Primary systemic amyloidosis (AL) is a plasma cell (PC) dyscrasia with clinical similarities to multiple myeloma (MM) and monoclonal gammopathy of undetermined significance (MGUS), but its molecular basis is poorly understood. Translocations at the immunoglobulin heavy-chain (IgH) locus, 14q32, are likely early genetic events in both MM and MGUS and involve several nonrandom, recurrent, partner chromosomes such as 11q13, 16q23, and 4p16.3. Given the similarities between MM, MGUS, and AL, bone marrow clonal PCs were evaluated in 29 patients with AL using interphase fluorescence in situ hybridization (FISH) combined with immunofluorescence detection of the cytoplasmic light-chain (cIg-FISH) for the presence of 14q32 translocations and the t(11;14)(q13;q32). Of 29 patients studied, 21 (72.4%) showed results compatible with the presence of a 14q32 translocation, and 16 (76.2%) of those had translocation (11;14)(q13;q32) for an overall prevalence of the abnormality of 55%. IgH translocations are common in AL, especially the t(11;14)(q13;q32).
Introduction
Amyloidosis (AL) is a plasma cell (PC) dyscrasia, similar to multiple myeloma (MM) and monoclonal gammopathy of undetermined significance (MGUS),1,2 characterized by the presence of clonal PCs in the bone marrow (BM) and extracellular deposition of amyloidogenic light chains with resultant organ dysfunction and an overall median survival of 1.5 years.1 2 The usually fatal outcome of the disease does not permit long-term observations of clonal growth.
Chromosomal translocations involving the immunoglobulin heavy-chain (IgH) locus may be initiating events in the development of several B-cell malignancies,3,4 including MGUS and MM.5 IgH translocations in MM involve a number of recurrent, nonrandom partner chromosomes including 11q13 and 4p16.3.14-16 Using the cytoplasmic light-chain (cIg)–fluorescence in situ hybridization (FISH) technique, we examined 29 patients with AL for the presence of IgH translocations, including t(11;14)(q13;q32).
Study design
Patient samples
Twenty-nine patients (10 women, 19 men; median age, 62 years) were studied. Institutional review board approval was obtained, and all patients gave written, informed consent for a research BM sample to be collected at the time of routine clinical procurement. Diagnostic criteria for AL included histologic proof of amyloid deposition in addition to the presence of a monoclonal protein or demonstration of monoclonal PCs. Five of the 29 patients had previously been treated with alkylating agents. None of the patients had clinical evidence of MM.
Cell enrichment and cIg-FISH
PCs were selected with CD 138 magnetic microbeads (Miltenyi Biotec, Sunnyvale, CA) after enrichment for mononuclear cells through centrifugation on a Ficoll-Hypaque gradient, density 1.076 (Pharmacia-Biotech, Uppsala, Sweden). Cytospin preparations were prepared according to a slight modification of our previously published cIg-FISH technique.6 We scored 100 PCs in all but 2 cases. One repeatedly failed to hybridize; in the other, we could only score 50 cells for t(11;14)(q13;q32), respectively. Each sample was scored by 2 independent examiners who had a high degree of agreement (r2 = 0.795).
FISH probes
Break-apart strategy.
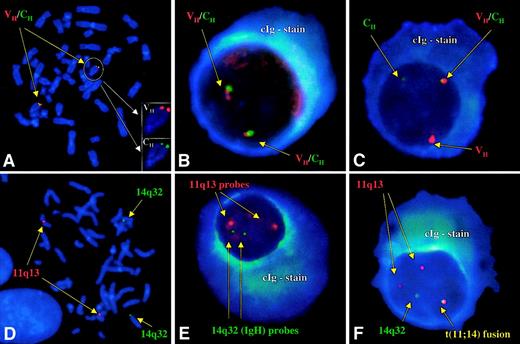
Probes that hybridize to the variable (VH; cosmid clone) and constant regions (CH; BAC clone) of the IgH locus7 were directly labeled with SpectrumRed and SpectrumGreen, respectively (Vysis, Downers Grove, IL). The presence of a 14q32 translocation was demonstrated by segregation of the signals (break-apart strategy; Figure 1). A normal pattern consisted of 2 pairs of closely associated red and green signals. An abnormal pattern was defined as a distance greater than 3 signal widths between probe pairs. An equivocal pattern was one in which there was loss of a probe signal (1 red-green pair/1 red or 1 red-green pair/1 green), representing deletion, an unbalanced translocation, or technical inability to discern the presence of a signal because of small size or background staining.
Normal and abnormal patterns of hybridization with both strategies.
(A) Hybridization of the VH and CH probes to the correct location at band 14q32 on normal human male metaphases. (B) Normal pattern for the VHCH probes with 2 pairs of closely associated signals. Note the intense blue fluorescence of the cytoplasmic light chain. (C) Cellular signal pattern indicative of a 14q32 translocation. The segregation of a pair of signals (break-apart) in a large percentage of cells allows for the identification of a patient with an IgH translocation. (D) Hybridization of the 14q32 and 11q13 probes to the correct locations on chromosomes 14 and 11, respectively, on normal human male metaphases. (E) Normal pattern for the 14q32/11q13 fusion strategy. There are no fusion signals observed, and there are 2 pairs of nonassociated green (14q32) and red (11q13) signals. (F) Abnormal comigration (fusion) of signals indicative of a t(11;14)(q13;q32). Because of the small signal size arising from the VHportion of the IgH probe (owing to the V/D/J recombination and DNA excision), the pattern most frequently observed is presumed to be that of fusions in the der14 and less commonly in the der11.
Normal and abnormal patterns of hybridization with both strategies.
(A) Hybridization of the VH and CH probes to the correct location at band 14q32 on normal human male metaphases. (B) Normal pattern for the VHCH probes with 2 pairs of closely associated signals. Note the intense blue fluorescence of the cytoplasmic light chain. (C) Cellular signal pattern indicative of a 14q32 translocation. The segregation of a pair of signals (break-apart) in a large percentage of cells allows for the identification of a patient with an IgH translocation. (D) Hybridization of the 14q32 and 11q13 probes to the correct locations on chromosomes 14 and 11, respectively, on normal human male metaphases. (E) Normal pattern for the 14q32/11q13 fusion strategy. There are no fusion signals observed, and there are 2 pairs of nonassociated green (14q32) and red (11q13) signals. (F) Abnormal comigration (fusion) of signals indicative of a t(11;14)(q13;q32). Because of the small signal size arising from the VHportion of the IgH probe (owing to the V/D/J recombination and DNA excision), the pattern most frequently observed is presumed to be that of fusions in the der14 and less commonly in the der11.
Fusion strategy.
For the detection of the t(11;14)(q13;q32), on separate experiments, we used the same IgH probes (labeled with SpectrumGreen) and a pool of cosmid and P1 clones encompassing all reported breakpoints in human MM cell lines with the t(11;14)(q13;q32)8-10 (labeled with SpectrumRed). The normal pattern was that of 2 pairs of nonassociated signals (2 red/2 green). Fusion of 2 signals in a cell was considered indicative of the translocation.
Statistical considerations
The normal range of PC patterns was documented previously using cIg-negative cells derived from patient samples and polyclonal PCs from BM donors. A patient was said to have an IgH translocation if the percentage of PCs exceeded the mean ± 3 SD.11 The upper limit of normal for the break-apart and fusion strategies was less than 10% for both. We arbitrarily chose an upper limit of normal of 25% for greater specificity.
Results and discussion
Break-apart strategy (VH/CHprobes)
Of these 29 patients, 16 (55%) had definitive evidence of an IgH translocation using the VH/CH strategy (Table 1). The median percentage of abnormal PCs (MAPC) was 55% (range, 27%-89%). Additionally, 5 (17%) patients had an equivocal pattern compatible with a possible IgH translocation (MAPC, 66%; range, 64%-83%). In total, 21 of 29 (72%) patients had evidence of an IgH translocation by the VH/CH probe patterns.
Patient clinical and laboratory features and associated FISH results
| No. . | Sex . | Age . | IgH status . | BM PC (%) . | κ:λ ratio . | Heavy chain . | Serum M spike (g/dL) . | Urine M spike (g/24 h) . | VH/CH . | t(11;14)(q13;q32) . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern . | Positive (%) . | Equivocal (%) . | Positive and equivocal . | Pattern . | Positive (%) . | |||||||||
| Newly diagnosed patients | ||||||||||||||
| 1 | M | 63 | t(11;14) | 19 | < 1:99 | ND | IF | 1.5 | 1F1R1G | 89 | 11 | 100 | 2F1G2R | 100 |
| 2 | F | 78 | t(11;14) | 8 | 1:40 | IgG | 0.6 | 0.9 | 1F1R1G | 84 | 8 | 92 | 1F1G2R | 86 |
| 3 | F | 51 | t(11;14) | 33 | < 1:99 | ND | IF | IF | 1F1R1G | 79 | 8 | 87 | 1F1G2R | 89 |
| 4 | F | 62 | t(11;14) | 12 | 1:2 | IgG | 0.4 | IF | 1F1R1G | 75 | 6 | 81 | 1F2R | 100 |
| 5 | M | 71 | t(11;14) | 4 | 1:4 | ND | ND | IF | 1F1R1G | 66 | 10 | 76 | 1F1R1G | 68 |
| 6 | M | 62 | t(11;14) | 13 | 1:54 | IgG | 0.8 | ND | 1F1R1G | 57 | 30 | 87 | 1F1R1G | 95 |
| 7 | M | 53 | t(11;14) | 22 | > 99:1 | ND | ND | IF | 1F1R1G | 55 | 31 | 86 | 1F1G2R | 85 |
| 8 | M | 57 | t(11;14) | 26 | > 99:1 | IgA | IF | IF | 1F1R1G | 54 | 36 | 90 | 1F2R | 85 |
| 9 | M | 77 | t(11;14) | 1 | 1:1 | IgG | ND | 0.7 (λ) | 1F1R1G | 53 | 19 | 72 | 1F1G1R | 71 |
| 10 | M | 65 | t(11;14) | 10 | 1:4 | ND | ND | ND | 1F1R1G | 38 | 30 | 68 | 1F1G1R | 95 |
| 11 | M | 63 | t(11;14) | 3 | < 1:99 | ND | IF | 3 | 1F1R1G | 30 | 59 | 89 | 1F1G3R | 93 |
| 12 | M | 75 | t(11;14) | 25 | > 99:1 | IgG | 1.6 | 1.5 | 1F1R1G | 28 | 68 | 96 | 1F2R1G | 72 |
| 13 | M | 40 | t(11;14) | 8.8 | 1:44 | ND | IF | IF | 1F1G | 21 | 69 | 90 | 1F2R | 68 |
| 14 | M | 41 | t(11;14) | 5 | 1:25 | ND | ND | 0.5 | 1F1G | 19 | 66 | 85 | 1F2R1G | 96 |
| 15 | M | 74 | 14q32+ | 10 | 1:60 | IgA | 0.7 | 1.2 | 1F1R1G | 41 | 35 | 76 | 2G2R | 15 |
| 16 | F | 67 | 14q32+ | 1 | 1:1 | ND | IF | IF (λ) | 1F1R1G | 38 | 14 | 52 | 2G2R | 19 |
| 17 | F | 57 | 14q32+ | 1 | < 1:99 | ND | ND | IF | 1F1R1G | 27 | 17 | 44 | 2G2R | 18 |
| 18 | F | 58 | Equivocal* | 3 | 1:13 | IgG | 1.7 | IF | 2F1G | 7 | 83 | 90 | 2G2R | 10 |
| 19 | M | 67 | Negative | 4.6 | < 1:99 | IgA | ND | IF | 2F | 18 | 14 | 32 | Fail | — |
| 20 | M | 64 | Negative | 9 | 1:23 | IgG | 0.9 | IF | 2F | 9 | 18 | 27 | 2G2R | 16 |
| 21 | F | 37 | Negative | 15 | 18:1 | ND | ND | IF | 2F | 23 | 19 | 42 | 2G2R | 15 |
| 22 | M | 65 | Negative† | 13 | 1:13 | ND | ND | ND | 2F | 20 | 20 | 40 | Fail | — |
| 23 | M | 60 | Negative | 38 | > 99:1 | IgA | 2.5 | ND | 2F | 19 | 24 | 43 | 2G2R | 13 |
| 24 | F | 70 | Negative | 34 | < 1:99 | IgA | 1.2 | IF | 2F | 14 | 11 | 25 | 2G2R | 23 |
| Patients with previous alkylator chemotherapy | ||||||||||||||
| 25 | M | 74 | t(11;14) | 5 | 4:1 | ND | ND | ND | 1F1G | 25 | 65 | 90 | 1F1G2R | 72 |
| 26 | F | 45 | t(11;14) | 17 | < 1:99 | IgG | IF | 0.05 | 2F1G | 12 | 64 | 76 | 1F1G2R | 80 |
| 27 | F | 55 | 14q32+ | 3 | < 1:99 | IgA | 0.5 | ND | 1F1R1G | 75 | 16 | 91 | 2G2R | 15 |
| 28 | M | 54 | Negative | 10 | 1:1 | IgM | 0.8 | 0.07 (λ) | 2F | 15 | 19 | 34 | 2G2R | 23 |
| 29 | M | 54 | Negative‡ | 22 | > 99:1 | IgG | 1 | 0.2 | 2F | 7 | 9 | 16 | Fail | — |
| No. . | Sex . | Age . | IgH status . | BM PC (%) . | κ:λ ratio . | Heavy chain . | Serum M spike (g/dL) . | Urine M spike (g/24 h) . | VH/CH . | t(11;14)(q13;q32) . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern . | Positive (%) . | Equivocal (%) . | Positive and equivocal . | Pattern . | Positive (%) . | |||||||||
| Newly diagnosed patients | ||||||||||||||
| 1 | M | 63 | t(11;14) | 19 | < 1:99 | ND | IF | 1.5 | 1F1R1G | 89 | 11 | 100 | 2F1G2R | 100 |
| 2 | F | 78 | t(11;14) | 8 | 1:40 | IgG | 0.6 | 0.9 | 1F1R1G | 84 | 8 | 92 | 1F1G2R | 86 |
| 3 | F | 51 | t(11;14) | 33 | < 1:99 | ND | IF | IF | 1F1R1G | 79 | 8 | 87 | 1F1G2R | 89 |
| 4 | F | 62 | t(11;14) | 12 | 1:2 | IgG | 0.4 | IF | 1F1R1G | 75 | 6 | 81 | 1F2R | 100 |
| 5 | M | 71 | t(11;14) | 4 | 1:4 | ND | ND | IF | 1F1R1G | 66 | 10 | 76 | 1F1R1G | 68 |
| 6 | M | 62 | t(11;14) | 13 | 1:54 | IgG | 0.8 | ND | 1F1R1G | 57 | 30 | 87 | 1F1R1G | 95 |
| 7 | M | 53 | t(11;14) | 22 | > 99:1 | ND | ND | IF | 1F1R1G | 55 | 31 | 86 | 1F1G2R | 85 |
| 8 | M | 57 | t(11;14) | 26 | > 99:1 | IgA | IF | IF | 1F1R1G | 54 | 36 | 90 | 1F2R | 85 |
| 9 | M | 77 | t(11;14) | 1 | 1:1 | IgG | ND | 0.7 (λ) | 1F1R1G | 53 | 19 | 72 | 1F1G1R | 71 |
| 10 | M | 65 | t(11;14) | 10 | 1:4 | ND | ND | ND | 1F1R1G | 38 | 30 | 68 | 1F1G1R | 95 |
| 11 | M | 63 | t(11;14) | 3 | < 1:99 | ND | IF | 3 | 1F1R1G | 30 | 59 | 89 | 1F1G3R | 93 |
| 12 | M | 75 | t(11;14) | 25 | > 99:1 | IgG | 1.6 | 1.5 | 1F1R1G | 28 | 68 | 96 | 1F2R1G | 72 |
| 13 | M | 40 | t(11;14) | 8.8 | 1:44 | ND | IF | IF | 1F1G | 21 | 69 | 90 | 1F2R | 68 |
| 14 | M | 41 | t(11;14) | 5 | 1:25 | ND | ND | 0.5 | 1F1G | 19 | 66 | 85 | 1F2R1G | 96 |
| 15 | M | 74 | 14q32+ | 10 | 1:60 | IgA | 0.7 | 1.2 | 1F1R1G | 41 | 35 | 76 | 2G2R | 15 |
| 16 | F | 67 | 14q32+ | 1 | 1:1 | ND | IF | IF (λ) | 1F1R1G | 38 | 14 | 52 | 2G2R | 19 |
| 17 | F | 57 | 14q32+ | 1 | < 1:99 | ND | ND | IF | 1F1R1G | 27 | 17 | 44 | 2G2R | 18 |
| 18 | F | 58 | Equivocal* | 3 | 1:13 | IgG | 1.7 | IF | 2F1G | 7 | 83 | 90 | 2G2R | 10 |
| 19 | M | 67 | Negative | 4.6 | < 1:99 | IgA | ND | IF | 2F | 18 | 14 | 32 | Fail | — |
| 20 | M | 64 | Negative | 9 | 1:23 | IgG | 0.9 | IF | 2F | 9 | 18 | 27 | 2G2R | 16 |
| 21 | F | 37 | Negative | 15 | 18:1 | ND | ND | IF | 2F | 23 | 19 | 42 | 2G2R | 15 |
| 22 | M | 65 | Negative† | 13 | 1:13 | ND | ND | ND | 2F | 20 | 20 | 40 | Fail | — |
| 23 | M | 60 | Negative | 38 | > 99:1 | IgA | 2.5 | ND | 2F | 19 | 24 | 43 | 2G2R | 13 |
| 24 | F | 70 | Negative | 34 | < 1:99 | IgA | 1.2 | IF | 2F | 14 | 11 | 25 | 2G2R | 23 |
| Patients with previous alkylator chemotherapy | ||||||||||||||
| 25 | M | 74 | t(11;14) | 5 | 4:1 | ND | ND | ND | 1F1G | 25 | 65 | 90 | 1F1G2R | 72 |
| 26 | F | 45 | t(11;14) | 17 | < 1:99 | IgG | IF | 0.05 | 2F1G | 12 | 64 | 76 | 1F1G2R | 80 |
| 27 | F | 55 | 14q32+ | 3 | < 1:99 | IgA | 0.5 | ND | 1F1R1G | 75 | 16 | 91 | 2G2R | 15 |
| 28 | M | 54 | Negative | 10 | 1:1 | IgM | 0.8 | 0.07 (λ) | 2F | 15 | 19 | 34 | 2G2R | 23 |
| 29 | M | 54 | Negative‡ | 22 | > 99:1 | IgG | 1 | 0.2 | 2F | 7 | 9 | 16 | Fail | — |
F indicates fusion; G, green; R, red; 14q32+, IgH translocation without partner identified; IF, immunofixation; ND, not detected.
Equivocal for 14q32 and negative for t(11;14).
Patient with monosomy of chromosome 14 in 55% of PCs.
Patient with monosomy of chromosome 14 in 76% of PCs.
t(11;14)(q13;q32)
Twelve of 16 (75%) patients with definitive IgH translocations by the VH/CH strategy had a t(11;14)(q13;q32) (MAPC, 87.5%; range, 68%-100%). In addition, 4 of the 5 (80%) patients with an equivocal pattern using the VH/CH strategy also had a t(11;14)(q13;q32) (MAPC, 76%; range, 68%-96%). Altogether, 16 of the 29 (55%) patients had evidence of a t(11;14)(q13;q32), accounting for 76% of all IgH translocations detected (16 of 21 patients). There was no statistically significant difference in the BM percentage PC between patients with and without the abnormality detected by either strategy (Wilcoxon signed-rank test, P > .2)
Other
Four patients had definitive evidence of an IgH translocation by the break-apart strategy that did not involve the 11q13 locus (MAPC, 64%; range, 44%-91%). One patient had a possible IgH translocation with an equivocal pattern by the VH/CH in 90% of PCs (not involving 11q13). Eight patients did not have evidence of IgH translocations, and 2 of those patients had only 1 pair of VH/CH probes in 55% and 76% of their cells, respectively, reflecting monosomy 14 or deletion of 14q32.
We have shown here that most patients with AL harbor IgH translocations in clonal PCs. In AL, the deposition of monoclonal light chains is the cause of the sickness and death associated with the disease, whereas clonal progression is usually not observed.12 Our results support the hypothesis that AL is the same disease entity as MGUS/SMM (at the clonal level), with the clinical differences (phenotype) arising from the amyloidogenic potential of the monoclonal light chain. Most patients with AL have a low degree of BM involvement and rarely have overt MM.13 We speculate the occasional patient with AL (approximately 5%), who also has coexistent MM, may have a lesser amyloidogenic variant of the light chain or a more rapid clonal progression.14 The frequency of t(11;14)(q13;q32) is much greater in AL than has been noted for MM15 or MGUS16-18 in some series but not in others.19Several facts allow us to conclude that the prevalence of the t(11;14)(q13;q32) we report in AL is correct. We used 2 independent scorers, and both concurred in the scoring of abnormal PCs in each sample with a high degree of agreement. The percentages of abnormal PCs by the break-apart and fusion strategies were similar in each patient, and the abnormalities were usually seen in most of the clonal PC. Finally, we also observed a higher proportion of t(11;14)(q13;q32) in patients with MGUS than in patients with MM.19 The frequencies of other partner chromosomes are being studied to assess the pathogenetic potential of these abnormalities, and we speculate that other partner chromosomes will also be involved.20-22
The results of our study identify cytogenetic lesions in the clonal PCs of patients with AL, similar to those observed in MGUS and MM. In addition, previous work has shown that VL gene sequences are highly somatically mutated, suggesting commonality among these dyscrasias.23 We have previously shown that aneuploidy is equally present in AL, MGUS, smoldering MM, and MM.24 Hallek3 proposes a step-wise transformation model for the development of MM from MGUS, and we propose the inclusion of AL in the model where AL is similar to MGUS and smoldering MM, albeit with an “unlucky” protein.
Supported in part by Public Health Service grant R01 CA83724-01 from the National Cancer Institute. P.R.G. is supported in part by research grant P01 CA62242 from the National Cancer Institute and by ECOG grant CA21115-25C from the National Cancer Institute. R.F. is a Leukemia and Lymphoma Society Translational Research Awardee. R.F. and P.R.G. are also supported by the Mayo Foundation and the CI-5 Cancer Research Fund, Lilly Clinical Investigator Award of the Damon Runyon-Walter Winchell Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Rafael Fonseca, Department of Hematology and Internal Medicine, 200 First St SW, Bldg W10B, Mayo Clinic, Rochester, MN 55905; e-mail: fonseca.rafael@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal