Abstract
The transcription factor C/EBPα is an important mediator of granulocyte differentiation and regulates the expression of multiple granulocyte-specific genes including the granulocyte–colony-stimulating factor (G-CSF) receptor, neutrophil elastase, and myeloperoxidase. Indeed C/EBPα knockout mice display a profound block in granulocyte differentiation. To study this block in granulocytic differentiation in more detail, retroviral vector-mediated transduction of a dominant-negative retinoic acid receptor was used to establish hematopoietic growth factor–dependent, lympho-myeloid progenitor cell lines from the fetal livers of both the C/EBPα knockout animals (C/EBPα(−/−)) and their heterozygous littermates (C/EBPα(+/−)). Surprisingly, the C/EBPα(−/−) cell lines displayed significant spontaneous granulocytic differentiation, and this differentiation was markedly enhanced when the cells were stimulated with granulocyte macrophage (GM)–CSF. This GM-CSF–mediated differentiation was associated with the up-regulation of G-CSF receptor mRNA, and the combination of GM-CSF and G-CSF generated more than 95% mature neutrophils in the C/EBPα(−/−) cultures. The addition of all-transretinoic acid also enhanced this granulocytic differentiation of the cultured C/EBPα(−/−) cells, indicating that the activated retinoic acid receptors can enhance granulocytic differentiation through a molecular pathway that is independent of C/EBPα. These studies clearly indicate that terminal granulocytic differentiation associated with the up-regulation of C/EBPα-responsive genes can occur in the absence of C/EBPα, and they indicate the existence of multiple independent molecular pathways potentially used by primitive hematopoietic precursors that can lead to the development of mature granulocytes.
Introduction
The regulation of cell differentiation invariably involves the alteration of gene expression patterns, and this is likely controlled by the activity of lineage-specific transcription factors. Such factors have been particularly well characterized in the multiple lineages that characterize hematopoiesis.1 The CCAAT enhancer binding protein alpha (C/EBPα) is a leucine zipper containing transcription factor involved in regulating the expression of specific target genes.2,3 C/EBPα is an important regulator of gene expression in both liver and adipose tissue.3,4 Moreover, several lines of evidence support a critical role for C/EBPα in regulating granulopoiesis. For example, in hematopoietic cells, C/EBPα expression occurs predominantly in granulocytes with markedly lower expression in monocytes and lymphocytes.5,6 C/EBPα binds to and activates the promoters of a number of granulocyte-specific genes, including the G-CSF receptor,7 neutrophil elastase,8 and myeloperoxidase.9 Moreover, induced expression of C/EBPα enhances the granulocytic differentiation of a number of different myeloid cell lines.6,10 11
Perhaps the most convincing evidence to support a role for C/EBPα in regulating granulopoiesis comes from observations using C/EBPα knockout mice. The homozygous mutant C/EBPα knockout animals die within 24 hours of birth, probably secondary to metabolic complications including reduced hepatic glycogen stores and hypoglycemia.12 These mice also exhibit a selective absence of peripheral blood neutrophils and eosinophils while displaying apparently normal development of other hematopoietic lineages.13 Interestingly, the fetal livers of these animals display reduced mRNA levels of granulocyte–colony-stimulating factor (G-CSF) receptor, an important regulator of granulocyte production and survival.14 These observations suggest that the selective defect in granulopoiesis displayed by the C/EBPα knockout animals may be secondary to reduced transcription of certain C/EBPα target genes critical for granulopoiesis, including the interleukin-6 (IL-6) and G-CSF receptors.13 15
We have previously observed that retroviral vector-mediated transduction of normal mouse bone marrow with a dominant-negative retinoic acid receptor alpha (RARα) generates immortalized hematopoietic growth factor–dependent cell lines that are frozen at different stages of hematopoietic development.16,17 These cell lines appear particularly valuable for assessing the molecular events regulating hematopoietic cell commitment and differentiation because they are derived from normal rather than leukemic cells, are dependent on physiologic hematopoietic growth factors for growth and differentiation, and display enhanced myeloid commitment and differentiation when exposed to relatively high pharmacologic (micromolar) concentrations of all-trans retinoic acid (ATRA). Indeed the in vitro manipulation of such cell lines has helped define changes in gene expression that accompany both hematopoietic lineage commitment18-20 and terminal granulocytic differentiation.21 Moreover, such cell lines have proved useful in functionally characterizing candidate mediators of hematopoietic lineage commitment22 and in identifying lineage-specific gene products that regulate hematopoiesis.23
In the current study, we describe the derivation and characteristics of hematopoietic cell lines from the fetal livers of both C/EBPα homozygous knockout mice and their heterozygous littermates. Surprisingly, the lympho-myeloid progenitor cell lines derived from the C/EBPα(−/−) mice display spontaneous granulocytic differentiation and a brisk differentiative response to physiologic agents including granulocyte macrophage (GM)–CSF, G-CSF, and ATRA. Moreover, the granulocytic differentiation in the C/EBPα(−/−) cell lines is accompanied by up-regulation of mRNA for the G-CSF receptor, a known target gene of C/EBPα. Our observations suggest that there are multiple molecular pathways involved in the generation of mature granulocytes, including some that are clearly C/EBPα independent.
Materials and methods
Genotyping of C/EBPα(+/+), (+/−), and (−/−) mice
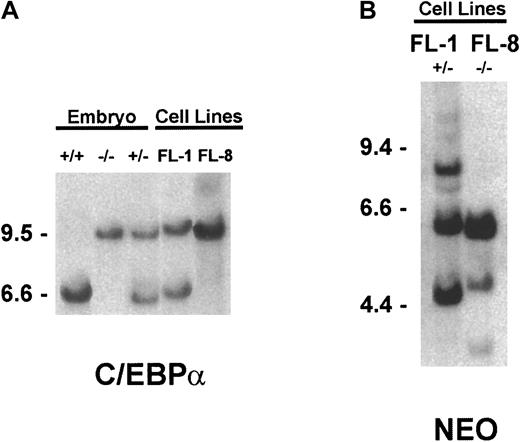
The derivation of the C/EBPα knockout mice has been previously described.12 For genotyping these animals, genomic DNA was extracted from mouse tail snips using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). DNA was digested withHincII and subjected to Southern blot analysis using a mouse genomic 1.8-kb HindIII-EcoR1 fragment immediately 3′ to the C/EBPα coding sequence. This fragment hybridizes to a 6-kb wild-type C/EBPα HincII fragment while identifying a 9.5-kb HincII fragment in the corresponding mutant “knockout” allele.12 Genotyping was also performed in the identical manner on embryonic DNA after removal of the fetal liver to identify the C/EBPα(+/−) versus the C/EBPα(−/−) embryos.
Retroviral vector integration sites
Genomic DNA was extracted from the cultured cell lines, digested with EcoR1, and subjected to Southern blot analysis using a 0.9-kb neo probe.16EcoR1 cuts only once within the LRARα403SN provirus upstream of theneo sequence; thus, the number of bands hybridizing to theneo probe on Southern blot analysis is proportional to the number of proviral integration sites.
Growth factors
Recombinant murine IL-3 and G-CSF were obtained from PeproTech (Rocky Hill, NJ). Sources of stem cell factor (SCF) included recombinant murine SCF (PeproTech) or conditioned media (10%-15%) from BHK cells transfected with an expression vector for the murine kit ligand gene (BHK/MKL). Sources of GM-CSF included recombinant murine GM-CSF (PeproTech) or conditioned media (5%) from cells transfected with an expression vector for the murine GM-CSF gene (HM5).
Generation of hematopoietic growth factor–dependent cell lines
Construction of the LRARα403SN retroviral vector, which harbors a truncated human RARα cDNA exhibiting dominant-negative activity, has been detailed.24 C/EBPα heterozygote mice were mated, and individual fetal livers were harvested on embryonic day 13.5. Fetal livers were minced to a single suspension and immediately frozen pending genotyping of the corresponding embryonic DNA. After genotyping, fetal liver cells from heterozygous (+/−) and homozygous knockout (−/−) embryos were thawed and immediately cocultured with ecotropic fibroblast producers (PE501) of the LRARα403 retroviral vector24 in the presence of SCF and IL-3 (5 ng/mL). After 2 days of coculture, the supernate cells were transferred to Dulbecco minimum essential medium supplemented with 10% fetal calf serum plus SCF and IL-3. Brisk in vitro growth of these hematopoietic cells was immediately observed. These cultured cells were genotyped and characterized within 8 to 12 weeks of retroviral vector transduction, and both the FL-1 (+/−) and FL-8 (−/−) cell lines have been continuously grown in liquid suspension for over 6 months in the presence of SCF and IL-3.
Surface antigen phenotyping
Cultured cells were stained with antibodies directly conjugated with fluorescein isothiocyanate or phycoerythrin and analyzed on a FACScan (Becton Dickinson). Antibodies used included anti-B220, anti-Gr-1, anti-CD11b, and anti-ter119 (Pharmingen, San Diego, CA) as well as anti F4/80 (Caltag, Burlingame, CA).
RNA isolation and Northern blot analysis
RNA was isolated using the TRIzol reagent and protocol supplied by Gibco BRL (Rockville, MD). For Northern blot hybridization, total cell RNA was electrophoresed through formaldehyde–agarose gels, and the gel was blotted to nylon membranes (Nytran Plus; Schleicher & Schuell, Keene, NH). Nylon membranes were hybridized to DNA probes that were labeled by nick translation. Probes included the above-described 1.8-kb HindIII-EcoR1 mouse genomic C/EBPα fragment, a 2-kb chick actin cDNA fragment, and a murine 325–base pair (bp) G-CSF receptor cDNA. The latter probe was prepared by polymerase chain reaction (PCR) amplification of cDNA synthesized from whole mouse embryo RNA. This cDNA synthesis was performed in 2 steps. The first reaction occurred in 36 μL with 80 ng/μL RNA template and 40 ng/μL oligo-dT. The reaction was incubated at 70°C for 10 minutes and then quickly chilled to 4°C. Next, the volume of the reaction was increased to 60 μL by adding the following components to the respective final concentrations: 1× buffer, 25 mM dithiothreitol, 1.25 mM dNTP, and SuperScript II RNase H-RT (GibcoBRL) at 25 U/μL. The reaction was incubated at 37°C for 1 hour and was followed by a PCR reaction performed in 25 μL 1× Gibco Taq polymerase buffer plus 1.5 mM MgCl2, 0.2 mM dNTP, 4 ng/μL forward and reverse primers, 0.1 U/μL Taq DNA polymerase, and 2 μL of the above cDNA synthesis reaction. Primers included GCSFR-1 (upstream) 5′ TCCGTCACCCTAAACATCTC-3′ and GCSFR-2 (downstream) 5′ TGGAAGGTTTCCTCTGTCAT-3′.
PCR cycle parameters included 94°C for 30 seconds, 63°C for 1 minute, and 72°C for 1 minute for 35 cycles. The amplified 325-bp G-CSF receptor cDNA fragment was purified from an ethidium-stained polyacrylamide gel, radiolabeled by nick translation, and used as a molecular probe for the Northern blots.
Results
Establishment of hematopoietic growth factor–dependent cell lines from C/EBP(+/−) and (−/−) mice
We have previously observed that retroviral vector-mediated transduction of a dominant-negative RARα construct into normal mouse bone marrow cells resulted in the reproducible establishment of hematopoietic growth factor–dependent cell lines blocked at different stages of differentiation.16,17 Because most of the C/EBPα knockout mice die within 24 hours after birth of metabolic dysfunction likely secondary to liver failure,12 we attempted to establish such hematopoietic cell lines from the knockout animals by transducing cells from fetal liver rather than bone marrow. C/EBP(+/−) heterozygotes were mated, and fetal livers were harvested on embryonic day 13.5 and genotyped as described in “Materials and methods” (Figure 1A). Fetal liver cells were infected with the dominant-negative RARα retroviral vector and then were cultured in a combination of SCF and IL-3. Brisk growth of the transduced fetal liver cells in this hematopoietic growth factor combination occurred almost immediately after transduction. With this approach, continuously proliferating hematopoietic growth factor–dependent fetal liver cell lines were reproducibly established with 100% efficiency from the C/EBPα(+/+), C/EBPα(+/−), and C/EBPα(−/−) fetal livers. At least 3 different cell lines have been established from the fetal livers of both the C/EBPα(+/−) and the C/EBPα(−/−) mice, and we describe here the characteristics of representative fetal liver cell lines (designated FL-1 and FL-8) derived from C/EBPα(+/−) and (−/−) mice, respectively.
Genotyping of the cultured fetal liver cell lines.
Genomic DNA was extracted from the indicated tissue and subjected to Southern blot hybridization, as detailed in “Materials and methods.” (A) To determine the C/EBPα genotype,HincII-digested DNA was hybridized with the C/EBPα genomic probe. (B) To assess the relative number of retroviral vector integration sites, EcoR1-digested DNA was hybridized to theneo-specific probe.
Genotyping of the cultured fetal liver cell lines.
Genomic DNA was extracted from the indicated tissue and subjected to Southern blot hybridization, as detailed in “Materials and methods.” (A) To determine the C/EBPα genotype,HincII-digested DNA was hybridized with the C/EBPα genomic probe. (B) To assess the relative number of retroviral vector integration sites, EcoR1-digested DNA was hybridized to theneo-specific probe.
Mixed lympho-myeloid characteristics of the fetal liver cell lines
Southern blot analysis of genomic DNA extracted from the embryos and cultured cell lines confirmed the genotype of the FL-1(+/−) and FL-8(−/−) cell lines (Figure 1A). As expected, Northern blot analysis revealed a C/EBPα band that was readily detected in the FL-1(+/−) RNA but not in the FL-8(−/−) RNA (not shown). Both these cultured cell lines are multiclonal since the proviral-specific neoprobe, when hybridized to genomic DNA, identifies multiple retrovirus integration sites in the FL-1 and FL-8 cell lines (Figure 1B).
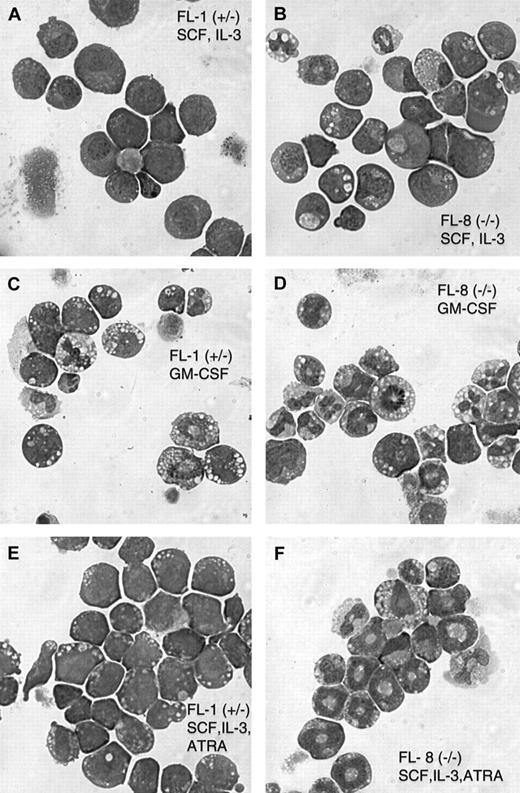
Both the FL-1(+/−) and FL-8(−/−) cell lines exhibit optimal proliferation in the combination of SCF and IL-3, with a doubling time of approximately 18 hours. Morphologically, the cell lines from both the C/EBPα(+/−) and (−/−) mice exhibit a mixture of immature blasts and more mature promyelocytes and maturing granulocytes (Table1 and Figure2A-B). Fluorescence-activated cell sorter (FACS) analysis indicates that both the FL-1 (C/EBPα(+/−)) and FL-8 (C/EBPα(−/−)) fetal liver cell lines harbor cells positive for both the B220 lymphoid antigen and the Gr-1 granulocyte marker (Table 2), indicating the mixed lympho-myeloid character of these cell lines. Unexpectedly, there was an increased number of maturing granulocytes in the SCF, IL-3-dependent FL-8(−/−) cell lines compared with the SCF, IL-3 dependent FL-1(+/−) cell lines as assessed by both morphology (Table 1) and Gr-1/CD11b surface antigen expression (Table 2, Figure3A,D).
Morphology of fetal liver cell lines under different culture conditions
| Cell types and conditions . | Blasts . | Promyelocytes . | Immature neutrophils . | Segmented neutrophils . |
|---|---|---|---|---|
| FL-1(+/−) SCF, IL-3 | 68 | 27 | 3 | 2 |
| FL-1(+/−) GM-CSF | 18 | 32 | 21 | 29 |
| FL-1(+/−) SCF, IL-3 + ATRA | 53 | 39 | 6 | 2 |
| FL-8(−/−) SCF, IL-3 | 43 | 33 | 13 | 11 |
| FL-8(−/−) GM-CSF | 15 | 22 | 25 | 38 |
| FL-8(−/−) SCF, IL-3 + ATRA | 29 | 20 | 30 | 21 |
| Cell types and conditions . | Blasts . | Promyelocytes . | Immature neutrophils . | Segmented neutrophils . |
|---|---|---|---|---|
| FL-1(+/−) SCF, IL-3 | 68 | 27 | 3 | 2 |
| FL-1(+/−) GM-CSF | 18 | 32 | 21 | 29 |
| FL-1(+/−) SCF, IL-3 + ATRA | 53 | 39 | 6 | 2 |
| FL-8(−/−) SCF, IL-3 | 43 | 33 | 13 | 11 |
| FL-8(−/−) GM-CSF | 15 | 22 | 25 | 38 |
| FL-8(−/−) SCF, IL-3 + ATRA | 29 | 20 | 30 | 21 |
Cytospins of the fetal liver cell lines cultured with the indicated growth factors were stained with Wright-Giemsa. Numbers are the percentages of cells of the indicated morphology with a minimum of 300 cells assessed for each culture. For GM-CSF induction, cells continuously growing in SCF and IL-3 were washed, resuspended in GM-CSF, and cultured for an additional 4 days. ATRA induction (10 μM) was for 5 days.
Immature neutrophils include myelocytes, metamyelocytes, and bands (“doughnut” nuclei).
Morphology of the cultured fetal liver cell lines derived from C/EBPα(+/−) and C/EBPα(−/−) mice.
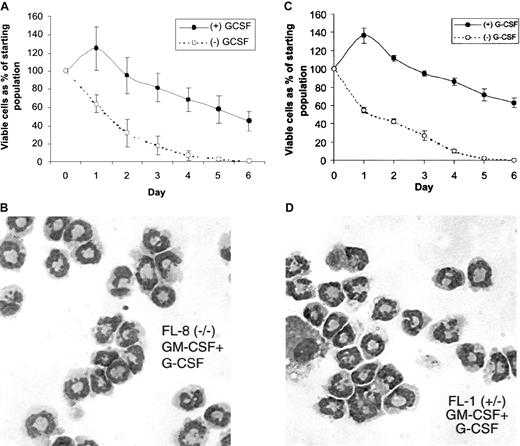
Cytospin preparations of the FL-1(+/−) and FL-8(−/−) cell lines cultured in the indicated growth factors were stained with Wright-Giemsa. Parental SCF + IL-3 cultured FL-1 and FL-8 (A,B). For GM-CSF stimulation (C, D), cells growing in SCF and IL-3 were washed and resuspended in GM-CSF alone and were cultured for 4 additional days. ATRA stimulation (10 μM) was for 5 days (E, F).
Morphology of the cultured fetal liver cell lines derived from C/EBPα(+/−) and C/EBPα(−/−) mice.
Cytospin preparations of the FL-1(+/−) and FL-8(−/−) cell lines cultured in the indicated growth factors were stained with Wright-Giemsa. Parental SCF + IL-3 cultured FL-1 and FL-8 (A,B). For GM-CSF stimulation (C, D), cells growing in SCF and IL-3 were washed and resuspended in GM-CSF alone and were cultured for 4 additional days. ATRA stimulation (10 μM) was for 5 days (E, F).
Surface phenotypes of fetal liver cell lines under different culture conditions
| Cell types and conditions . | Percentage of cells positive . | |||
|---|---|---|---|---|
| B220 . | Gr-1 . | CD11b . | F4/80 . | |
| FL-1(+/−) SCF, IL-3 | 95 ± 2 | 30 ± 10 | 5 ± 1 | 2 ± 1 |
| FL-1(+/−) GM-CSF | 63 ± 3 | 67 ± 7 | 51 ± 5 | 3 ± 1 |
| FL-1(+/−) SCF, IL-3 + ATRA | 94 ± 2 | 34 ± 6 | 4 ± 2 | ND |
| FL-8(−/−) SCF, IL-3 | 37 ± 2 | 35 ± 3 | 45 ± 3 | 7 ± 2 |
| FL-8(−/−) GM-CSF | 11 ± 3 | 92 ± 2 | 91 ± 3 | 2 ± 1 |
| FL-8(−/−) SCF, IL-3 + ATRA | 35 ± 1 | 47 ± 5 | 65 ± 3 | ND |
| Cell types and conditions . | Percentage of cells positive . | |||
|---|---|---|---|---|
| B220 . | Gr-1 . | CD11b . | F4/80 . | |
| FL-1(+/−) SCF, IL-3 | 95 ± 2 | 30 ± 10 | 5 ± 1 | 2 ± 1 |
| FL-1(+/−) GM-CSF | 63 ± 3 | 67 ± 7 | 51 ± 5 | 3 ± 1 |
| FL-1(+/−) SCF, IL-3 + ATRA | 94 ± 2 | 34 ± 6 | 4 ± 2 | ND |
| FL-8(−/−) SCF, IL-3 | 37 ± 2 | 35 ± 3 | 45 ± 3 | 7 ± 2 |
| FL-8(−/−) GM-CSF | 11 ± 3 | 92 ± 2 | 91 ± 3 | 2 ± 1 |
| FL-8(−/−) SCF, IL-3 + ATRA | 35 ± 1 | 47 ± 5 | 65 ± 3 | ND |
Fetal liver cell lines cultured under the indicated conditions were harvested, incubated with phycoerythrin or fluorescein isothiocyanate–conjugated antibodies to the indicated antigens, and subjected to FACS analysis. For GM-CSF induction, cells continuously growing in SCF and IL-3 were washed, resuspended in GM-CSF alone, and cultured for an additional 4 days. ATRA induction (10 μM) was for 5 days.
ND indicates not determined.
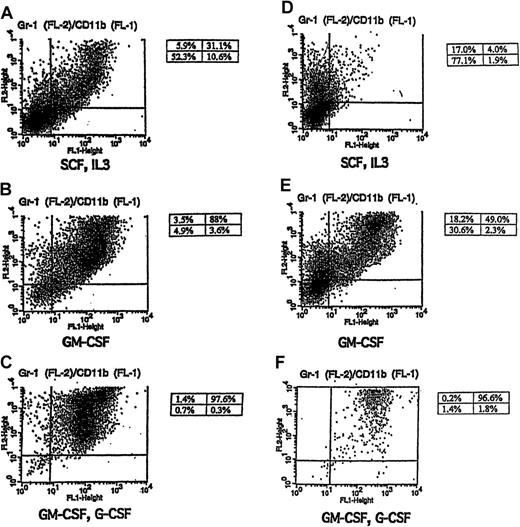
Gr-1 and CD11b expression in cytokine-stimulated FL-8(−/−) and FL-1(+/−) cells.
Shown are FACS-generated histograms of Gr-1 and CD11b expression in FL-8(−/−) (A-C) and FL-1(+/−) (D-F) cells cultured in the indicated cytokines including (A, D) SCF plus IL-3, (B, E) GM-CSF alone for 4 days, and (C, F) GM-CSF for 2 days, followed by G-CSF (5 ng/mL) for 4 days.
Gr-1 and CD11b expression in cytokine-stimulated FL-8(−/−) and FL-1(+/−) cells.
Shown are FACS-generated histograms of Gr-1 and CD11b expression in FL-8(−/−) (A-C) and FL-1(+/−) (D-F) cells cultured in the indicated cytokines including (A, D) SCF plus IL-3, (B, E) GM-CSF alone for 4 days, and (C, F) GM-CSF for 2 days, followed by G-CSF (5 ng/mL) for 4 days.
GM-CSF induced granulocytic differentiation of the C/EBPα(+/−) and (−/−) cell lines
We wanted to determine whether the spontaneous granulocytic differentiation noted in the fetal liver cell lines could be enhanced with different hematopoietic growth factors. Indeed, we observed that when the cultured cells were switched from the SCF, IL-3 combination to GM-CSF alone, there was a rapid (3- to 5-day) induction of terminal granulocytic differentiation in the cultures. This was indicated by morphologic granulocytic differentiation in the GM-CSF–treated cultures (Table 1, Figure 2C-D) and by the acquisition of surface membrane antigens characteristic of more mature granulocytes (Gr-1+/CD11b+) (Table 2, Figure 3B,E). This brisk granulocytic differentiation was noted in both the FL-1(+/−) and the FL-8(−/−) cell lines, and, again, the granulocytic differentiation was more pronounced in the C/EBPα(−/−) cells.
GCSF receptor mRNA expression in the C/EBPα(+/−) and (−/−) cell lines
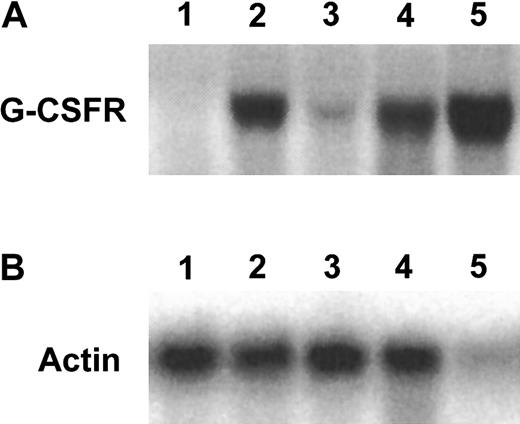
C/EBPα binds to the promoter of the G-CSF gene and enhances its expression.7 Previous studies had indicated a reduction in G-CSF receptor mRNA levels in the fetal livers of the C/EBPα(−/−) mice compared with the C/EBPα(+/−) mice, suggesting that the in vivo block to granulocytic differentiation displayed by these C/EBPα-deficient animals may be directly related to this G-CSF receptor down-regulation.15 Therefore, we were interested in determining whether the FL-8(−/−) cell line also displayed down-regulation of C/EBPα target genes such as the G-CSF receptor. We used Northern blot hybridization to compare levels of expression of the G-CSF receptor mRNA in the fetal liver cell lines derived from the C/EBPα(+/−) compared with the C/EBPα(−/−) mice. As a positive control, we used mRNA derived from the IL-3–dependent 32D cells, which undergo granulocytic differentiation in response to G-CSF. We noted a slight increase in G-CSF receptor mRNA in the FL-8 (C/EBPα(−/−)) cell line compared with the FL-1 (C/EBPα(+/−)) cells when both were cultured in SCF and IL-3 (Figure 4; compare lanes 1 and 3). Of note, the GM-CSF–induced granulocytic differentiation of these cells resulted in a significant up-regulation of the G-CSF receptor mRNA levels in both the C/EBPα(+/−) and the (−/−) cell lines (Figure 4). Our observation with the C/EBPα(−/−) cells indicates that GM-CSF can induce enhanced G-CSF receptor mRNA expression in the absence of C/EBPα.
GM-CSF–mediated up-regulation of G-CSF receptor RNA in the fetal liver cell lines.
Total RNA extracted from the indicated cell types were subjected to Northern blot hybridization (8 μg/lane) with the indicated probes. Lanes 1 to 5 harbor RNAs extracted from (1) FL-1(+/−) cultured in SCF and IL-3, (2) FL-1(+/−) cultured for 4 days in GM-CSF alone, (3) FL-8(−/−) cultured in SCF and IL-3, (4) FL-8(−/−) cultured in GM-CSF alone for 4 days, and (5) 32D cells cultured in IL-3 alone.
GM-CSF–mediated up-regulation of G-CSF receptor RNA in the fetal liver cell lines.
Total RNA extracted from the indicated cell types were subjected to Northern blot hybridization (8 μg/lane) with the indicated probes. Lanes 1 to 5 harbor RNAs extracted from (1) FL-1(+/−) cultured in SCF and IL-3, (2) FL-1(+/−) cultured for 4 days in GM-CSF alone, (3) FL-8(−/−) cultured in SCF and IL-3, (4) FL-8(−/−) cultured in GM-CSF alone for 4 days, and (5) 32D cells cultured in IL-3 alone.
Functional activity of the up-regulated G-CSF receptors
To determine whether this GM-CSF–mediated up-regulation of G-CSF receptor mRNA observed in the C/EBPα(−/−) cell lines was associated with the acquisition of functional G-CSF receptors, we treated the SCF, IL-3–dependent, FL-8(−/−) cells, and control FL-1(+/−) cells with GM-CSF alone for 2 days to induce G-CSF mRNA up-regulation, then washed and further cultured the cells in either no growth factor or G-CSF alone. Both the GM-CSF–induced FL-8(−/−) and the FL-1(+/−) cells rapidly died in the absence of any added growth factor (Figure5A,C). In contrast, survival was significantly prolonged in the FL-8(−/−) and the FL-1(+/−) cultures incubated with G-CSF (Figure 5A,C), and both these cells morphologically differentiated to mature granulocytes as evidenced by morphology (Figure 5B,D) and surface antigen (Gr-1, Cd11b) expression (Figure 3C,F). Thus, G-CSF exerts considerable influence on both the survival and differentiation of the GM-CSF–induced FL-8(−/−) cells, indicating that these GM-CSF–treated FL-8(−/−) cells, which exhibit enhanced expression of G-CSF receptor mRNA (Figure 4), express functionally active G-CSF receptors.
Response of the fetal liver cell lines to G-CSF.
Cells from the fetal liver cell lines continuously growing in SCF and IL-3 were washed and resuspended for 2 days in media containing GM-CSF alone to induce G-CSF receptor mRNA expression (Figure 4). These cells were then washed and resuspended (105/mL) in culture media in the presence or absence of G-CSF (5 ng/mL). At the indicated time after culture initiation, the viable cell number was determined using trypan blue dye exclusion for the (A) FL-8(−/−) and (C) FL-1(+/−) cells. Wright-Giemsa stains of cytospin preps are shown for the (B) FL-8(−/−) and (D) FL-1(+/−) cells that were cultured in GM-CSF alone for 2 days, followed by G-CSF (5 ng/mL) alone for 4 additional days.
Response of the fetal liver cell lines to G-CSF.
Cells from the fetal liver cell lines continuously growing in SCF and IL-3 were washed and resuspended for 2 days in media containing GM-CSF alone to induce G-CSF receptor mRNA expression (Figure 4). These cells were then washed and resuspended (105/mL) in culture media in the presence or absence of G-CSF (5 ng/mL). At the indicated time after culture initiation, the viable cell number was determined using trypan blue dye exclusion for the (A) FL-8(−/−) and (C) FL-1(+/−) cells. Wright-Giemsa stains of cytospin preps are shown for the (B) FL-8(−/−) and (D) FL-1(+/−) cells that were cultured in GM-CSF alone for 2 days, followed by G-CSF (5 ng/mL) alone for 4 additional days.
Response of the FL-1 and FL-8 cell lines to ATRA
We also tested the effect of ATRA on inducing granulocytic differentiation of these cells. We had previously observed that in GM-CSF–dependent cell lines expressing the dominant-negative RARα construct, high pharmacologic concentrations (1-10 μM) of ATRA would overcome the effect of the dominant-negative RARα and induce terminal granulocytic differentiation of these cells.16Surprisingly, we observed that ATRA (10 μM), when added to the SCF, IL-3–dependent cell lines, enhanced the granulocytic differentiation of the FL-8(−/−) cells while inducing little change in the FL-1(+/−) cells (Tables 1 and 2; Figure 2E-F).
Discussion
Studies involving in vitro cultured cell lines and knockout mice have implicated a number of different transcription factors as important regulators of granulopoiesis, including PU.1, acute myelogenous leukemia 1 (AML-1), Myb, C/EBPα, and retinoic acid receptors.25 Of these, C/EBPα appears particularly important given that C/EBPα(−/−) knockout mice display virtually absent granulopoiesis.13 To more thoroughly dissect the nature and characteristics of this block to differentiation displayed by the C/EBPα(−/−) hematopoietic cells, we used dominant-negative RAR retroviral constructs to establish hematopoietic growth factor–dependent cell lines from the fetal livers of the C/EBPα(−/−) knockout embryos. Unexpectedly, we observed that the SCF and IL-3–dependent lympho-myeloid precursors that we established from these C/EBPα(−/−) animals underwent brisk granulocytic differentiation in liquid suspension culture in response to a number of different physiologic factors, including GM-CSF, G-CSF, and ATRA. Indeed, the granulocytic differentiation observed in the cell lines from the fetal livers of the C/EBPα(−/−) mice was even more prominent than the differentiation observed in the cell lines derived from their heterozygous (C/EBPα(+/−)) littermates. Thus, the C/EBPα(−/−) knockout mice exhibit a profound block in granulocytic differentiation in vivo,13 and yet lympho-myeloid cell lines reproducibly derived from the hematopoietic cells of these mice readily differentiate in vitro after induction with physiologic agents.
How can we explain this marked discrepancy between the in vivo and in vitro behavior of these C/EBPα(−/−) granulocyte precursors? One possibility is that the granulocyte maturation defect in the C/EBPα(−/−) animals lies not in the granulocytic precursors themselves but in an accessory cell that might be involved in supporting or inhibiting granulopoiesis in vivo and whose function may be altered in the C/EBPα(−/−) animals. However, this model is unlikely because it has been previously demonstrated that fetal liver cells from the C/EBPα(−/−) mice, when transplanted into a congenic strain of C/EBPα(+/+) mice, also exhibit defective granulopoiesis indicating that the defect is intrinsic to the granulocyte precursors.13 Another possibility is that a mutation(s) arose in the cultured cells from the C/EBPα(−/−) mice that somehow accelerates granulopoiesis. This also seems unlikely because these cell lines were established by selecting for cells that actively proliferate rather than differentiate. Moreover, the cell lines exhibit multiple proviral integration sites, indicating that they are multicellular rather than clonal in origin (Figure 1B) and making it improbable that cells harboring rare mutations have evolved in these cultures.
It appears that in the derivation of these cell lines, our selective culture conditions expanded SCF/IL-3–responsive granulocyte precursors from the C/EBPα(−/−) animals that have the clear potential to generate mature granulocytes but for unclear reasons undergo limited differentiation in vivo. Indeed a previous study indicated the presence in C/EBPα(−/−) mice of IL-3–responsive precursors that underwent distinct granulocyte differentiation after stimulation by IL-3/GM-CSF in semisolid medium.15 The SCF/IL-3–dependent cell lines we have derived may be the functional equivalent of such previously observed IL-3–responsive granulocyte precursors. However, the IL-3–responsive cells previously described in the C/EBPα(−/−) mice exhibited limited G-CSF receptor expression.15 In contrast in our own SCF/IL-3–dependent C/EBPα(−/−) cell lines, G-CSF receptor mRNA was not only readily detected but was also significantly up-regulated as the cells were induced to differentiate with GM-CSF (Figure 4). Thus, although C/EBPα binds to and activates the G-CSF receptor promoter,7 our observations indicate that the GM-CSF–mediated enhanced expression of the G-CSF receptor can occur independently of C/EBPα, and they suggest that other transcription factors are involved in the up-regulation of G-CSF receptor expression in the C/EBPα(−/−) cell lines.
Retinoic acid receptors, particularly RARα, are important regulators of granulopoiesis. Retinoic acid is a clinically useful compound that induces terminal granulocyte differentiation of most acute promyelocytic leukemia cells, and mice genetically deficient in both RARα and RARγ display an in vitro block in granulocyte differentiation.26 The immediate downstream targets of the activated RARs that trigger this differentiation are still uncertain. However, our observation that ATRA enhances the granulocytic differentiation of the IL-3–dependent C/EBPα(−/−) hematopoietic cells (Tables 1, 2, Figure 2F) suggests that RAR can trigger granulocytic differentiation using a molecular pathway that is independent of C/EBPα.
Our observations strongly support a previously proposed model, suggesting that there may be multiple molecular pathways involved in granulocyte differentiation, some that are C/EBPα dependent and others are C/EBPα independent.15 Indeed, in certain in vitro–cultured myeloid precursors, enhanced C/EBPα expression promotes granulocytic differentiation,6,10 11 yet our own observations indicate that granulocytic differentiation can be triggered in the complete absence of C/EBPα. Thus, in some myeloid precursors, C/EBPα is “sufficient” to trigger granulocyte differentiation, whereas in other precursors C/EBPα is clearly not “necessary” to induce such differentiation.
Our observations suggesting that multiple independent molecular pathways may stimulate granulocyte differentiation indicate that the regulation of granulopoiesis involves a striking degree of redundancy and robustness. Perhaps this is not surprising given the critical need for animals to generate a brisk neutrophilia in response to a variety of different endogenous and environmental microorganisms. That is, immature hematopoietic precursors may use distinctly different surface membrane receptors and cytokine–growth factor combinations to enhance granulocyte differentiation and production. Such robustness in neutrophil generation has been recently demonstrated at the membrane receptor level in G-CSF knockout mice. G-CSF is an important regulator of granulopoiesis, yet G-CSF knockout mice generate a brisk neutrophilia in response to Candida albicansinfection.27 This indicates that the immature hematopoietic precursors in these animals can use membrane receptors other than G-CSF receptors to generate viable, mature granulocytes. Our own observations suggest that hematopoietic precursors may use not only different membrane receptors but also different combinations of transcription factors to generate mature granulocytes.
We thank LeMoyne Mueller and Jutta Fero for excellent technical assistance and Alan Friedman for supplying the 32D cells.
Supported by National Institutes of Health grant CA58292 (S.J.C.). L.E.P. is a Special Fellow of the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Steven J. Collins, Human Biology Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: scollins@fhcrc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal