0.09 | Regular | Feb 1995 | Failed to engraft UD BMT; died 9 months later |
| 9 | S | Jul 1995 | < 0.1 | Regular | Dec 1999 | Remission, 4 months after UD BMT |
| 10 | S | Jan 1987 | 0.04 | (Regular) | Now on intermittent G-CSF | |
| 11 | S | Mar 1999 | 0.13 | Regular | ||
| 12 | S | Feb 1996 | < 0.1 | Regular | ||
| 13 | S | Aug 1987 | 0.1 | (Regular) | Now off G-CSF; neutrophils 0.5 × 109/L | |
| 14 | S | Nov 1999 | 0.0 | Regular | ||
| 15 | S | Oct 1987 | 0.0 | Regular | ||
| 16 | S | Oct 1993 | 0.26 | (Regular) | Now on infrequent G-CSF | |
| 17 | S | May 1989 | 0.1 | Infrequent | Needle phobia; would probably benefit from regular G-CSF | |
| 18 | S | Aug 1986 | 0.1 | Infrequent |
| Patient . | Inheritance . | Date of birth . | Baseline neutrophil count × 109/L . | G-CSF administration . | Date of transformation to AML . | Comment . |
|---|---|---|---|---|---|---|
| 1 | AR | Dec 1993 | 0.15 | Never | ||
| 2 | AR | Apr 1988 | 0.0 | Regular | Dec 1998 | Relapsed 7 months after UD BMT |
| 3 | AR | May 1999 | 0.05 | Regular | ||
| 4 | AD | Sep 1975 | 0.2 | Regular | ||
| 5* | AD | May 1989 | 0.1 | Regular | ||
| 6* | AD | Dec 1966 | 0.1 | (Regular) | Poor patient compliance | |
| 7† | AD | Dec 1988 | 0.0 | Regular | ||
| 8† | AD | Dec 1988 | 0.09 | Regular | Feb 1995 | Failed to engraft UD BMT; died 9 months later |
| 9 | S | Jul 1995 | < 0.1 | Regular | Dec 1999 | Remission, 4 months after UD BMT |
| 10 | S | Jan 1987 | 0.04 | (Regular) | Now on intermittent G-CSF | |
| 11 | S | Mar 1999 | 0.13 | Regular | ||
| 12 | S | Feb 1996 | < 0.1 | Regular | ||
| 13 | S | Aug 1987 | 0.1 | (Regular) | Now off G-CSF; neutrophils 0.5 × 109/L | |
| 14 | S | Nov 1999 | 0.0 | Regular | ||
| 15 | S | Oct 1987 | 0.0 | Regular | ||
| 16 | S | Oct 1993 | 0.26 | (Regular) | Now on infrequent G-CSF | |
| 17 | S | May 1989 | 0.1 | Infrequent | Needle phobia; would probably benefit from regular G-CSF | |
| 18 | S | Aug 1986 | 0.1 | Infrequent |
AR indicates autosomal recessive; UD BMT, unrelated donor bone marrow transplantation; AD, autosomal dominant; (Regular), previous prophylactic G-CSF usage, now superseded by intermittent use only; S, sporadic.
Members of family A.
Members of family B.
Eighteen patients were screened. Eight were familial cases from 6 different families, 3 with an apparent autosomal recessive pattern of inheritance and 3 with an apparent autosomal dominant pattern. Ten were unrelated sporadic cases. In the first apparently recessive family (patient 1; Table 1), the parents are second cousins of Sri Lankan origin. The parents of patient 2 are first cousins of Pakistani origin; they had 2 other children with SCN who died of infective complications in early childhood. The parents of patient 3 are also first cousins from an inter-married Pakistani family, and both are first cousins to a 10-year-old child with SCN. In the apparently autosomal dominant families, at least 2 members of 2 or more consecutive generations were affected, but there was no evidence of consanguinity.
Patients 2, 8, and 9 developed acute myeloid leukemia. Patient 2 had a relapse and died 7 months after undergoing fully engrafted haplo-identical bone marrow transplantation. Patient 8 did not achieve durable myeloid engraftment from unrelated donor bone marrow transplantation and died 9 months later of the complications of gut graft-versus-host disease and recurrent SCN. Patient 9 is alive and well 4 months after unrelated donor bone marrow transplantation.
Mutational analysis
DNA was extracted from peripheral blood leukocytes or bone marrow mononuclear cells using standard techniques. Three fragments of DNA covering all 5 exons of ELA2 and at least 15 bases of the flanking regions were amplified using polymerase chain reaction (PCR) and the primers in Table 2. Each product was prepared in a 25 μL reaction with 0.625 U BiotaqTaq polymerase (Bioline, London, United Kingdom), proprietary buffer, 1 mM MgCl2, 200 μΜ dNTPs, and 0.5μΜ concentrations of each primer. PCR was run at 95°C for 5 minutes before the Taq was added and then at 35 cycles of 95°C for 30 seconds, 68°C for 30 seconds, 72°C for 1 minute, and a final extension of 72°C for 5 minutes. Products were cleaned using the Promega (Madison, WI) Wizard PCR Preps system and were directly sequenced in both directions using the same primers and additional internal sequencing primers as appropriate (Table 2). An Applied Biosystems (Foster City, CA) 310 Analyzer using BigDye version 2 terminator chemistry was used for all sequencing.
PCR primer sequences
| . | Start* . | Forward primer . | Start . | Reverse primer . | Annealing temperature . |
|---|---|---|---|---|---|
| DNA fragment | |||||
| 1 | 1252 | 5′-CGGAGGGGCAGAGACCCCGGA-3′ | 1992 | 5′-AGACCGGGACGCGGGGTCCGA-3′ | — |
| Sequencing primer | 1768 | 5′-CACCCGGTGTGTCCCCAGGCA-3′ | — | — | — |
| 2 | 2065 | 5′-CTCGAGCACCTTCGCCCTCAG-3′ | 2517 | 5′-TCAACGGCCCATGGCGGGTAT-3′ | — |
| 3 | 4363 | 5′-CCTGCCCTGCAGGATCCCAGA-3′ | 5133 | 5′-GGAGAGTGTGGGTGTGGGCAG-3′ | — |
| Sequencing primers | 4599 | 5′-AGGAACCGTGGGATCGCCAGC-3′ | 4680 | 5′-TGCAGACGTTGCTGCGACGGC-3′ | — |
| Promoter region | 997 | 5′-CCTGGGCGATAGAGCAAGACT-3′ | 1399 | 5′-AACCCGGGACAGAGGGAGCAG-3′ | — |
| RNA fragment | |||||
| Complete cDNA | 1252 | 5′-CGGAGGGGCAGAGACCCCGGA-3′ | 5133 | 5′-GGAGAGTGTGGGTGTGGGCAG-3′ | — |
| Sequencing primers | 1791 | 5′-GCGCTGGCCTCGGAGATTGTG-3′ | 1930 | 5′-CAGTGCGCGGCCGACATGACG-3′ | — |
| 2240 | 5′-GTGTTCGCCGTGCAGCGCATC-3′ | 2288 | 5′-AGTTTACGGGGTCGTAGCCGT-3′ | — | |
| 4622 | 5′-CCTGCAGGAGCTCAACGTGAC-3′ | 4680 | 5′-TGCAGACGTTGCTGCGACGGC-3′ | — | |
| Polymorphic marker | |||||
| KB9† | — | 5′-TGCAAAGGCTTGGAGGGCTGATG-3′ | — | 5′-ATCTCGGACAACAGCAGGCCTCG-3′ | 67°C |
| D19S886‡ | — | 5′-TGGATCTACACTCCGGC-3′ | — | 5′-ATTTTACTGGCTGGCACTTG-3′ | 57°C |
| D19S814‡ | — | 5′-AGACCAGCCTGGGTAACATG-3′ | — | 5′-AGAAGGCTGAGTTGGTACAGG-3′ | 55°C |
| . | Start* . | Forward primer . | Start . | Reverse primer . | Annealing temperature . |
|---|---|---|---|---|---|
| DNA fragment | |||||
| 1 | 1252 | 5′-CGGAGGGGCAGAGACCCCGGA-3′ | 1992 | 5′-AGACCGGGACGCGGGGTCCGA-3′ | — |
| Sequencing primer | 1768 | 5′-CACCCGGTGTGTCCCCAGGCA-3′ | — | — | — |
| 2 | 2065 | 5′-CTCGAGCACCTTCGCCCTCAG-3′ | 2517 | 5′-TCAACGGCCCATGGCGGGTAT-3′ | — |
| 3 | 4363 | 5′-CCTGCCCTGCAGGATCCCAGA-3′ | 5133 | 5′-GGAGAGTGTGGGTGTGGGCAG-3′ | — |
| Sequencing primers | 4599 | 5′-AGGAACCGTGGGATCGCCAGC-3′ | 4680 | 5′-TGCAGACGTTGCTGCGACGGC-3′ | — |
| Promoter region | 997 | 5′-CCTGGGCGATAGAGCAAGACT-3′ | 1399 | 5′-AACCCGGGACAGAGGGAGCAG-3′ | — |
| RNA fragment | |||||
| Complete cDNA | 1252 | 5′-CGGAGGGGCAGAGACCCCGGA-3′ | 5133 | 5′-GGAGAGTGTGGGTGTGGGCAG-3′ | — |
| Sequencing primers | 1791 | 5′-GCGCTGGCCTCGGAGATTGTG-3′ | 1930 | 5′-CAGTGCGCGGCCGACATGACG-3′ | — |
| 2240 | 5′-GTGTTCGCCGTGCAGCGCATC-3′ | 2288 | 5′-AGTTTACGGGGTCGTAGCCGT-3′ | — | |
| 4622 | 5′-CCTGCAGGAGCTCAACGTGAC-3′ | 4680 | 5′-TGCAGACGTTGCTGCGACGGC-3′ | — | |
| Polymorphic marker | |||||
| KB9† | — | 5′-TGCAAAGGCTTGGAGGGCTGATG-3′ | — | 5′-ATCTCGGACAACAGCAGGCCTCG-3′ | 67°C |
| D19S886‡ | — | 5′-TGGATCTACACTCCGGC-3′ | — | 5′-ATTTTACTGGCTGGCACTTG-3′ | 57°C |
| D19S814‡ | — | 5′-AGACCAGCCTGGGTAACATG-3′ | — | 5′-AGAAGGCTGAGTTGGTACAGG-3′ | 55°C |
Nucleotide position corresponds to GenBank entry AC Y00477.
Taken from reference 4.
Sequences from the Genome Database (http://www.gdb.org).
In the patients in whom no mutation was identified, the 5′ promoter region from one member of each kindred was screened using the technique above; primers are detailed in Table 2. The MgCl2concentration was optimized to 0.6 mM, and the annealing temperature was optimized to 64°C for the initial PCR.
RNA was extracted from bone marrow mononuclear cells using Trizol (Gibco BRL, Paisley, United Kingdom). Complementary DNA (cDNA) for the complete ELA2 coding region was produced by reverse transcription and used as a template for PCR with the specified primers (Table 2). Bidirectional sequencing was performed as above. Patients 1, 2, 7, 8, 10, 12, and 17 were only screened using RNA.
Confirmatory tests
The presence of each mutation was confirmed using restriction endonuclease digestion of the relevant PCR fragment (Table3). The mutation in patient 14 required the use of a mismatch reverse primer 5′-TGCCCAGAAGGCCCCAGCCCATGGCCAGAC-3′) (the mismatch is underlined). For each mutation found, at least 50 healthy controls (and parents, if available) were screened using PCR and restriction endonuclease digestion. Positive screening results were confirmed by bidirectional sequencing using the method detailed above.
Results of mutational analysis by inheritance in severe congenital neutropenia
| Patient . | Inheritance . | Intron/exon . | DNA change3-150 . | RE used . | Predicted protein change3-151 . | Parents . | Healthy controls . |
|---|---|---|---|---|---|---|---|
| 1 | AR | No mutation | |||||
| 2 | AR | No mutation | |||||
| 3 | AR | No mutation | |||||
| 4 | AD | Intron 4 | G4716A (ivs4 + 1sd) | BsiWI | del Val161-Phe170 | N/A | 0/50 |
| 53-152 | AD | No mutation | |||||
| 63-152 | AD | No mutation | |||||
| 73-153 | AD | No mutation | |||||
| 83-153 | AD | No mutation | |||||
| 9 | S | Exon 2 | G1882A3-155 | MwoI | Cys26Tyr | W/T | 0/57 |
| 10 | S | Exon 3 | G2202A3-155 | BsaJI | Gly56Glu | W/T | 0/52 |
| 11 | S | Intron 3 | C4477A (ivs3-8sa) | AciI | insProGln94 | W/T | 0/57 |
| 12 | S | Exon 4 | C4495T | MscI | Ser97Leu | N/A | 0/57 |
| 13 | S | Exon 4 | C4495T | MscI | Ser97Leu | N/A | 0/57 |
| 14 | S | Exon 4 | T4569A3-155 | HinfI | Cys122Ser | W/T | 0/55 |
| 15 | S | Exon 5 | C4898G3-155 | StyI | Pro176Arg | N/A | 0/51 |
| 16 | S | Exon 5 | C5054T3-155 | BsoBI | Pro228Leu | Father C5054T | 1/110 |
| 17 | S | No mutation | |||||
| 18 | S | No mutation |
| Patient . | Inheritance . | Intron/exon . | DNA change3-150 . | RE used . | Predicted protein change3-151 . | Parents . | Healthy controls . |
|---|---|---|---|---|---|---|---|
| 1 | AR | No mutation | |||||
| 2 | AR | No mutation | |||||
| 3 | AR | No mutation | |||||
| 4 | AD | Intron 4 | G4716A (ivs4 + 1sd) | BsiWI | del Val161-Phe170 | N/A | 0/50 |
| 53-152 | AD | No mutation | |||||
| 63-152 | AD | No mutation | |||||
| 73-153 | AD | No mutation | |||||
| 83-153 | AD | No mutation | |||||
| 9 | S | Exon 2 | G1882A3-155 | MwoI | Cys26Tyr | W/T | 0/57 |
| 10 | S | Exon 3 | G2202A3-155 | BsaJI | Gly56Glu | W/T | 0/52 |
| 11 | S | Intron 3 | C4477A (ivs3-8sa) | AciI | insProGln94 | W/T | 0/57 |
| 12 | S | Exon 4 | C4495T | MscI | Ser97Leu | N/A | 0/57 |
| 13 | S | Exon 4 | C4495T | MscI | Ser97Leu | N/A | 0/57 |
| 14 | S | Exon 4 | T4569A3-155 | HinfI | Cys122Ser | W/T | 0/55 |
| 15 | S | Exon 5 | C4898G3-155 | StyI | Pro176Arg | N/A | 0/51 |
| 16 | S | Exon 5 | C5054T3-155 | BsoBI | Pro228Leu | Father C5054T | 1/110 |
| 17 | S | No mutation | |||||
| 18 | S | No mutation |
RE indicates restriction enzyme; ivs, intervening sequence; sa, splice acceptor; sd, splice donor; W/T, wild type; N/A, not available; for other abbreviations, see Table 1.
Nucleotide position corresponds to GenBank entry AC Y00477. Splice site numbering is relative to the intron/exon boundary.
Amino acid number one is the first after the presignal peptide.
Members of family A.
Members of family B.
Novel mutation.
Linkage analysis
Linkage analysis was performed using 3 polymorphic markers on chromosome 19p13.3: KB9, D19S886, and D19S814. KB9 had the highest peak 2-point lod score of 13.11 in the original study on cyclical hematopoiesis.4 D19S886 and D19S814 are centromeric and telomeric to ELA2, respectively.
One primer of each pair (Table 2) was γ-[32P]–ATP end-labeled, and PCR was performed using the method above with the following modifications: 0.2 μΜ concentrations of each primer for D19S814, annealing temperatures as indicated in Table 2, cycle extension times of 30 seconds, total of 25 cycles. Products were run on a 6% denaturing polyacrylamide gel and were visualized using autoradiography.
Results
Mutational analysis
No mutations were found in the patients with presumed autosomal recessive disease. In the 5 patients from 3 autosomal dominant families, only one mutation was found. This was a splice donor variant (G4716A) resulting in the deletion of Val161 to Phe170. The G4716A mutation and others leading to this deletion have been described in 7 patients with familial cyclical hematopoiesis, 3 patients with sporadic cyclical hematopoiesis, and 2 patients with SCN.4 5
Seven different heterozygous base substitutions were identified in 8 of 10 patients with sporadic SCN (Table 3). They were found in all exons except exon 1. Six were point substitutions that would lead to the following amino acid changes: Cys26Tyr, Gly56Glu, Ser97Leu (2 patients), Cys122Ser, Pro176Arg, and Pro228Leu. The remaining mutation was at a splice acceptor site and would lead to the insertion of 2 amino acids—insProGln94. All were confirmed by restriction digestion; a typical example is shown in Figure 1A. Both parents of 5 patients were available for study. Parents of patients 9 to 11 and patient 14 were shown to have the wild-type allele at the appropriate locus. However, the substitution in patient 16 leading to Pro228Leu (initially identified by analysis of bone marrow-derived cDNA and later confirmed by analysis of genomic DNA) was subsequently found in the hematologically healthy father and in one of 110 healthy control samples. It was thus most likely a novel low-frequency polymorphism. None of the other mutations were found in at least 50 healthy controls.
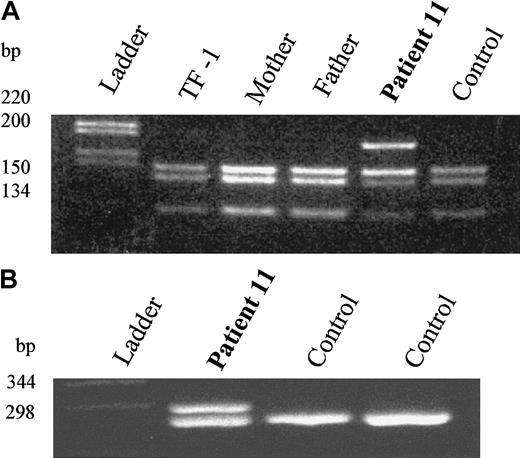
Molecular analysis of DNA and RNA from an SCN patient with an
ELA2 mutation. (A) AciI restriction enzyme digestion demonstrating heterozygosity in patient 11 and wild-type alleles in the parents. C4477A mutation removes a cleavage site for the enzyme AciI. (B) RT-PCR analysis demonstrating equivalent amounts of the normal and mutant mRNA. Splice acceptor mutation leads to insertion of 6 bp in the mRNA transcript.
Molecular analysis of DNA and RNA from an SCN patient with an
ELA2 mutation. (A) AciI restriction enzyme digestion demonstrating heterozygosity in patient 11 and wild-type alleles in the parents. C4477A mutation removes a cleavage site for the enzyme AciI. (B) RT-PCR analysis demonstrating equivalent amounts of the normal and mutant mRNA. Splice acceptor mutation leads to insertion of 6 bp in the mRNA transcript.
For the 2 splice variants, cDNA was produced from bone marrow RNA, and the appropriate fragment was amplified by PCR. Wild-type and mutant cDNAs were present in approximately equal quantities; an example is shown in Figure 1B. The 2 bands were separated on low-melting-point agarose. Sequencing confirmed the predicted changes in both.
Four of the 7 mutations (seen in patients 9, 10, 14, and 15) and the polymorphism (seen in patient 16) have not been previously described.
In the patients in whom no mutation was found in the coding region, the 5′ promoter region (starting at −240 and including all of exon 1) was sequenced bidirectionally in at least one member of each kindred (including the 3 patients with sporadic disease). This fragment includes the critical region between −106 and −76 that appears to be responsible for the myeloid specificity of neutrophil elastase transcription8 and all the other possible promoter sites identified by these authors. A C1063A (nucleotide −199) base substitution was identified in patient 1 (homozygous), patients 3 and 16 (heterozygous), and the unaffected mother of patient 16 (heterozygous) (data not shown). However, this is a recognized single nucleotide polymorphism (http://www.snp.cshl.org/db/snp/snp?name=TSC0102675) and the fact that the relative of patient 3 with SCN does not have this single nucleotide polymorphism argues strongly against a role for this particular substitution in the pathogenesis of SCN.
Linkage analysis
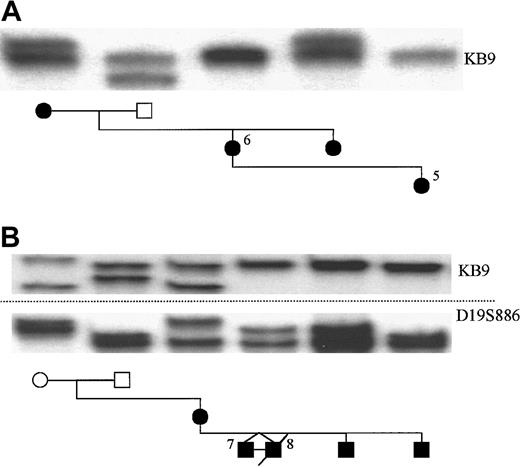
Linkage analysis was performed in the 2 autosomal dominant families without mutations in ELA2 using the 3 polymorphic markers described above. In family A, samples were available from affected relatives spanning 3 generations (Figure2A). Using the KB9 marker, the results showed that the 2 affected daughters (IIi and IIii) inherited different alleles from their affected mother (Ii), thus excluding linkage to chromosome 19p13.3 in this family. Corroborating evidence was also provided by identical results with the other 2 markers and by data from 3 unaffected half-siblings (data not shown), making recombination most unlikely. Analysis of X chromosome inactivation patterns using theHUMARA gene9 did not show any evidence of skewing in this family, thus excluding an X-linked disorder with extreme lyonization (data not shown).
Linkage analysis in the autosomal dominant families without ELA2 mutations.
In family A, which includes patients 5 and 6, the KB9 marker is shown not to segregate with the disease phenotype. In family B, which includes patients 7 and 8, linkage cannot be excluded by the use of the markers KB9 (upper) and D19S886 (lower).
Linkage analysis in the autosomal dominant families without ELA2 mutations.
In family A, which includes patients 5 and 6, the KB9 marker is shown not to segregate with the disease phenotype. In family B, which includes patients 7 and 8, linkage cannot be excluded by the use of the markers KB9 (upper) and D19S886 (lower).
In family B, the mother (IIi) and all 4 of her male children are affected, her parents are not. Results using KB9, D19S886 (Figure 2B), and D19S814 (data not shown) showed that the mother had passed her paternal chromosome 19 to all her children. Her 61-year-old father had a normal neutrophil count whenever tested and did not experience an excess of infective illnesses. Spontaneous mutation in the mother and linkage to chromosome 19p13.3 cannot, therefore, be excluded in this family. X-linked disease with extreme lyonization in the mother has been excluded by the demonstration of balanced expression of her polymorphic p55 transcripts10 (data not shown).
Too few family members were available to perform linkage analysis in the autosomal recessive families.
Discussion
In the current study, a mutation in the ELA2 gene was detected in 1 of 3 apparently autosomal dominant kindreds with familial SCN. No mutations were identified in the apparently autosomal recessive families. These results are compatible with those published previously showing that mutations were found in 5 of 5 autosomal dominant families,5 but they suggest that not all autosomal dominant SCN is caused by mutations in ELA2. The original disorder described by Kostmann,1 infantile genetic agranulocytosis, was an autosomal recessive disease. The current study is the first report in which mutations in ELA2 have been sought in patients with autosomal recessive SCN; none were found. This is perhaps not surprising in that a single heterozygous mutation that appears to act in a dominant fashion does not fit with an autosomal recessive pattern of inheritance in which, by definition, both copies of a gene are defective. Nevertheless, it is possible that total loss of a putative neutrophil elastase inhibitor (likely to be a recessive event) might lead to a similar disease.
Although we only studied 3 families with the autosomal dominant pattern of inheritance, using the KB9 marker linkage to chromosome 19p13.3 could be excluded in 1 of these families, effectively ruling out the involvement of mutations in ELA2 and any immediate promoter-enhancer regions in the pathogenesis of the neutropenia in this kindred. Linkage analysis was unhelpful in the remaining autosomal dominant family. Furthermore, sequencing of the 5′ promoter region in all 8 kindreds without a coding region mutation (ie, 3 families with autosomal recessive, 2 families with autosomal dominant, and 3 patients with sporadic disease) only demonstrated a previously identified single nucleotide polymorphism.
The results presented in this study confirm the high frequency of heterozygous mutations in the neutrophil elastase gene in sporadic SCN that has been previously reported.5 Four novel mutations and a low-frequency polymorphism were detected. The control group used in this study was predominantly of white United Kingdom origin, whereas the patient with the polymorphism was of mixed race, and the incidence of the polymorphism in the population of origin could be higher than the 1 in 110 found in our study. These data further strengthen the argument for the role of neutrophil elastase mutations in the pathogenesis of SCN. It should be noted that the patients with apparently sporadic disease without ELA2 mutations (patients 16-18) could actually have autosomal recessive disease and that nearly all cases of sporadic SCN may result from de novo heterozygous mutations inELA2.
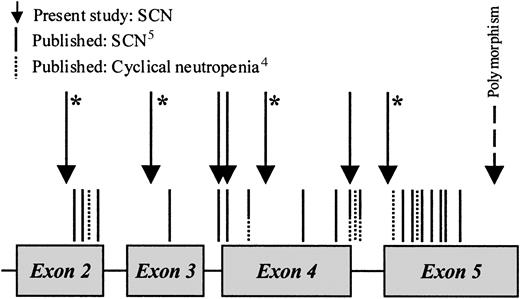
Figure 3 shows schematically the location of published mutations and those from the current study in SCN and cyclical neutropenia. To date, we have only studied 2 cases of classical cyclical neutropenia; in one there was no mutation and in the other there was the previously described Ala32Val (coded for by C1900T in exon 2; data not shown). It can be seen that in SCN the mutations are scattered throughout the molecule. There is also clear overlap between the 2 diseases, particularly in the cluster of mutations at the intron 4 splice donor site, all of which led to the loss of the last 10 residues of exon 4. The latter mutations have now been described in 7 families with cyclical neutropenia, 3 patients with sporadic cyclical neutropenia, and 3 patients with sporadic congenital neutropenia.4 5
Schematic diagram showing the location of mutations in the
ELA2 gene in severe congenital and cyclical neutropenia. An asterisk denotes a novel mutation.
Schematic diagram showing the location of mutations in the
ELA2 gene in severe congenital and cyclical neutropenia. An asterisk denotes a novel mutation.
It remains difficult to explain the different phenotypes and clinical severity of cyclical and severe congenital neutropenia, in particular the different risks for malignant transformation. To date, secondary leukemia has not developed in any patient with cyclical neutropenia (though one patient with a clonal cytogenetic abnormality has recently been described in the literature11), whereas in SCN there is a 1 in 10 risk of developing myelodysplasia or acute myeloid leukemia.2 Furthermore, the presence of a mutant neutrophil elastase does not preclude the development of leukemia (as evidenced by patient 9 in the current study) nor is it an essential prerequisite (patients 2 and 8).
Interestingly, there may be variation in phenotype within SCN patients with identical ELA2 mutations. Patient 12 (now aged 5) with the Ser97Leu substitution has continuing severe neutropenia and remains on G-CSF therapy with only a modest response. In contrast, patient 13 (now aged 13) had severe neutropenia (less than 0.1 × 109/L) and recurrent infections until he started G-CSF at the age of 4. He responded well and needed only a small maintenance dose of 2 μg/kg per 24 hours. G-CSF was discontinued when he was 8; he has remained free of major infections and now has a neutrophil count of approximately 0.5 × 109/L. This may reflect the influence of other inherited modifying factors, and it may suggest a more multigenic pathogenesis for SCN.
The pathogenicity of mutations in ELA2 remains mysterious. Neutrophil elastase has no known role in myelopoiesis. Interestingly, heterozygous and homozygous ELA2 knockout mice have normal neutrophil counts, and only the latter have a demonstrable increase in susceptibility to infection.12,13 Experiments in which the mutant enzyme has been transfected into cell lines (rat basophilic cells [RBL-1] and murine myeloblasts [32D]) have shown reduced levels of elastase activity in some patients.14 However, other investigators have reported reduced elastase activity as a more general phenomenon in congenital neutropenia15 with or without elastase mutations, implying that this is a general feature of SCN rather than one that is related to ELA2 mutations and disease pathogenesis. Therefore, it appears likely that the pathogenesis is due to a gain of function in the mutant protein, perhaps allowing the enzyme to bind a novel substrate involved in myelopoiesis or changing its binding with an intracellular inhibitor. An alternative explanation may relate to changes in intracellular packaging. During normal myeloid differentiation, the granule matrix immobilizes the digestive enzymes in the azurophilic granules.6 Should the ELA2 mutations lead to significant shape change of the mature protein, this immobilization may no longer be possible and the developing myeloid cells may be killed by autodigestion. Finally, recent evidence shows increased susceptibility to apoptosis of myeloid precursors in SCN,16 but the link between this and mutant neutrophil elastase has not been established.
We thank the families for kindly providing material, we thank the following for their assistance in sample and data collection: Brady Baxter, Great Ormond Street Hospital, London; Peter Arkwright, Department of Child Health, Manchester; Paula Bolton-Maggs, Alder Hey Hospital, Liverpool; Carole Edwards and Sally Kinsey, SCN International Registry, Leeds; Brenda Gibson, Royal Hospital for Sick Children, Glasgow; Diab Haddad and Tanya Bernard, St Peter's Hospital, Chertsey; Judith Marsh, St George's Hospital, London; Janice Simpson and James Nicholson, Addenbrooke's Hospital, Cambridge; and Graham Smith, Royal United Hospital, Bath, United Kingdom.
Supported by the Roald Dahl Foundation, Great Missenden (Bucks, United Kingdom) (P.A.) and by an unrestricted educational grant from Amgen (Cambridge, United Kingdom) (P.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Phil Ancliff, Department of Haematology, University College London, 98 Chenies Mews, London, WC1E 6HX, United Kingdom; e-mail: p.ancliff@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal