Abstract
Hepatitis C virus (HCV) binds to platelets in chronically infected patients where free HCV constitutes only about 5% of total circulating virus. Free HCV preferentially binds to human mononuclear cell lines but free and complexed virus binds equally to platelets. The extent of free HCV binding to human Molt-4 T cells (which express CD81) and to human promonocytic U937 cells or to platelets (which do not express CD81) was similar. The binding of free HCV to the cell lines was saturated at a virus dose of 1 IU HCV RNA per cell but binding to platelets was not saturable. Human anti-HCV IgG, but not anti-CD81, markedly inhibited HCV binding to target cells in a dose-dependent manner. Human antibodies to HCV hypervariable region 1 of E2 glycoprotein partially inhibited viral binding to target cells. Recombinant E2 also inhibited viral binding to target cells in a dose-dependent manner, with the efficacy of this decreasing in the rank order of Molt-4 cells more than U937 cells more than platelets. In contrast to HCV, recombinant E2 bound to Molt-4 cells to an extent markedly greater than that apparent with U937 cells or platelets. These results suggest that the binding of HCV to blood cells is mediated by multiple cell surface receptors and that recombinant E2 binding may not be representative of the interaction of the intact virus with target cells.
Introduction
Human platelets bind many viruses, including vaccinia virus, human immunodeficiency virus, Epstein-Barr virus, dengue 2 virus, influenza virus, cytomegalovirus, and hantavirus.1-12 However, the possible interaction between platelets and hepatitis C virus (HCV) has not been previously investigated. HCV, the principal causative agent of non-A, non-B hepatitis,13,14 is an enveloped virus classified in the Flaviviridae family.15,16 Most patients infected with HCV are unable to clear the virus as a result of insufficient humoral and cellular immunity,17-19 and many such individuals develop chronic liver disease. The structural proteins of HCV include a nucleocapsid subunit and 2 putative envelope glycoproteins, E1 (polyprotein residues 192-383) and E2 (residues 384-746).20-22 When expressed in cultured cells, E1 and E2 interact noncovalently to form a stable complex whose size is consistent with that of an E1-E2 heterodimer.23 24
Viral glycoproteins exposed on the surface of enveloped viruses mediate viral attachment and subsequent entry into target cells. The mechanism by which HCV enters its target cells remains unknown, and studies of infection have been hampered by the lack of an efficient and reliable cell culture system that supports the replication of this virus. The E2 glycoprotein is thought to be responsible for initiating virus attachment to a receptor on a target cell.25 The NH2-terminus of E2 contains a hypervariable region known as HVR1. Although hypervariable, this 27-amino acid region includes 2 conserved amino acids at positions 23 and 26 that are important for the antigenic and functional activity of E2.25-27 HVR1 is an important neutralization determinant.28-35 Cell surface-expressed human CD81 binds E2,36 suggesting a role for this protein as a cellular receptor for HCV. CD81 is a member of the tetraspanin family of membrane proteins that contain 4 transmembrane domains, a short intracellular domain, and 2 extracellular loops37,38 and that are widely expressed in most human tissues. The binding of E2 appears specific to human CD81,36,39 and it has been suggested that no additional human cell-specific factors are required for such binding.39
The biologic significance of the E2-CD81 interaction in HCV infection remains controversial, however. The relevance of E2-CD81 binding is suggested by the observation that sera (containing antibodies to E2) from chimpanzees immunized with HCV envelope glycoprotein are able to prevent HCV infection in vivo40and to inhibit the interaction of E2 with CD81 in vitro.36However, the E2-CD81 interaction does not correlate with species permissiveness to infection, given that tamarin (nonpermissive to HCV infection) CD81 binds E2 with greater affinity than does human CD81.41,42 The low-density lipoprotein (LDL) receptor has also been proposed to function as a receptor for HCV-LDL complexes at the target cell surface.43-45
We have now examined the putative role of human CD81 as a receptor for HCV by studying the binding of free HCV to normal human platelets as well as to CD81-expressing and nonexpressing human mononuclear cells.
Materials and methods
Cell lines
Human Molt-4 T cells and U937 promonocytic cells were grown under an atmosphere of 5% CO2 at 37°C in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS).
Platelets
Platelets were prepared from platelet-rich plasma that was obtained from prospective blood donors at the East Anglia Blood Centre. They were counted with the use of a flow cytometer (Sysmex K-1000, Milton Keynes, United Kingdom), centrifuged, and washed with platelet-washing buffer (PWB: phosphate-buffered saline [PBS, pH 6.8] containing 10 mM EDTA and 0.25% bovine serum albumin). Platelets were also prepared from whole blood of 30 HCV-seropositive and HCV RNA-positive patients and of 5 healthy controls. After washing, platelet suspensions were incubated with magnetic beads coated with antibodies to CD45 (Dynal, Oslo, Norway) to deplete residual lymphocytes.
Plasma
Plasma from an individual (X) with congenital agammaglobulinemia who was infected with HCV of genotype 1a was collected at The Bristol Blood Centre by T. Wallington. The viral load in this plasma was estimated as 1.7 × 106 IU/mL, and the virus was designated X. HCV-infected plasma from S5, S7, S8, S9, and Af were from blood donors and collected at The East Anglia Blood Centre. Patients' samples were provided by Dr G. J. M. Alexander, (Hepatology Clinic, Department of Medicine, University of Cambridge Medical School, United Kingdom).
Peptides
Peptides based on the HVR1 sequences of 2 variants of virus X (CSGLAGLFTPGAKQNI [X1] and CSGLVGLFTPGAKQNI [X2]) and on the corresponding sequence (TSGLVSLLAPGAKQN [E2 peptide]) of the prototype HCV from which recombinant E2 glycoprotein (rE2) was derived40 were synthesized by Severn Biotech (Kidderminster, United Kingdom). A cysteine residue was added to the NH2-terminus of each peptide to facilitate presentation of coated peptides. Peptides were dissolved in 4% dimethyl sulfoxide at a concentration of 2 mg/mL.
Assay of HCV binding to cells
For binding experiments, platelets (1 × 106) or Molt-4 or U937 cells (1 × 105) were washed with PBS, and the cell pellet was incubated for 1 hour at 37°C with 100 μL undiluted X plasma. The cells were then washed 4 times with 10 mL ice-cold PBS and once with 500 μL PBS, and this final wash was saved for RNA extraction. To investigate the effects of various specific antibodies (hIgG anti-HCV and HVR1-specific IgG) on the interaction of HCV with cells, we incubated 100 μL undiluted X plasma for 1 hour at 37°C with the antibodies before adding the plasma to cells. The effect of E2 on viral binding was investigated by incubating cells or platelets for 1 hour at 37°C with rE2 (genotype 1a; kindly provided by M. Houghton, Chiron, Emeryville, CA) and washing 3 times with PBS before the addition of X plasma.
Purification of human IgG specific for X peptides
Samples of plasma from HCV-infected blood donors were obtained from the East Anglia Blood Centre and tested for reactivity with a mixture of X1 and X2 peptides by enzyme-linked immunosorbent assay (ELISA). A highly reactive plasma (S5) was used for purification of total IgG with a HiTrap protein G column (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). To isolate IgG specific for X peptides, we subjected the total IgG fraction to further immunopurification with X peptide-conjugated thiopropyl Sepharose 6B (Amersham Pharmacia Biotech). The X peptides (1 mg) were cross-linked to the gel through the added NH2-terminal cysteine residues. Total IgG (25 mg) in 5 mL binding buffer (PBS containing 10 mM EDTA) was applied to the affinity column in a closed circulating system for 4 hours. The nonretained material was saved for testing, and the column was washed and then subjected to elution with 0.1 M glycine-HCl (pH 2.5). The eluate was neutralized with 1 M Tris-HCl (pH 9.0) and then assayed together with the nonbound fraction, for protein and reactivity to X peptides with the BCA reagent (Pierce, Tattenhall, United Kingdom) and ELISA, respectively.
Peptide ELISA
The reactivity of human IgG with X or E2 peptides was determined by ELISA as previously described.29
Antibodies
A mouse monoclonal antibody (mAb) to E2 (clone 291A2, IgG1) was kindly provided by S. Abrignani (IRIS Research Center, Siena, Italy). Fluorescein isothiocyanate (FITC)–conjugated rabbit F(ab′)2 to mouse IgG was obtained from Dako (Ely, United Kingdom). Human IgG specific for rE2 and X peptides were purified from the plasma of HCV-infected patients S8 and S5, respectively, with a HiTrap protein A column (Amersham Pharmacia Biotech). Normal human IgG and normal mouse IgG1-κ were from Sigma (Gillingham, United Kingdom), and a mouse mAb (IgG1-κ) to human CD81, a phycoerythrin (PE)–conjugated mouse mAb (IgG1-κ) to human CD81, and an FITC-conjugated mouse mAb (IgG1-κ) to human CD9 were obtained from Pharmingen (Oxford, United Kingdom).
Flow cytometry
The interaction between rE2 and platelets or cultured cells was analyzed by flow cytometry. In brief, Molt-4 and U937 cells were washed twice in PBS containing 1% FBS and 0.05% NaN3 (cell washing buffer [CWB]), whereas platelets were washed twice in PWB. Cells (2 × 105 in 0.25 mL CWB) or platelets (1 × 106 in 0.1 mL PWB) were incubated with rE2 (1.4 μg) for 1 hour at room temperature. The unbound rE2 was removed by washing the cultured cells or platelets (centrifugation at 900 or 3400 rpm, respectively, for 5 minutes) 3 times with the corresponding wash buffer. Bound rE2 was then detected by incubation of the cultured cells or platelets in a final volume of 200 μL wash buffer for 1 hour at room temperature first with a mouse mAb to E2 (5 μg/mL) and then with FITC-conjugated rabbit F(ab′)2 to mouse IgG (5 μg/mL); unbound antibodies from each incubation were removed with 3 washes. Normal mouse IgG1-κ was used as negative control for rE2 and the mAb to E2, respectively. The expression of CD81 on cultured cells and platelets was evaluated by flow cytometry with a PE-conjugated mouse mAb to human CD81 and an FITC-conjugated mouse mAb to human CD9 as positive control, respectively. Finally, labeled targets were resuspended in 0.5 mL wash buffer and subjected to flow cytometry. Median fluorescence intensity was determined using a Beckman Coulter XL flow cytometer running System II software (Beckman Coulter, High Wycombe, United Kingdom). The effect of antibodies to HCV E2 on the binding of rE2 to target cells was examined by incubating human IgG specific for HCV E2 with rE2 for 1 hour at 37°C in 50 μL PBS containing 1% fetal bovine serum before addition of the mixture to platelets or cultured cells as described above.
Detection of CD81 messenger RNA
Total RNA was extracted from Molt-4 or U937 cells (RNeasy Mini Kit, Qiagen, Crawley, United Kingdom) and subjected to reverse transcription (RT) followed by the polymerase chain reaction (PCR) with Amplitaq DNA polymerase (PerkinElmer, Warrington, United Kingdom) and primers specific for human CD81 messenger RNA (mRNA) (forward, 5′-CCAAGTGCATCAAGTACCTGCTC-3′; reverse, 5′-CACCTCACAGGCAAACAGGATG-3′). A plasmid encoding human CD81 (kindly provided by S. Levy, Stanford University Medical Center, Palo Alto, CA) was used as a positive control for PCR.
Real-time quantitative RT-PCR analysis of HCV RNA and CD45 mRNA
Total RNA was extracted from cells with an RNeasy Mini Kit (Qiagen) or from plasma and wash buffer with a Qiaamp Viral RNA Mini Kit (Qiagen). HCV RNA and CD45 mRNA were measured by real-time quantitative RT-PCR with a PE Applied Biosystems Prism model 7700 sequence detection instrument (ABI, Warrington, United Kingdom). The forward and reverse primers for the noncoding region of HCV RNA were 5′-TCTGCGGAACCGGTGAGTA-3′ and 3′-CGGGTTGATCCAAGAAAGGA-5′, respectively; those for CD45 mRNA were 5′-GGAAGTGCTGCAATGTGTCATT-3′ and 3′-TGACATCAGATAATAGTATGCATGTCAAG-5′, respectively. The TaqMan fluorogenic probes used for quantitation of HCV RNA and CD45 mRNA were 5′-6FAM-CCGGAATTGCCAGGACGACCG-3′ and 5′-6FAM-ACAACTAAAAGTGCTCCTCCAAGCCAGGTCT-3′, respectively. The probe contains a fluorescent reporter 6-carboxyfluorescein (FAM) at the 5′ end and a fluorescent quencher 6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The Ct value, which correlates inversely with the concentration of target RNA, was determined as the cycle number at which the fluorescence emission of the reporter probe increases above a threshold level. As an internal control, 18S ribosomal RNA (rRNA) was simultaneously amplified and quantitated in the same reaction wells with the use of a kit (PerkinElmer). Each sample was analyzed in triplicate and the results were averaged.
Results
Binding of HCV to human platelets in vivo
To determine whether HCV is bound to platelets in vivo, we isolated these cells by centrifugation from whole blood of 30 HCV-seropositive and HCV RNA-positive patients and 5 healthy controls. After washing, the platelet suspension was incubated with magnetic beads coated with antibodies to CD45 to deplete residual lymphocytes. Total RNA was then extracted from patient and control plasma and platelets; the buffer was used for the final wash of platelets and was analyzed for the presence of HCV RNA by quantitative real-time RT-PCR. The absence of lymphocytes in the platelet preparations was confirmed by the failure of RT-PCR analysis to detect CD45 mRNA in the total RNA fractions isolated from platelets (data not shown). Whereas HCV RNA was not detected in the wash buffer of any of the platelet preparations or in plasma or platelets from control individuals, it was detected in plasma and platelets from all HCV-infected patients (data not shown). These observations thus indicate that HCV is bound to platelets in chronically infected individuals.
Effect of immunoglobulin depletion on the binding of HCV in plasma to human cell lines and platelets in vitro
HCV particles appear to circulate as complexes with lipoproteins46-49 or with immunoglobulin.49-52We therefore investigated the ability of human platelets and cell lines to bind native and free HCV present in plasma. Plasma samples from 2 HCV-infected individuals (S7 and Af) were selected as representative of medium and high viral load (1.7 × 105 and 2.7 × 107 IU/mL, respectively). The plasma samples were depleted of immune complexes by passage through a protein G column; the absence of IgG in the column flow-through fraction was confirmed with a sandwich ELISA (data not shown). RNA was extracted from the plasma samples before and after chromatography for determination of viral load. The viral load of S7 and Af plasma samples after chromatography (6.6 × 103 and 1.4 × 106 IU/mL, respectively) revealed that the proportion of free HCV in the original specimens was 3.9% and 5.2% of total virus, respectively.
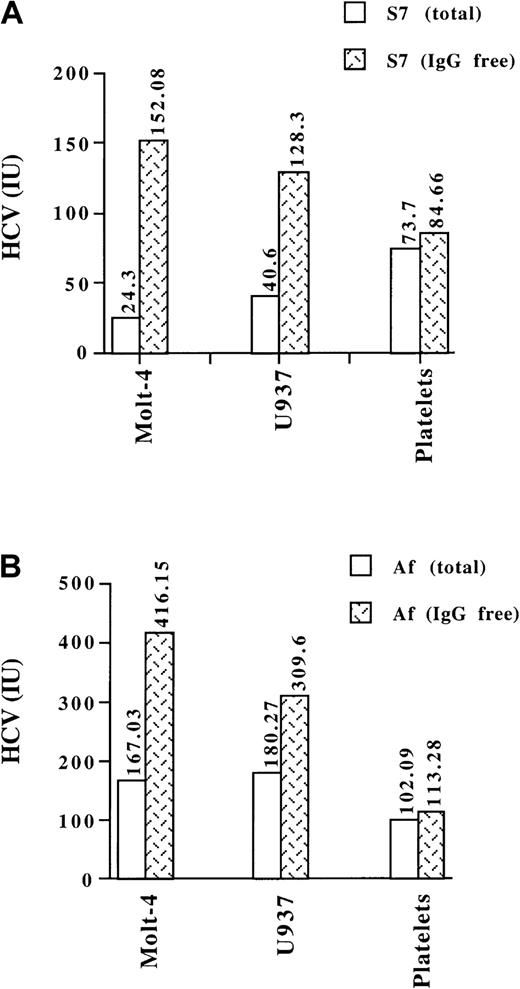
The ability of normal human platelets as well as of Molt-4 and U937 cells to bind HCV in S7 and Af plasma before and after depletion of immune complexes was determined by quantitative real-time RT-PCR analysis. Incubation of equivalent concentrations of native or free virus with cultured cells revealed that the extent of binding observed with free virus was 1.7 to 6 times that apparent with native virus (Figure 1), suggesting that Molt-4 and U937 cells preferentially interact with uncomplexed HCV. In contrast, with human platelets, the extent of binding of free virus was similar to that observed for native virus.
Binding of total and free HCV to human cell lines and platelets.
Molt-4 or U937 cells (1 × 105) or platelets (1 × 106) were incubated with untreated or IgG-depleted plasma samples from HCV-infected patients S7 (A) or Af (B). The amount of HCV added to the incubations was adjusted to 5.7 × 103 and 6.7 × 104 IU for S7 and Af samples, respectively. The extent of viral binding to the target cells was determined by quantitative real-time RT-PCR analysis. The exact amounts of HCV (IU) bound are shown at the top of each column. Data are means of triplicates from a representative experiment.
Binding of total and free HCV to human cell lines and platelets.
Molt-4 or U937 cells (1 × 105) or platelets (1 × 106) were incubated with untreated or IgG-depleted plasma samples from HCV-infected patients S7 (A) or Af (B). The amount of HCV added to the incubations was adjusted to 5.7 × 103 and 6.7 × 104 IU for S7 and Af samples, respectively. The extent of viral binding to the target cells was determined by quantitative real-time RT-PCR analysis. The exact amounts of HCV (IU) bound are shown at the top of each column. Data are means of triplicates from a representative experiment.
Role of CD81 in the binding of HCV to human platelets or cultured cells
For further investigation of the interaction of HCV with platelets and cell lines, plasma was obtained from an individual (X) with congenital agammaglobulinemia who was infected with HCV of genotype 1a. Given that viral infectivity is related to the concentration of free virus,49-52 we chose to use a model of HCV binding in which the virus is present as IgG-free (uncomplexed) particles. Total RNA was extracted from X plasma and the viral load was quantified by real-time RT-PCR. A standard curve was constructed with serial dilutions of viral RNA extracted from a calibrated reference plasma (National Institute for Biological Standards and Control). The average Ct values were plotted against the logarithm of the amount of input genome (expressed in IUs). The viral load of X plasma was thus determined to be 1.7 × 106 IU/mL.
Given that CD81 has been described as a cell surface receptor for HCV and that this protein is not expressed on the platelet surface,37 we investigated the binding of free HCV to cells expressing (Molt-4 T cells) or not expressing (U937 promonocytic cells and platelets) CD81. We first verified the CD81 phenotypes of the chosen cell types. The various cells were labeled with a PE-conjugated mAb to human CD81 and subjected to flow cytometric analysis. The results confirmed that CD81 was expressed on the surface of Molt-4 cells and that it was not detected on either U937 cells or platelets (Figure 2A). Expression of the CD81 gene in U937 and Molt-4 cells was also examined by RT-PCR analysis of total RNA. Again, CD81 mRNA was detected in Molt-4 cells but not in U937 cells (Figure 2B).
Expression of CD81 in human cell lines and platelets.
(A) Flow cytometric analysis of CD81 expression by Molt-4 cells, U937 cells, and human platelets. Fluorescence profiles are shown as open for cells labeled with a mAb to CD81 and as shaded for cells labeled with an IgG isotype control mAb. Results are representative of 3 to 5 separate experiments. (B) RT-PCR analysis of CD81 mRNA in Molt-4 (lane 1) and U937 (lane 2) cells. Total RNA from the 2 cell types was also analyzed with primers specific for β-actin mRNA as an internal control; given the similar molecular sizes of the CD81 and β-actin amplicons, 2 separate amplifications were performed with the same cDNA sample. Lanes labeled −ve and +ve represent reaction mixtures in which total RNA was omitted or replaced with a plasmid encoding human CD81, respectively. The leftmost and rightmost lanes contain molecular size markers.
Expression of CD81 in human cell lines and platelets.
(A) Flow cytometric analysis of CD81 expression by Molt-4 cells, U937 cells, and human platelets. Fluorescence profiles are shown as open for cells labeled with a mAb to CD81 and as shaded for cells labeled with an IgG isotype control mAb. Results are representative of 3 to 5 separate experiments. (B) RT-PCR analysis of CD81 mRNA in Molt-4 (lane 1) and U937 (lane 2) cells. Total RNA from the 2 cell types was also analyzed with primers specific for β-actin mRNA as an internal control; given the similar molecular sizes of the CD81 and β-actin amplicons, 2 separate amplifications were performed with the same cDNA sample. Lanes labeled −ve and +ve represent reaction mixtures in which total RNA was omitted or replaced with a plasmid encoding human CD81, respectively. The leftmost and rightmost lanes contain molecular size markers.
The ability of normal human platelets as well as Molt-4 and U937 cells to bind HCV in X plasma was quantitatively examined by real-time RT-PCR. Platelets (1 × 106) or cultured cells (1 × 105) were incubated with either PBS or X plasma (1.7 × 105 IU HCV). The amount of HCV RNA in total RNA subsequently extracted from the cells and from the buffer used for the final wash of cells was then determined with specific primers and a fluorogenic 6FAM-labeled probe. As an internal control, 18S rRNA was simultaneously quantified in the same reaction wells with a corresponding pair of primers and a probe labeled with a different fluorogenic group (VIC). The CD81-expressing (Molt-4) and nonexpressing (U937) cell lines and normal human platelets all bound HCV in X plasma to similar extents, whereas washing buffer and cells incubated with PBS were negative for HCV RNA (Figure 3A). The amount of cell-bound virus ranged between 0.6% and 1.1% of the total HCV in the incubation mixture (Figure 3B). The amount of HCV bound per cell was 0.019 (1 of 53), 0.017 (1 of 59), and 0.001 (1 of 919) IU for Molt-4 cells, U937 cells, and platelets, respectively.
Quantitative real-time RT-PCR analysis of the binding of HCV in X plasma to human platelets and cell lines.
Human platelets (1 × 106) or Molt-4 or U937 cells (1 × 105) were incubated with 100 μL PBS or of X plasma (1.7 × 105 IU HCV). The amounts of HCV RNA and 18S rRNA (internal control) were subsequently determined in total RNA extracted from cells or from the buffer used for the final wash of cells. Data are presented as Ct values (aCt value of 40 indicates no amplification; decreasing Ct values indicate increasing concentrations of amplicon) in panel A and as absolute total amounts of HCV RNA (IU) in panel B, and are means ± SD of 5 separate experiments.
Quantitative real-time RT-PCR analysis of the binding of HCV in X plasma to human platelets and cell lines.
Human platelets (1 × 106) or Molt-4 or U937 cells (1 × 105) were incubated with 100 μL PBS or of X plasma (1.7 × 105 IU HCV). The amounts of HCV RNA and 18S rRNA (internal control) were subsequently determined in total RNA extracted from cells or from the buffer used for the final wash of cells. Data are presented as Ct values (aCt value of 40 indicates no amplification; decreasing Ct values indicate increasing concentrations of amplicon) in panel A and as absolute total amounts of HCV RNA (IU) in panel B, and are means ± SD of 5 separate experiments.
The specificity of viral binding was evaluated by construction of dose-response curves for HCV binding (1.7 × 104-3.4 × 105 IU X virus) with a constant number of cells. The binding of HCV to both Molt-4 and U937 cells increased with increasing viral concentration and achieved saturation at approximately 100 000 IU/1 × 105 cells (Figure4).
Saturation analysis of HCV binding to human cell lines and platelets.
Molt-4 (A) or U937 (B) cells (1 × 105) or platelets (1 × 106) (C) were incubated with PBS or various amounts of X plasma (1.7 × 104-3.4 × 105 IU HCV), after which the abundance of cell-associated HCV RNA and 18S rRNA was quantitated by real-time RT-PCR as described in Figure 3. Data are presented as Ct values and are means ± SD from 3 independent experiments.
Saturation analysis of HCV binding to human cell lines and platelets.
Molt-4 (A) or U937 (B) cells (1 × 105) or platelets (1 × 106) (C) were incubated with PBS or various amounts of X plasma (1.7 × 104-3.4 × 105 IU HCV), after which the abundance of cell-associated HCV RNA and 18S rRNA was quantitated by real-time RT-PCR as described in Figure 3. Data are presented as Ct values and are means ± SD from 3 independent experiments.
Inhibition of X virus binding to cells by antibodies to HCV or to HVR1
We next examined the effect of human IgG specific for HCV on the binding of HCV in X plasma to human cell lines and platelets. X plasma was incubated with various concentrations of human anti-HCV IgG or of control IgG from individuals negative for HCV before its addition to Molt-4 or U937 cells or platelets. The binding of HCV to all 3 target cells was inhibited by human IgG to HCV in a dose-dependent manner (Figure 5A); the maximal extent of inhibition ranged from 27% to 42%, depending on the target cell. No inhibition of HCV binding was apparent with control human IgG.
Inhibition by antibodies to HCV and to HVR1 of the binding of HCV in X plasma to human cell lines and platelets.
(A) X plasma (1.7 × 105 IU HCV) was incubated with various concentrations of human anti-HCV IgG or control human IgG before addition to Molt-4 cells, U937 cells, or platelets. The binding of HCV to cells was then quantitated by real-time RT-PCR analysis of HCV RNA. Data are expressed as percentage inhibition of HCV binding and are means ± SD of values from 3 to 5 independent experiments. (B) IgG from an HCV-infected patient was purified on a protein G column. Anti-X HVR1 peptide was then purified by affinity chromatography. Anti-X HVR1 (eluted fraction), the corresponding nonretained IgG fraction, and control human IgG were incubated at a concentration of 5 μg/mL with X plasma prior to addition to target cells. The binding of HCV to cells was then quantified by real-time PCR analysis of HCV RNA. Data are expressed as percentage inhibition and are means ± SD of values from 5 independent experiments.
Inhibition by antibodies to HCV and to HVR1 of the binding of HCV in X plasma to human cell lines and platelets.
(A) X plasma (1.7 × 105 IU HCV) was incubated with various concentrations of human anti-HCV IgG or control human IgG before addition to Molt-4 cells, U937 cells, or platelets. The binding of HCV to cells was then quantitated by real-time RT-PCR analysis of HCV RNA. Data are expressed as percentage inhibition of HCV binding and are means ± SD of values from 3 to 5 independent experiments. (B) IgG from an HCV-infected patient was purified on a protein G column. Anti-X HVR1 peptide was then purified by affinity chromatography. Anti-X HVR1 (eluted fraction), the corresponding nonretained IgG fraction, and control human IgG were incubated at a concentration of 5 μg/mL with X plasma prior to addition to target cells. The binding of HCV to cells was then quantified by real-time PCR analysis of HCV RNA. Data are expressed as percentage inhibition and are means ± SD of values from 5 independent experiments.
To investigate the effect of human IgG specific for HVR1 of the X virus E2 glycoprotein, we selected a plasma specimen from an HCV-infected patient (S5) that contained IgG able to recognize X virus HVR1 peptides used as capture antigen in an ELISA. Total IgG from S5 plasma was purified with a protein G column, and the antibodies specific for X virus HVR1 peptides were then further purified with an immunoaffinity column. The reactivity of the specific (eluted) IgG fraction with X peptides as measured by ELISA was markedly greater than that of the nonretained IgG fraction (absorbance at 405 nm: 2.19 and 0.28, respectively). Both the eluted and nonretained IgG fractions, but not control human IgG, inhibited the binding of X virus to Molt-4 cells, U937 cells, and platelets (Figure 5B). At similar IgG concentrations, the inhibitory effect of the antibodies specific for HVR1 with Molt-4 and U937 cells was approximately half of that of the nonretained antibody fraction; the difference in inhibitory activity between the 2 fractions was much less pronounced with platelets. These results suggest that HVR1 plays a role in virus attachment to all 3 types of target cells.
Effect of antibodies to CD81 on HCV binding to Molt-4 cells
The role of CD81 in the binding of HCV to Molt-4 cells was further investigated by incubating the cells with various concentrations of antibodies to human CD81 (1, 10, 50, 100, or 200 μg/mL) before the addition of X virus. The amount of cell-associated HCV RNA was not affected by pretreatment of the cells with antibodies to CD81 (data not shown), suggesting that HCV particles are able to bind to Molt-4 cells by CD81-independent mechanisms.
Binding of rE2 to human cell lines and platelets
We next examined the ability of rE2 to bind to Molt-4 cells, U937 cells, and platelets with the use of flow cytometric analysis (Figure6A). The extent of rE2 binding to Molt-4 cells was about 4 times that apparent with U937 cells and 2 times that observed with platelets (Figure 6B). To assess the specificity of rE2 binding, we examined the effect of IgG purified from the plasma of an HCV-infected patient (S8) that was highly reactive with rE2 (but not with a peptide based on the corresponding COOH-terminal region of HVR1) by enzyme immunoassay. Pretreatment of rE2 with the human anti-E2 IgG (70 μg/mL) markedly inhibited the subsequent binding of rE2 to Molt-4 cells (62.1% inhibition) and to U937 cells (44.7% inhibition), but not that to platelets (Figure 6B).
Flow cytometric analysis of the binding of rE2 to human cell lines and platelets.
(A) Molt-4 cells, U937 cells, and platelets were incubated with rE2, and cell-bound protein was detected by flow cytometry with a mouse mAb to E2 and FITC-conjugated rabbit antibodies to mouse IgG (shaded profiles); as a control, the mouse mAb to E2 was replaced with an IgG isotype control (open profiles). (B) The recombinant E2 protein was incubated in the absence or presence of E2-reactive IgG from an HCV-infected patient or of control human IgG before addition to target cells. The binding of rE2 to cells was then determined by flow cytometry as in panel A. Data are expressed as median fluorescence intensity and are representative of 3 to 5 separate experiments.
Flow cytometric analysis of the binding of rE2 to human cell lines and platelets.
(A) Molt-4 cells, U937 cells, and platelets were incubated with rE2, and cell-bound protein was detected by flow cytometry with a mouse mAb to E2 and FITC-conjugated rabbit antibodies to mouse IgG (shaded profiles); as a control, the mouse mAb to E2 was replaced with an IgG isotype control (open profiles). (B) The recombinant E2 protein was incubated in the absence or presence of E2-reactive IgG from an HCV-infected patient or of control human IgG before addition to target cells. The binding of rE2 to cells was then determined by flow cytometry as in panel A. Data are expressed as median fluorescence intensity and are representative of 3 to 5 separate experiments.
The effect of rE2 on HCV binding to human cells was also investigated. Human cell lines or platelets were incubated with various concentrations of rE2 before the addition of X plasma. The recombinant E2 protein markedly inhibited HCV binding to Molt-4 cells in a concentration-dependent manner (Figure7); the inhibition by rE2 of virus binding was less pronounced with U937 cells and even less so with platelets.
Effect of rE2 on HCV binding to human target cells.
Molt-4 or U937 cells (1 × 105) or platelets (1 × 106) were incubated with various concentrations of rE2 before the addition of X plasma (1.7 × 105 IU HCV) and quantitation of HCV binding by real-time RT-PCR. Data are expressed as percentage inhibition of HCV binding and are means ± SD of values from 2 to 5 separate experiments.
Effect of rE2 on HCV binding to human target cells.
Molt-4 or U937 cells (1 × 105) or platelets (1 × 106) were incubated with various concentrations of rE2 before the addition of X plasma (1.7 × 105 IU HCV) and quantitation of HCV binding by real-time RT-PCR. Data are expressed as percentage inhibition of HCV binding and are means ± SD of values from 2 to 5 separate experiments.
Discussion
The selective interaction of viruses with specific cell surface receptors is an essential step in the initiation of infection. This process often determines the host range and cellular tropism of a virus. However, the interaction of a virus with a cell does not always result in productive infection because the cells may not be permissive for viral gene expression, genome replication, or assembly. Human platelets associate with and bind several viral agents.1-12 Our observations on the compartmentalization of HCV in the blood of chronically infected individuals now indicate that, in addition to the expected presence of virus in the cellular and plasma fractions, HCV RNA is also associated to a substantial extent with platelets. These observations were confirmed experimentally by measurement of the direct binding of free and IgG-complexed HCV to normal human platelets. The HCV-platelet interaction is not mediated by CD81, given that this molecule is not expressed on the surface of platelets. The possibility that other molecules specifically mediate HCV binding is further suggested by the observation that the promonocytic cell line U937, which does not express CD81, binds HCV to an extent similar to that observed with CD81-expressing Molt-4 cells. It is also possible that the spongelike morphology of platelets results in the nonspecific adsorption or entrapment of free and complexed HCV on the cell surface. Platelets might play a role in the clearance of HCV through uptake of complexed virus for elimination by the reticuloendothelial system. Alternatively, the binding of free HCV to the platelet membrane may facilitate virus dissemination and persistence, possibly by sheltering and protecting the virus from neutralizing antibodies. Only the second of these 2 roles is likely for mononuclear cells, given that these cells preferentially bind free virus.
Consistent with previous observations,53 we showed that U937 cells do not express CD81 on the cell surface or contain CD81 mRNA. The similar levels of HCV binding observed with Molt-4 and U937 cells indicate that CD81 is not the only cell membrane component that mediates viral attachment. Two lines of evidence support the specificity of the attachment of free HCV to human T and promonocytic cell lines. First, increasing concentrations of X virus resulted in saturation of binding to Molt-4 and U937 cells at a viral dose of about 100 000 IU/100 000 cells. The binding of increasing concentration of HCV to platelets followed different kinetics from mononuclear cells. The binding curve appears monophasic with no clear plateau. Second, human anti-HCV IgG inhibited viral binding in a dose-dependent manner, irrespective of the expression of CD81. The partial nature of this inhibition might be attributable to an insufficient concentration of IgG, to the use of cross-reactive antibodies from patient S5, or to a certain extent of nonspecific binding of antibodies to promonocytic (U937) cells and platelets. The shape of the binding inhibition curve for Molt-4 cells suggests that the inhibition at the highest IgG concentration tested was almost maximal.
In infected individuals, HCV circulates as a heterogeneous population of related variants (quasi-species). The envelope glycoproteins (E1 and E2) exhibit the highest degree of genetic heterogeneity, especially in the 27 amino acids of HVR1 located at the NH2-terminus of E2.54,55 Previous studies suggested that HVR1 contributes to the neutralization of HCV, given that it contains linear epitopes that are recognized by patient antibodies.56-62 Antibodies to HVR1 inhibit the binding of rE225 and virus35 to cells, and rabbit hyperimmune serum generated in response to the COOH-terminal 21 amino acids of HVR1 neutralizes the infectivity of HCV in chimpanzees28,30 and inhibits viral binding to cells.29 Our observation that immunopurified IgG specific for the linear HVR1 sequence of the X virus partially inhibited viral binding suggests that the binding of HCV to human cells might be mediated in part by HVR1. Another possibility is that HVR1 does not participate directly in viral attachment to the cell surface, but that antibody binding to this region of E2 induces a conformational change that hinders access to the cell receptors. The observation that the IgG fraction depleted of anti-HVR1 also inhibited HCV binding suggests that ligands other than HVR1 contribute to viral attachment. The lack of effect of antibodies to CD81 on HCV binding to Molt-4 cells is consistent with the notion that other specific molecules mediate this binding.
Both rE2 and a truncated, secreted version of E2 have previously been shown to bind to human cell lines expressing CD81.25,36,39,42,63,64 Moreover, the regions of E2 that mediate binding of the protein to the large extracellular loop of CD81 were mapped to amino acids 480 to 493 and 544 to 551.39Our observation that the extent of rE2 binding to Molt-4 cells was 4 and 2 times that apparent with U937 cells and platelets is consistent with previous data36 showing that the level of rE2 binding correlates with CD81 expression. The discrepancy between viral and rE2 binding to CD81-expressing Molt-4 cells and nonexpressing U937 cells and platelets is not likely related to viral genotype, given that both the X virus and rE2 are of genotype 1a. These data therefore suggest that ligands other than E2 may be required for viral binding. Recent data indicate that both E1 and E2 are required for the pH-dependent fusion activity of HCV envelope glycoproteins.65 Our proposal is also consistent with evidence for the formation by E1 and E2 of a noncovalent heterodimer both in vitro and in vivo.23,24,66 67
The differences observed between the binding of free HCV and that of rE2 suggest that rE2 is not truly representative of HCV in terms of viral interaction with target cells, especially with those that do not express CD81. Indeed, a recent study68 showed that HCV binding to hepatocytes may not depend entirely on CD81. In addition, the binding of rE2 to CD81 does not correlate with species permissiveness to infection, given that rE2 binds to the nonpermissive cotton-top tamarin cell line B95-8.41 42
Alternative mechanisms of HCV-cell interaction may involve either direct ligands, such as E1 or E1-E2 complexes, or indirect ligands, such as LDL-HCV.43-45 HCV particles are associated with β-lipoproteins,47 and endocytosis of HCV is mediated by LDL receptors.45 The level of cell surface expression of the LDL receptor was shown to be directly correlated with the number of HCV-positive cells in vitro.47 In addition, transfection of COS-7 cells with an expression vector for the LDL receptor confers the ability to bind HCV,43 suggesting that HCV particles associated with LDL bind to the LDL receptor. A recent study of the binding of HCV and rE2 to human cells showed that, although rE2 binds CD81, the extent of viral entry correlated with LDL receptor abundance and was independent of CD81 expression69; interaction of rE2 with LDL or the LDL receptor was not detected. These observations further suggest that ligands other than E2 may be required for viral binding. In the present study, we did not explore the potential role of HCV-LDL complexes in HCV binding to platelets. Such a role therefore cannot be excluded.
We thank G. Alexander, who provided blood samples from infected patients to study HCV binding to platelets. We also thank M. Houghton for providing rE2, S. Abrignani for providing the mAb to E2, S. Levy for providing the plasmid encoding human CD81, T. Wallington for providing plasma from patient E.H., M. Duc Dodon for providing the U937 cell line, as well as P. Smethurst, S. Garner, C. Howard, and C. Hurd for technical assistance.
Supported in part by grant 97/13 from the National Blood Authority, England, and the National Blood Service, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Jean-Pierre Allain, Division of Transfusion Medicine, East Anglia Blood Centre, Long Rd, Cambridge, CB2. 2PT, United Kingdom; e-mail: jpa1000@cam.ac.uk.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal