Abstract
Chronic myelogenous leukemia (CML) is commonly characterized by the presence of the p210Bcr-Abl oncoprotein. Many downstream effectors of Bcr-Abl have been described, including activation of the Grb2-SoS-Ras–MAP kinase (Erk) pathway. The precise contributions of these signal-transduction proteins in CML blast cells in human patients are not yet well defined. To gain further insight into the importance of Grb2 for CML, peptides that disrupt Grb2-SoS complexes were tested. These high-affinityGrb2-binding peptides (HAGBPs) can autonomously shuttle into cells and function by binding to the N-terminal SH3 domain of Grb2. The HAGBPs were analyzed for their effects on Bcr-Abl–expressing cell lines and freshly isolated CML blast cells from patients. They induced a dramatic decrease in the proliferation of CML cell lines. This was not observed with point-mutated control peptides with abolished Grb2SH3(N) binding. As expected, Grb2-SoS complexes were greatly diminished in the HAGBP-treated cells, and MAP kinase activity was significantly reduced as determined by an activation-specific phospho-MAPK antibody. Furthermore, cell fractions that are enriched for blast cells from CML patients with active disease were also incubated with the Grb2 blocker peptides. The HAGBPs led to a significant proliferation reduction of these cells in the majority of the isolates, but not in all patients' cells. These results show that, in addition to the direct targeting of Bcr-Abl, selective inhibition of Grb2 protein complexes may be a therapeutic option for a significant number of CML patients.
Introduction
Bcr-Abl proteins result from chromosomal translocations that lead to the fusion of parts from thebreakpoint cluster region (bcr) gene on chromosome 22 to sequences of the c-abl gene on chromosome 9. Although both genes encode for protein kinases,1-3 it is thought that mainly the activation of the tyrosine kinase activity encoded in the fragment of the c-abl gene is important for the development of a human disease termed chronic myelogenous leukemia (CML).4-6 The bcr portion of thebcr-abl fusion gene is thought to contribute to the course of the disease by providing an oligomerization domain, which resides in a coiled-coil region at the N-terminus of the Bcr protein,7 but also by antagonizing the Abl kinase activity.8
State-of-the-art therapies for CML patients currently include cytostatic drugs, α-interferon and its derivatives, as well as bone marrow or stem cell transplantation for younger patients.6 9 However, the current treatment options are not yet satisfactory with respect to the long-term survival of the patients.
During the last decade, a tremendous number of studies have unraveled many aspects of the intracellular signal-transduction events resulting from the chromosomal translocations that lead to the formation of Bcr-Abl proteins. It is therefore hoped that specific signal-transduction inhibitors will eventually complement the spectrum of therapeutic choices for CML.10 As in many other cancers, the central mitogenic cascade, which leads via the small GTPase Ras to the activation of mitogen-activated protein kinases (MAPKs),11 has gained much attention in this respect. Activation of Ras and MAPK was also observed for the Tel-Abl (ETV6-Abl) fusion protein, a leukemic protein similar to Bcr-Abl, which uses instead of Bcr the Ets family protein Tel (ETV6) to oligomerize the Abl kinase.12 13
Numerous reports link Bcr-Abl directly to the adapter proteins Shc and Grb2, which are signal transducers upstream of Ras and are known to recruit the Ras-activating exchange factor SoS toward the intracellular sites where Ras is located.14-19
Despite the large number of studies obtained with different established cell lines, no clear picture of how these data relate to the situation in human patients' cells has yet emerged. This is in part because, despite considerable efforts, so far it has not been possible to generate animal models that convincingly mimic human CML.20 21
Signal-transduction–based strategies, which are often still in an experimental state, have so far mainly targeted Bcr-Abl on the protein or RNA level. A prominent example is the new tyrosine kinase inhibitor compound CGP57148B (recently renamed STI571), which showed promising results in first clinical trials.22
Despite this positive result, it is already obvious that the development of resistance to STI571 will become a problem at least in some cases.23 To overcome this obstacle, in CML therapy as well as in many other cancers, it may be better to use cocktails of compounds directed against several cellular targets relevant to the disease mechanism.21
In this report, we tested whether cell-penetrating high-affinity peptides that block the N-terminal SH3 domain of Grb2 (high-affinityGrb2-binding peptides; HAGBPs) have an effect on CML cell lines and primary blast cells from CML patients with active disease. Indeed, it is possible to strongly inhibit CML cell proliferation with these peptides. Uncoupling of the Ras activator SoS from Grb2 as well as a reduction of MAPK activity was observed. While this manuscript was being prepared, a study published in this journal by Million and Van Etten24 showed that in a CML mouse model, Grb2 binding to Bcr-Abl is required for the induction of the disease, strongly supporting our findings with human cells. Together with our data, these results encourage future tests of inhibitors developed to block Grb2 protein interactions in the context of Bcr-Abl–dependent disease.
Materials and methods
Peptide synthesis
Synthesis was carried out by the Fmoc (fluorenylmethoxycarbonyl)/t-butyl–based solid-phase peptide chemistry method on SMPS 350 (Zinsser Analytic, Frankfurt/Main, Germany) and ABI 433A (PerkinElmer, Rodgau, Germany) synthesizers. The peptides were deprotected and cleaved from the resin with trifluoroacetic acid/ethanedithiol/water, 94:3:3, for 120 minutes. After filtration, precipitation with cold ter-butyl methyl ether, and lyophilization, the crude peptides were purified by reverse-phase high-performance liquid chromatography (HPLC) on preparative Vydac C18 columns with linear gradients of 80% acetonitrile plus 0.05% trifluoroacetic acid versus 0.07% aqueous trifluoroacetic acid. Correct mass was checked by electrospray mass spectrometry (Sciex API III; Perkin Elmer). Pure fractions were pooled, and the final product was analyzed by reverse-phase HPLC on a Vydac C18 column (250 × 4.6 mm) and capillary zone electrophoresis (Biofocus 3000; Bio-Rad, Munich, Germany). Disulfide-containing peptides were generated as described previously.25
Fluorescence spectrometry
Measurement of binding affinities based on the interaction of the peptides with aromatic residues (predominantly W; single-letter amino acid code) in the SH3 domains was done as described previously26 on an LS50B spectrometer (Perkin Elmer) with a water-cooled cuvette chamber. We were unable to obtain usable results with the W-containing peptides because of the high background fluorescence of the peptides. These peptides were therefore initially characterized by functional assays described previously.26The disulfide bond–containing peptides (Table1) could be measured only before coupling to the W-containing Antp sequence.
Sequences and SH3 affinity measurements with W fluorescence of peptides used in this study
| Nomenclature . | Sequence . | Kd (μM) Grb2SH3(N) . |
|---|---|---|
| P1 (hu. SoS1 aa 1152-1162) | VPPPVPPRRRP-amide | 11.63 ± 2.44 |
| P2 (HAGBP) | PPPPLPPRRRR-amide | 2.72 ± 0.18 |
| P3 (Antp) | GTERQIKIWFPNRRMKWKKEN | NA |
| P4 (Antp-HAGBP) | GPPPPLPPRRRR-GTERQIKIWFPNRRMKWKKEN-amide | NA |
| P5 (Antp-Ctrl. LL) | GPPPPLPPLLRR-GTERQIKIWFPNRRMKWKKEN-amide | NA |
| P6 (Antp-SS-HAGBP) | KKWKMRRNPFWIKIQRC-CGIRVVDNSPPPPLPPRRRRSAPSPTRV-amide | 0.78 ± 0.03* |
| P7 (Antp-SS-Ctrl. GGLL) | KKWKMRRNPFWIKIQRC-CGIRVVDNSPPGALGPLLRRSAPSPTRV-amide | NB* |
| Nomenclature . | Sequence . | Kd (μM) Grb2SH3(N) . |
|---|---|---|
| P1 (hu. SoS1 aa 1152-1162) | VPPPVPPRRRP-amide | 11.63 ± 2.44 |
| P2 (HAGBP) | PPPPLPPRRRR-amide | 2.72 ± 0.18 |
| P3 (Antp) | GTERQIKIWFPNRRMKWKKEN | NA |
| P4 (Antp-HAGBP) | GPPPPLPPRRRR-GTERQIKIWFPNRRMKWKKEN-amide | NA |
| P5 (Antp-Ctrl. LL) | GPPPPLPPLLRR-GTERQIKIWFPNRRMKWKKEN-amide | NA |
| P6 (Antp-SS-HAGBP) | KKWKMRRNPFWIKIQRC-CGIRVVDNSPPPPLPPRRRRSAPSPTRV-amide | 0.78 ± 0.03* |
| P7 (Antp-SS-Ctrl. GGLL) | KKWKMRRNPFWIKIQRC-CGIRVVDNSPPGALGPLLRRSAPSPTRV-amide | NB* |
HAGBP indicates high-affinity Grb2-binding peptide; NA, not applicable (W in shuttle); NB, no binding.
Measured without W-containing shuttle sequence.
Cell culture and protein complex formation inhibition
Human Bcr-Abl(p210)–positive CML cells K562 (American Type Culture Collection) and BV173 were cultured in RPMI 1640 with 5% fetal bovine serum (FBS). B15 cells (Bcr-Abl–positive human acute lymphoblastic leukemia [ALL]27) were supplemented with 15% FBS. 32D (mouse myeloid cells) and 32D-p210Bcr-Ablcells28 were cultured with 10% FBS. Ba/F3 and 32D cells were also supplemented with interleukin (IL)-3–containing conditioned WEHI-3 medium. Cell extracts were prepared by solubilization in mixed-micelle RIPA buffer (20 mM Tris HCl, pH 7.5, 100 mM NaCl, 1 mM Na2EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecylsulfate [SDS]) containing protease and phosphatase inhibitors (20 μg/mL aprotinin, 10 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 10 μM antipain, 10 μM pepstatin, 1 mM sodium molybdate, and 1 mM sodium orthovanadate).
NIH3T3 fibroblast cells expressing p210Bcr-Abl14 (gift of J. Y. J. Wang, University of California San Diego [UCSD]) or v-Raf (cells stably expressing EHneoGag–v-Raf; kindly provided by J. Troppmair, Würzburg) were grown in Dulbecco modified Eagle medium containing 5% FBS. Petri dishes and flasks for culture of NIH3T3 cells expressing p210Bcr-Abl were coated with poly-D-lysine (Roche, Mannheim, Germany) according to the manufacturer's instructions. Therefore, poly-D-lysine powder was dissolved in 1× phosphate-buffered saline (PBS) at 10 μg/mL. For coating, the suspension (50 μL per 1 cm2) was incubated at room temperature for 5 minutes. After aspiration of the unbound poly-D-lysine, the coated surfaces were washed once with PBS.
For inhibition assays with living cells, K562 and 32D cells were washed and resuspended in medium. Cells at 1 × 105 were seeded into 24-well plates in 0.2 mL medium. The peptide solution, which had been mixed with 0.2 mL of medium, was then slowly added to the cells. On the next day, cells were again treated with peptide. Evaluation on day 3 was done by counting the viable cells after staining with trypan blue dye. Activity of MAPK was evaluated with activation-specific anti–phospho-MAPK (#9101; New England Biolabs) following the manufacturer's instructions. Total MAPK protein was visualized with a polyclonal anti-MAPK rabbit antiserum (anti-Erk1, sc-93; Santa Cruz, Heidelberg, Germany).
Inhibition assays with adherent NIH3T3 cells were in principle performed after the assays described for K562 and 32D cells. Cells at 1 × 105 were seeded into 24-well plates (in the case of Bcr-Abl–expressing cells, wells were precoated with poly-D-lysine) in 0.5 mL medium. The next day, the cells were washed once with 0.5 mL PBS, and peptide application was carried out as before.
Immunoprecipitation and immunoblotting
Grb2 was immunoprecipitated from whole-cell lysates (500 μg) by incubation with 4 μg of an anti-Grb2 rabbit polyclonal antibody (sc-255; Santa Cruz) in IP buffer (20 mM Tris HCl, pH 7.5, 1 mM Na2EDTA, 100 mM NaCl, 5% glycerol, 0.1% Tween 20) containing protease and phosphatase inhibitors (described earlier) for 2 hours at 4°C. The immune complexes were precipitated by incubation with protein A–sepharose for 1 hour at 4°C. The precipitates were washed 3 times with 0.5% Triton X-100/Tris-buffered saline (20 mM Tris, pH 7.5, 150 mM NaCl) and resuspended in sample buffer. After SDS-polyacrylamide gel electrophoresis (PAGE) and blotting, p210Bcr-Abl, SoS, and Grb2 were detected with monoclonal antibodies for Abl (8E9; gift of J. Y. J. Wang, UCSD), SoS (S15520; Transduction Laboratories), or Grb2 (G16720; BD, Heidelberg, Germany).
In vitro kinase assays
K562 cells were treated with peptide as described earlier. On day 3, the cells were lysed in mixed-micelle RIPA buffer (20 μg/mL aprotinin, 10 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 10 μM antipain, 10 μM pepstatin, 1 mM sodium molybdate, 1 mM sodium orthovanadate, 50 mM sodium fluoride). MAPK was immunoprecipitated from 250 μg cell lysate with 1 μg of an anti-Erk rabbit polyclonal antibody (sc-154; Santa Cruz) in IP buffer with protease inhibitors (described earlier) and phosphatase inhibitors (1 mM sodium orthovanadate, 1 mM sodium fluoride, 3 mM β-glycerophosphate) overnight at 4°C and subsequently by incubation with protein A–sepharose for 1 hour at 4°C. The precipitated kinase was washed twice with IP buffer and once with kinase buffer (5 mM MgCl2, 5 mM MnCl2, 20 mM HEPES, pH 7.5, 100 mM NaCl, 20 mM sodium β-glycerophosphate, 1 mM dithiothreitol, 1 mM sodium orthovanadate). The samples were then incubated with 20 μg myelin basic protein (MBP; Life Technologies, Karlsruhe, Germany) as kinase substrate in the presence of 500 nM adenosine triphosphate (ATP), 5 μCi γ-32P-ATP (3000 Ci/mmol), and kinase buffer in a total volume of 50 μL for 20 minutes at 30°C. Reactions were stopped by the addition of SDS-PAGE sample buffer. After SDS-PAGE (12% gel), kinase activity was quantified with the Bio-Imaging BAS 2000 analyzer (Fuji, Düsseldorf, Germany) and calculated with the software TINA 2.09d (Raytest Isotopenmeßgeräte GmbH; Germany). Subsequently, the gel was subjected to autoradiography with an X-ray film.
Differentiation of K562 cells with Ara-C
K562 cells were washed, and 2.5 × 104 cells were seeded into 24-well plates in 0.4 mL medium. Thereafter, arabinosylcytosine (Ara-C) was added to a final concentration of 0.70 μM. On days 3 and 5, cells were diluted with 0.4 mL medium and again treated with Ara-C at 0.70 μM. On day 6, hemoglobin-producing cells were scored by benzidine staining.29 For this, 20 μL of cell suspension was diluted with 35 μL medium, and 10 μL of a benzidine solution (benzidine dihydrochloride 2 mg/mL in 0.5% acetic acid) mixed with 5 μL of a 30% hydrogen peroxide solution was added. For determining the influence of the peptides on erythroid differentiation, we treated K562 cells (2.5 × 104) with the peptides P6 (Antp-SS-HAGBP) and P7 (Antp-SS-Ctrl GGLL) at a daily concentration of 5 or 10 μM for up to 10 days.
Isolation and peptide treatment of CML patient cell preparations enriched for blast cells
Peripheral blood from CML patients with active disease was diluted with one volume of PBS. Ten milliliters of this diluted blood was layered onto 5 mL of Histopaque-1077 (#1077-1; Sigma, Deisenhofen, Germany). A low-density gradient was obtained by centrifugation in a swing-out rotor (400g, 20 minutes, 20°C). The low-density cell fraction enriched for blasts was aspirated with a pipette. Cells were then washed twice with PBS (2000g, 10 minutes, 4°C) and incubated with DNase I (20 μg/mL, D4513; Sigma) for 30 minutes at 37°C in RPMI 1640 with 10% FBS to fragment DNA from dead cells, which may bind with residual affinity to the Antp peptides. Cell-penetrating peptides (20 μM) were then added as described earlier for K562 cells.
RNA extraction and reverse transcription–polymerase chain reaction for Bcr-Abl transcripts
Total RNA was isolated using Trizol Reagent (Life Technologies, Gaithersburg, MD) following the manufacturer's directions. RNA was quantitated, aliquoted, and stored at −70°C. A total of 19 μL (1.5-6.5 μg) of RNA was heated to 65°C for 10 minutes and then placed on ice. A total of 21 μL of reverse transcription reagents, including 95 mM Tris (pH 8.3); 142.5 mM KCl; 5.7 mM MgCl2; 19 mM dithiothreitol; 19 mM each of dATP, dCTP, dTTP, and dGTP; 200 μg/mL random hexamers pdN6; 1.4 × 104 U/mL M-MLV reverse transcriptase (Life Technologies); and 1400 U/mL Rnasin (Roche); were added to the samples and incubated at 37°C for 2 hours. Heating to 65°C for 10 minutes terminated the reaction, and cDNA was stored at −20°C until assayed.
The Bcr-Abl(p210) competitor was kindly provided by Drs Cross and Feng.30 Dilutions were made in the range from 107 to 10 molecules per 2.5 μL, with steps at every half order of magnitude on a logarithmic scale. Competitive nested polymerase chain reaction (PCR) to amplify b2a2, b3a2, and e19a2 Bcr-Abl types was also performed as described previously by Cross et al.30 c-Abl mRNA was quantitated in all samples as an internal standard to assess the amount of cDNA and the quality of RNA. Reaction products were subjected to electrophoresis on a 1.5% agarose gel, and bands were visualized under ultraviolet light with ethidium bromide staining. Because of the size difference between the competitor and Bcr-Abl, the number of molecules of competitor at the equivalence point was multiplied by 1.2 (559 bp/458 bp) for b2a3 and 1.5 (559 bp/383 bp) for b2a2 to derive the number of Bcr-Abl molecules in the sample. The results were expressed as the percentage ratio between Bcr-Abl and c-Abl.
Results
Generation of cell-penetrating high-affinity Grb2SH3(N) blocking peptides (HAGBPs)
The disruption of SH3 domain–dependent protein complexes is thought to be an important strategy to inhibit specific signal-transduction pathways, and various laboratories including ours have done molecular studies in this context (4,26,31-33and references therein). One prominent target is the Grb2-SoS complex, which is located upstream of the small GTPase Ras in the central mitogenic cascade. Ras has been implicated in Bcr-Abl signaling by many studies,13,18 34-36 although its role in human CML is not fully clarified. In this study, we investigated whether uncoupling of Grb2-SoS complexes affects the proliferation of CML cells.
A general problem when using short peptides to block SH3 domains is the low affinity of the naturally occurring binding motifs. This problem can be overcome by the use of nonnatural amino acid analogues33 or the introduction of suitable mutations in the binding peptide sequences.26 Using the latter strategy, we could show that mutation of the naturally occurring Grb2SH3(N) binding motif of the Ras-specific guanine nucleotide exchange factor SoS (designated P1, Table 1) results in a peptide (P2) with markedly improved affinity. The binding of this high-affinity Grb2SH3(N) binding peptide (HAGBP) can be further increased by elongating the peptide chain (compare P2 and P6, Table 1), presumably allowing additional contacts of the peptide with the Grb2SH3(N) domain. The details of this flank elongation have been described elsewhere.37 The affinities were measured by tryptophan fluorescence spectrometry, a method frequently used to quantify SH3 domain interactions.26,31 38 Because SH3 domains often have conserved W residues, which contact the specific binding peptide, this method is widely applicable. However, it is feasible only for peptides void of W residues.
Because most peptides are unable to penetrate cellular membranes, it is necessary to attach the Grb2SH3(N) blocker peptide to a shuttle system that allows the efficient accumulation of the peptide in the desired compartment of the target cells. Probably the best-characterized system that has been successfully used by numerous groups in recent years involves peptides derived from the third helix of the transcription factor Antennapedia.39 The wild-type form of this peptide accumulates strongly in the cell nucleus because helix 3 is a DNA-binding motif. By mutating a single residue in this sequence from Q to P, the DNA binding is largely abolished and the resulting peptide accumulates prominently in the cytoplasm. Because Grb2-SoS complexes are largely cytosolic, for this study the P-mutant Antp was used (P3, Table 1). Two different types of coupling between the shuttle and the SH3 blocker peptide were done and subsequently compared for biologic activity. Peptides P4 and P5 were both synthesized as a continuous peptide chain, whereas peptides P6 and P7 were generated through coupling of shuttle and blocker peptide via a disulfide bond. Point-mutant control peptides with abolished Grb2SH3(N) binding (P5 and P7) were also made, based on the detailed ultrastructural studies available.40-42 It is noteworthy that the Antp sequence can be used in forward and reverse orientation without loss of biologic activity.39 Our initial studies were carried out with a 21–amino acid Antp sequence (P3-5). However, because it was later shown that 16 amino acids are fully sufficient for Antp-shuttle activity,39 this shorter Antp form, plus a C necessary for coupling of the cargo peptide, was subsequently used (P6 and P7).
HAGBPs inhibit proliferation of K562 CML cells
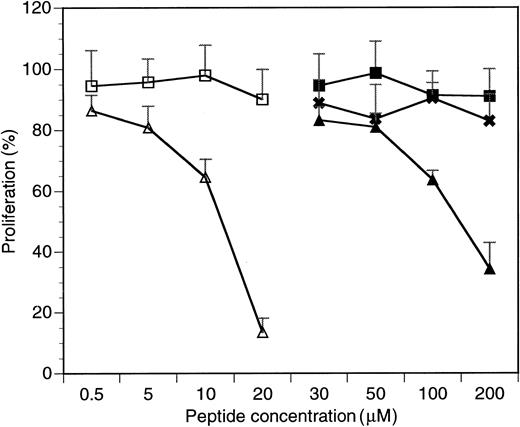
To study the biological activity of the cell-penetrating HAGBPs, K562 cells,43 a well-studied CML cell line expressing p210Bcr-Abl, was initially chosen. Peptides were added onto growing K562 cells twice, in each case followed by a 24-hour incubation period. The stabilities of SH3 domain blocker peptides in cells have been tested previously37 and range between 12 and 72 hours, depending on the cell type analyzed. After a total incubation time of 48 hours, viable cells were counted. The results are summarized in Figure 1. Cells grown without peptide were set to 100% proliferation. The results show that Antp peptide alone (P3) or point-mutated control peptides (P5, P7) do not significantly affect the proliferation of K562 cells. By contrast, the Antp-coupled Grb2SH3(N) blocker peptides (P4 and P6) strongly reduced K562 cell proliferation. Similar effects were also seen with the CML and ALL cell lines BV173 and B15 (data not shown).
Inhibitory effects of HAGBPs on proliferation of the K562 CML blast cell line.
Proliferation without peptide addition was set to 100%. Peptides were added as detailed in Materials and methods. Continuous peptides without disulfide bond (P3-P5) were effective only above 50 μM. Therefore, concentrations below 30 μM are not shown here. Disulfide-bridged peptides were much more potent and therefore extensively tested in a concentration range between 0.5 and 20 μM as shown. Error bars indicate SEM (n = 6 to 9). ■ indicates P7 (Antp-SS-Ctrl GGLL); ▵, P6 (Antp-SS-HAGBP); ▪, P3 (Antp); ✖, P5 (Antp-Ctrl LL); and ▴, P4 (Antp-HAGBP).
Inhibitory effects of HAGBPs on proliferation of the K562 CML blast cell line.
Proliferation without peptide addition was set to 100%. Peptides were added as detailed in Materials and methods. Continuous peptides without disulfide bond (P3-P5) were effective only above 50 μM. Therefore, concentrations below 30 μM are not shown here. Disulfide-bridged peptides were much more potent and therefore extensively tested in a concentration range between 0.5 and 20 μM as shown. Error bars indicate SEM (n = 6 to 9). ■ indicates P7 (Antp-SS-Ctrl GGLL); ▵, P6 (Antp-SS-HAGBP); ▪, P3 (Antp); ✖, P5 (Antp-Ctrl LL); and ▴, P4 (Antp-HAGBP).
A remarkable difference in K562 cell growth inhibition potency was also observed between peptides P4 and P6. The disulfide bond–coupled Antp-SS–HAGBP (P6) was already highly effective at 20 μM, whereas with the continuous-chain peptide Antp-HAGBP (P4), 10-fold higher concentrations were required to achieve a strong inhibitory effect. Therefore, the disulfide-coupled peptides P6 and P7 were used in all further studies. A simple explanation for this observed difference could be that the continuous-chain peptides (P4 and P5) permanently shuttle in and out of the cells, generating a concentration equilibrium of the SH3 blocker peptide between cell culture medium and cytoplasm. By contrast, the disulfide bond in peptides P6 and P7 can open up under the reducing conditions found inside cells, still allowing the shuttle peptide to leave but trapping its cargo inside the cell. This probably leads to an intracellular accumulation of HAGBPs above the concentration in the culture medium.
During the course of the experiments, we did not observe signs of increased apoptosis by cell morphology criteria (eg, membrane blebbing). Annexin V staining experiments for apoptosis-related cell membrane changes were also negative (data not shown).
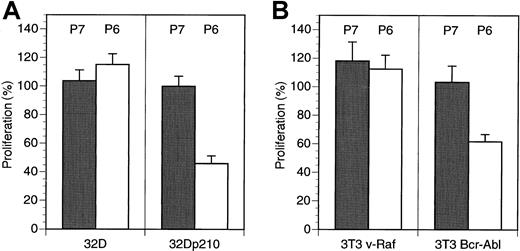
To further confirm the specific nature of the antiproliferative effects seen with the HAGBPs, we analyzed p210Bcr-Abl-expressing 32D cells and parental 32D cells. 32D cells normally require IL-3 for survival and proliferation. By contrast, p210Bcr-Abl-expressing cells grow in the absence of IL-3 because the oncoprotein generates a strong proliferative stimulus, which is thought to result from the activation of the Ras-MAPK (Erk) pathway.13,18 34-36 Incubation of 32D-p210Bcr-Abl cells (grown without IL-3) but not of parental 32D cells (grown with IL-3) with Antp-SS–HAGBP (P6) led to a strong reduction of proliferation. This effect was not seen with the control peptide (P7), which does not bind to the Grb2SH3(N) domain (Figure 2A).
HAGBP affects Bcr-Abl–dependent proliferation of 32D and NIH3T3 cells, but not the proliferation of 32D cells in the presence of IL-3 or v-Raf–expressing NIH3T3 cells.
(A) A total of 105 IL-3–dependent 32D mouse myeloid cells grown in the presence of IL-3 or the same number of IL-3–independent Bcr-Abl–expressing 32D cells (32D-p210Bcr-Abl) grown without IL-3 were incubated with 20 μM inactive control peptide (P7) or Antp-SS–HAGBP (P6) as indicated. Peptide was applied twice, once initially and once after 24 hours. Viable cells were counted after 48 hours. Error bars indicate SEM (n = 6). One hundred percent corresponds to approximately 6.5 × 105 cells obtained in the absence of exogenously added peptide at the end of the incubation period. (B) A total of 105 3T3 v-Raf cells or 3T3 Bcr-Abl cells were grown in the presence of 5% FBS without peptide addition or with 20 μM of the peptides indicated (P6 = Antp-SS–HAGBP; P7 = control peptide). One hundred percent corresponds to approximately 2 × 105 untreated 3T3 v-Raf cells or approximately 3.5 × 105 3T3 Bcr-Abl cells at the end of the incubation period.
HAGBP affects Bcr-Abl–dependent proliferation of 32D and NIH3T3 cells, but not the proliferation of 32D cells in the presence of IL-3 or v-Raf–expressing NIH3T3 cells.
(A) A total of 105 IL-3–dependent 32D mouse myeloid cells grown in the presence of IL-3 or the same number of IL-3–independent Bcr-Abl–expressing 32D cells (32D-p210Bcr-Abl) grown without IL-3 were incubated with 20 μM inactive control peptide (P7) or Antp-SS–HAGBP (P6) as indicated. Peptide was applied twice, once initially and once after 24 hours. Viable cells were counted after 48 hours. Error bars indicate SEM (n = 6). One hundred percent corresponds to approximately 6.5 × 105 cells obtained in the absence of exogenously added peptide at the end of the incubation period. (B) A total of 105 3T3 v-Raf cells or 3T3 Bcr-Abl cells were grown in the presence of 5% FBS without peptide addition or with 20 μM of the peptides indicated (P6 = Antp-SS–HAGBP; P7 = control peptide). One hundred percent corresponds to approximately 2 × 105 untreated 3T3 v-Raf cells or approximately 3.5 × 105 3T3 Bcr-Abl cells at the end of the incubation period.
Moreover, NIH3T3 mouse fibroblasts stably expressing a constitutively active v-Raf kinase protein were insensitive to Antp-SS–HAGBP (P6) and control peptide (P7). This can be expected if HAGBP indeed primarily interferes with the Grb2-SoS-Ras-c-Raf-1 signaling cascade (Figure 2B, left side). Interestingly, NIH3T3 cells stably expressing the active p210Bcr-Abl kinase were at least partially dependent on Bcr-Abl for proliferation under conditions in which serum growth factors were present in suboptimal amounts (Figure 2B, right side). This is in agreement with previously published results documenting the dependence of p210Bcr-Abl-expressing NIH3T3 cells on Grb2 for efficient proliferation.14
HAGBPs disrupt Grb2-SoS complexes and lead to reduced MAPK activity
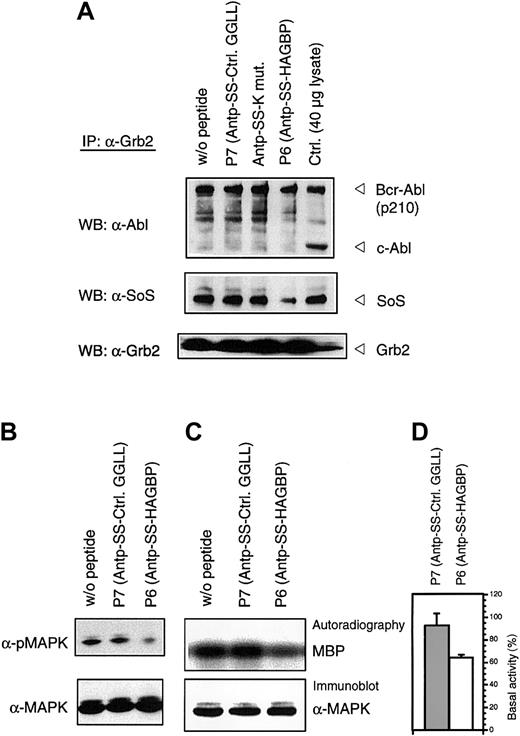
To verify that the HAGBPs reach their target in the cells, we performed coimmunoprecipitation studies. K562 cells were incubated with 10 μM peptide for 48 hours as indicated. This lower concentration was used because a 20-μM application of HAGBP (P6) leads to a complete block of proliferation, resulting in very little protein lysate for the analyses. Grb2-SoS complexes were precipitated from the lysates as described previously.44 Incubation of cells with 10 μM HAGBP led to a 35% reduction in proliferation (Figure 1) and a significant reduction of detectable Grb2-SoS complexes (Figure 3A, middle panel). The control peptide P7 (Table 1) or a single-point–mutant peptide (Antp-SS–K mut) in which the crucial R in the P-x-x-P-x-R Grb2SH3(N) core binding motif is replaced by K did not significantly affect the Grb2-SoS complexes. This mutation to K abolishes the binding to the Grb2SH3(N) domain.26 As expected, complexes between Grb2 and Bcr-Abl that depend on the SH2 domain of Grb2 and Y177 in the Bcr portion of the Bcr-Abl protein14 45 were not affected (Figure 3A, top panel), indicating the specificity of the peptide effect observed. Moreover, we noted a clear reduction of phosphorylated, active MAPK in HAGBP-treated cells but not in control cells (Figure 3B).
HAGBP disrupts Grb2-SoS complexes and reduces MAPK activity.
K562 cells were incubated as detailed in Materials and methods but with only 10 μM of the peptide indicated. This leads to a significant reduction of proliferation in the case of peptide P6 (HAGBP; Figure 1) but still allows some growth, which is essential to yield enough protein for the precipitation experiments. (A) Immunoprecipitation (IP) and Western blot (WB) were carried out with the antibodies indicated as described in Materials and methods. A total of 40 μg of total K562 lysate from untreated cells was also loaded as a control. Note that complexes of Grb2 are detectable with p210Bcr-Abl, but not with the normal cellular p145c-Abl protein. (B) Total lysates of K562 cells treated as in (A) were separated by SDS-PAGE, blotted, and probed with anti-MAPK or activation-specific phospho-MAPK antibodies, as described in Materials and methods. (C) A total of 250 μg of K562 total cell protein from cells treated with 10 μM of peptide, where indicated, was precipitated with anti-MAPK antibody in the presence of protease and phosphatase inhibitors to maintain the kinase activity state from the time point of cell lysis. After precipitating anti-MAPK with protein A–sepharose and removing unbound proteins by several wash steps, samples were analyzed by in vitro kinase assay with MBP as substrate. A representative experiment is shown. (D) Statistical analysis of 4 independent in vitro kinase experiments with lysates of cells treated with 10 μM of the indicated peptides. Incorporated radioactivity was determined by phosphoimager and quantified using the TINA 2.09d software.
HAGBP disrupts Grb2-SoS complexes and reduces MAPK activity.
K562 cells were incubated as detailed in Materials and methods but with only 10 μM of the peptide indicated. This leads to a significant reduction of proliferation in the case of peptide P6 (HAGBP; Figure 1) but still allows some growth, which is essential to yield enough protein for the precipitation experiments. (A) Immunoprecipitation (IP) and Western blot (WB) were carried out with the antibodies indicated as described in Materials and methods. A total of 40 μg of total K562 lysate from untreated cells was also loaded as a control. Note that complexes of Grb2 are detectable with p210Bcr-Abl, but not with the normal cellular p145c-Abl protein. (B) Total lysates of K562 cells treated as in (A) were separated by SDS-PAGE, blotted, and probed with anti-MAPK or activation-specific phospho-MAPK antibodies, as described in Materials and methods. (C) A total of 250 μg of K562 total cell protein from cells treated with 10 μM of peptide, where indicated, was precipitated with anti-MAPK antibody in the presence of protease and phosphatase inhibitors to maintain the kinase activity state from the time point of cell lysis. After precipitating anti-MAPK with protein A–sepharose and removing unbound proteins by several wash steps, samples were analyzed by in vitro kinase assay with MBP as substrate. A representative experiment is shown. (D) Statistical analysis of 4 independent in vitro kinase experiments with lysates of cells treated with 10 μM of the indicated peptides. Incorporated radioactivity was determined by phosphoimager and quantified using the TINA 2.09d software.
The reduction of MAPK activity indicated by the phospho-specific antibodies was further analyzed by in vitro kinase assay with MBP as substrate (Figure 3C,D). Again, a highly reproducible reduction of MAPK activity was observed.
Minimal effect of HAGBPs on erythroid differentiation of K562 cells
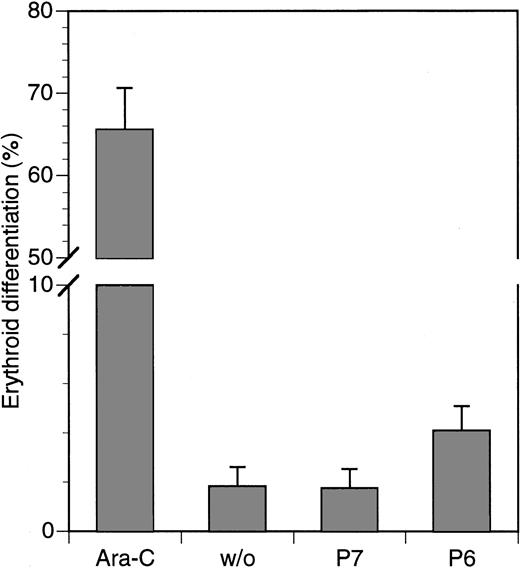
It has been reported previously that the inhibition of the Ras-MAPK pathway in K562 cells can lead to their differentiation along the erythroid lineage (29 and references therein). To determine whether this occurs with the HAGBPs, we treated cells for 5 days with HAGBP (P6) or control peptide (P7) at a concentration of 5 μM. In parallel, 0.70 μM Ara-C, a strong inducer of erythroid differentiation in K562 cells, which is also clinically relevant for CML therapy, was used as a positive control. Cells were stained with benzidine for hemoglobin, and positive cells were counted. Among Ara-C–treated cells, 70% were positive for hemoglobin, whereas only 2% of the untreated cells or cells treated with control peptide (P7) were positive. Of the HAGBP-treated cells, approximately 4% stained positive (Figure 4). Hence, only a slight but statistically significant elevation of differentiating cells was observed under these experimental conditions. In additional experiments, various other concentrations or longer incubation periods were tested. We were, however, unable to find any major effect of HAGBP or control peptide on erythroid differentiation of K562 cells (data not shown).
Marginal effect of HAGBP (P6) on erythroid differentiation of K562 cells.
Cells were incubated with 5 μM of peptides indicated or Ara-C, a well-known inducer of erythroid differentiation in K562 cells, for 5 days with daily application of the peptides. Ara-C was added on days 1, 3, and 5. Hemoglobin-positive cells were detected by benzidine stain. Error bars indicate SEM (n = 6).
Marginal effect of HAGBP (P6) on erythroid differentiation of K562 cells.
Cells were incubated with 5 μM of peptides indicated or Ara-C, a well-known inducer of erythroid differentiation in K562 cells, for 5 days with daily application of the peptides. Ara-C was added on days 1, 3, and 5. Hemoglobin-positive cells were detected by benzidine stain. Error bars indicate SEM (n = 6).
HAGBPs reduce proliferation of primary CML cells in the majority of patients
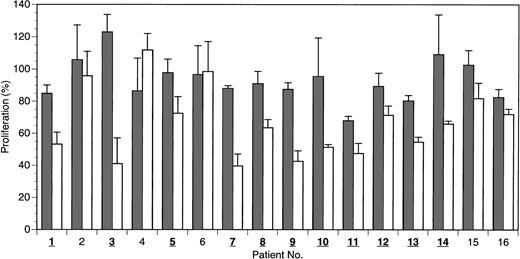
Because it is not obvious how well CML cell lines in long-term culture reflect the conditions found in CML cells from patients, a subsequent set of experiments was conducted using freshly isolated cell preparations from patients with active disease, which were enriched for CML blast cells. The patients were mostly undergoing different standard or experimental treatments, as indicated in Table2. Most patients had been under therapy for several years. The presence of the bcr-abloncogene was clearly confirmed by nested PCR in most cases. Leukocyte counts of the patients were generally elevated, ranging from 4300 (nearly normal) to 80 000/μL. A blast-enriched cell fraction was isolated by density gradient centrifugation from whole blood and equilibrated in culture medium under standard conditions (37°C, 5% CO2) for 12 to 24 hours before peptide application. Then 20 μM of HAGBP (P6), control peptide (P7), or no peptide was added. Viable cells were counted after 48 hours. The results are summarized in Figure 5. A significant reduction of the proliferation rate with HAGBP versus control peptide was observed in most but not all patients. For example, cells from patient no. 6 were clearly fully resistant to treatment with HAGBP, whereas cells from patients 3, 7, and 9 showed a dramatic decrease of cell proliferation. These individually different outcomes may reflect the molecular heterogeneity of the CML cells in the late stages of the disease.5
Characterization of CML patients
| Patient no. . | Therapy . | Bcr-Abl transcripts* (nested PCR) . | Fusion transcripts (multiplex PCR) . | Leukocytes (cells/μL) . |
|---|---|---|---|---|
| 1 | No tolerance of HU and Ara-C (currently w/o therapy) | 1.70 × 105 | b2a2+ | 26 000 |
| 2 | IFN | 1.27 × 105 | b3a2+ | NA |
| 3 | IFN + Ara-C | 1.70 × 105† | b2a2+ | NA |
| 4 | PEG IFN → hepatic toxicity | 5.48 × 104 | b3a2+ | 5 800 |
| 5 | PEG IFN | 2.40 × 105 | b3a2+ | 51 000 |
| 6 | PEG IFN | ND | ND | 6 800 |
| 7 | PEG IFN | 9.90 × 104 | b2a2+/− | 11 000 |
| 8 | PEG IFN | 2.99 × 105 | b3a2+ | 61 300 |
| 9 | PEG IFN + Ara-C | 3.97 × 105 | b3a2+ | 20 600 |
| 10 | No tolerance of IFN → PEG IFN → no effect | 3.60 × 105 | b3a2+ | NA |
| 11 | Resistance to IFN → PEG IFN | 2.20 × 104 | b3a2+ | 4 300 |
| 12 | No tolerance of IFN → PEG IFN | 3.10 × 105 | b3a2+ | 11 800 |
| 13 | HU | 6.78 × 104 | b2a2+ | 10 000 |
| 14 | HU | 4.03 × 105 | b3a2+ | 6 900 |
| 15 | Busulfan | 2.17 × 105 | b3a2+ | 80 000 |
| 16 | CGP57148B (STI571) → no effect | 9.03 × 104 | b3a2+ | 41 000 |
| Patient no. . | Therapy . | Bcr-Abl transcripts* (nested PCR) . | Fusion transcripts (multiplex PCR) . | Leukocytes (cells/μL) . |
|---|---|---|---|---|
| 1 | No tolerance of HU and Ara-C (currently w/o therapy) | 1.70 × 105 | b2a2+ | 26 000 |
| 2 | IFN | 1.27 × 105 | b3a2+ | NA |
| 3 | IFN + Ara-C | 1.70 × 105† | b2a2+ | NA |
| 4 | PEG IFN → hepatic toxicity | 5.48 × 104 | b3a2+ | 5 800 |
| 5 | PEG IFN | 2.40 × 105 | b3a2+ | 51 000 |
| 6 | PEG IFN | ND | ND | 6 800 |
| 7 | PEG IFN | 9.90 × 104 | b2a2+/− | 11 000 |
| 8 | PEG IFN | 2.99 × 105 | b3a2+ | 61 300 |
| 9 | PEG IFN + Ara-C | 3.97 × 105 | b3a2+ | 20 600 |
| 10 | No tolerance of IFN → PEG IFN → no effect | 3.60 × 105 | b3a2+ | NA |
| 11 | Resistance to IFN → PEG IFN | 2.20 × 104 | b3a2+ | 4 300 |
| 12 | No tolerance of IFN → PEG IFN | 3.10 × 105 | b3a2+ | 11 800 |
| 13 | HU | 6.78 × 104 | b2a2+ | 10 000 |
| 14 | HU | 4.03 × 105 | b3a2+ | 6 900 |
| 15 | Busulfan | 2.17 × 105 | b3a2+ | 80 000 |
| 16 | CGP57148B (STI571) → no effect | 9.03 × 104 | b3a2+ | 41 000 |
HU indicates hydroxyurea; Ara-C, arabinosylcytosine; IFN, interferon-alpha; NA, not available; PEG, pegylated; ND, not detectable (fewer than 103 transcripts); CGP57148B (STI571), Abl kinase inhibitor.
Transcripts per microgram RNA.
Detected with multiplex PCR.
HAGBP inhibits proliferation of freshly isolated preparations enriched with blast cells from human CML patients.
Blast cells were isolated from 16 patients with active disease by density gradient centrifugation and equilibrated for 16 to 24 hours in culture medium at 37°C before peptide application. Peptides were added initially at a concentration of 20 μM, and the same dose was added again after 24 hours. Viable blast cells were counted after 48 hours. Blast cell number without peptide addition equals 100% proliferation. Error bars indicate SEM (n = 3 to 5). Isolates in which a significant inhibitory effect is seen on proliferation with HAGBP (P6; white bars) versus control peptide (P7; gray bars) are indicated by bold and underlined numbers.
HAGBP inhibits proliferation of freshly isolated preparations enriched with blast cells from human CML patients.
Blast cells were isolated from 16 patients with active disease by density gradient centrifugation and equilibrated for 16 to 24 hours in culture medium at 37°C before peptide application. Peptides were added initially at a concentration of 20 μM, and the same dose was added again after 24 hours. Viable blast cells were counted after 48 hours. Blast cell number without peptide addition equals 100% proliferation. Error bars indicate SEM (n = 3 to 5). Isolates in which a significant inhibitory effect is seen on proliferation with HAGBP (P6; white bars) versus control peptide (P7; gray bars) are indicated by bold and underlined numbers.
At this point, we cannot fully exclude the possibility that impurities from nonblast white blood cells in the blast-enriched cell preparations tested here reduce the observed antiproliferative effect of HAGBP, leading to an underestimation of its biologic activity. The number of postmitotic, differentiated nonblast cells is usually small in patients with leukocyte counts much higher than 5000/μL, but in some of the patient samples (for leukocyte counts see Table 2), we cannot rule out that significant numbers of nonblast cells were present in the blast-enriched cell preparations. However, because of very limited availability of CML patient blood, we were unable to investigate homogeneous blast cell preparations. Future studies with pure blast cells are therefore highly desirable.
Discussion
In this study, we documented that cell-penetrating peptides that bind to the first SH3 domain of Grb2 can strongly inhibit the proliferation of CML blast cells. Analyzing freshly isolated blast cell preparations from patients, we observed that many, but not all, patients' cells were responsive to these peptides. This is not surprising if one takes into consideration the heterogeneous nature of CML.5 During the accelerated phase of CML and in the late stages of the disease, it can be expected that patients acquire multiple mutations in clones of their blast cells, which can, for example, affect components of the central mitogenic cascade SoS-Ras-Raf-MEK-MAPK/Erk. In such a case, the inability of a peptide that disrupts Grb2-SoS complexes to have a biologic effect on proliferation is no surprise.
Besides the expected down-regulation of Grb2-SoS complexes and MAPK activity, it was observed that v-Raf–transformed NIH3T3 mouse fibroblast cells were not proliferation-inhibited in the presence of HAGBP. This was also expected because Raf is a downstream target of Ras. Moreover, Bcr-Abl expression in NIH3T3 cells rendered these cells partially sensitive to HAGBPs under suboptimal concentrations of serum growth factors, a finding again expected from previous reports in the literature.14
It is very clear from a large body of literature that the Grb2-SoS-Ras pathway is used not only by the bcr-abloncogene, but also by many growth factor receptors. The Grb2SH3 domain blocker peptides tested here are therefore likely to affect other signaling systems present in CML cells that also depend on the Grb2-SoS interaction. Cell type–specific targeting of Grb2-SoS inhibitors into CML or other cancer cells may thus be a necessity for future blocker molecules to have clinical value.
Grb2SH2 and -SH3 domain blocking peptides or compounds have already shown biologic activity in other systems and are useful tools for basic research studies in living cells. Several of these studies have used cell-penetrating shuttle peptides to transport the domain-specific blocker peptides into cells, rather than using microinjection techniques, in which only small numbers of cells can be analyzed. For example, Rojas et al46 have used a Grb2SH2 domain blocker phosphopeptide, which mimics the Y1068 region of the activated epidermal growth factor (EGF) receptor, to inhibit the interaction of Grb2 with this receptor in fibroblasts overexpressing the receptor. Peptide application also led to a strong reduction of EGF-induced Ras activation.46 Intracellular delivery of the Grb2SH2 blocker peptide in that case was accomplished with a shuttle peptide derived from Kaposi fibroblast growth factor.47
Similarly, in myoblast cells, the EGF receptor Y1068-derived phosphopeptide inhibited MAPK activation.48 More recently, the Grb2SH2 blocker compound CGP78850 was shown to inhibit fibroblast proliferation.49
A blocker peptide thought to bind to both SH3 domains of Grb2 with an in vitro affinity of 40 nM was recently reported to inhibit the growth of Her2-transformed NIH3T3 fibroblasts in soft agar.32 In that report, 2 repeats of a single-site binding peptide corresponding to the naturally occurring Grb2SH3(N) binding motif of the SoS protein were linked by a short spacer. The single motif binds with an affinity of 16 μM to Grb2.32 This is relatively low compared with the Grb2SH3(N) binding peptides used in our study (Table 1). In addition, replacement of P residues in the core motif P-x-x-P crucial for the binding of most SH3 domains by nonnatural amino acid derivatives allows one to greatly improve the binding affinity for the Grb2SH3(N) domain.33
There is now evidence that protein sequences exist that bind with high affinity very selectively to the C-terminal Grb2 domain.57 Therefore, we should be able to create in the near future novel peptides and peptoids that bind both SH3 domains of Grb2 strongly and simultaneously, resulting in affinities in the picomolar range.
Our study is also another step toward the characterization of the molecular nature of CML in individual patients. Such analyses have not yet been done to an extent that a clear picture of the molecular events in an individual CML case can be obtained. Part of the problem is the current lack of convincing animal models for CML by transgenic expression of Bcr-Abl. These transgenic animal models usually show ALL-like phenotypes and also do not reflect the molecular heterogeneity of the naturally occurring disease. Recently improved engraftment techniques of patient CML blasts using nonobese diabetic/severe combined immunodeficiency mice (50 and references therein) are therefore more likely to be useful in this context.
Based on our current results, many interesting questions can be addressed in future studies. It will be useful to determine whether the type of blast cell in a patient (eg, lymphoid versus myeloid) or other molecular characteristics correlate with the sensitivity to Grb2SH3 blocker peptides. Furthermore, differentiation effects of the SH3 blocker peptides could be further analyzed because our current results do not allow us to absolutely exclude that under certain application conditions, a more pronounced effect can be accomplished. Such a differentiation effect could be highly beneficial for clinical therapy.
An interesting finding from our experiments, which is not directly linked to Bcr-Abl and CML, was the observation that the parental, IL-3–dependent 32D cells were apparently insensitive to Grb2-SoS blockers (Figure 2). This is suggestive of a pathway independent of Ras, which regulates proliferation in this particular cell line. One candidate pathway is the PI3 kinase-PKCε-Raf-MEK-MAPK cascade, thought to play a role in the signal transduction of erythropoietin, which is similar to IL-3 signaling in many respects.51
The long-term potential of peptide- or peptoid-based therapeutics that target intracellular signal-transduction components is still largely unclear, although successes can be expected in at least some cases.52-54 There are several methods that should help to develop the peptides currently at hand into clinically valuable molecules. One such strategy (already briefly mentioned) is cell type–specific targeting of the inhibitory peptides. This should greatly improve their biologic activities and at the same time reduce undesired effects, which could for example result from targeting the central mitogenic cascade. Cell type–specific targeting of biologically active compounds using short peptide sequences is clearly possible. Prominent examples are the tumor vasculature–specific peptides recently developed by Ruoslahti and coworkers.55 56
The value of peptide inhibitors may in some cases become most apparent when they are used in combination with other therapeutics. Several drugs, including interferon, hydroxyurea, and Ara-C, are currently in use in clinical CML therapy. Additional compounds are in clinical trials or various experimental stages6,9 and can be tested in combination with the inhibitory peptides. Some of these drugs, for example CGP57148B (STI571),22 target the tyrosine kinase activity of Bcr-Abl. By contrast, the protein–protein interaction inhibiting peptides are directed against different target sites. A combination of these inhibitors with different targets may result in synergistic therapeutic effects. As pointed out before, Grb2SH3 domain blocker peptides so far do not show activity in some CML patients' blast cells. It therefore seems likely that future therapy schedules that selectively target the individual molecular aberrations underlying each CML case will be preceded by an ex vivo sensitivity test to determine whether a Grb2SH3 domain blocker is a useful component of the therapeutic cocktail.
We are indebted to M. Warmuth (Munich) for providing the 32D-p210Bcr-Abl cells, to S. Frühauf (Heidelberg) for BV173 cells, to J. Y. J. Wang (UCSD) for Abl monoclonal antibody 8E9 and Bcr-Abl–expressing NIH3T3 cells, to J. Troppmair for v-Raf NIH3T3, and to Marc P. A. Lewitzky and Jan Voss for critical reading of the manuscript.
Supported by grants from Wilhelm-Sander-Stiftung and Deutsche Forschungsgemeinschaft (DFG-FE439).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephan M. Feller, Cell Signalling Laboratory, Imperial Cancer Research Fund, Institute of Molecular Medicine, John Radcliffe Hospital, Headley Way, Oxford, OX3 9DS, United Kingdom; e-mail: s.feller@icrf.icnet.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal