This report describes an unusual extramedullary hematologic malignancy in an 18-month-old child who presented with a capillary leak syndrome that evolved into hyperleukocytosis with malignant cells. The circulating tumor cells did not express an antigen profile typical of any subtype of leukemia commonly observed in children. Tumor cells were CD3−/CD56+; had germline TCRgenes; and strongly expressed CD30, epithelial membrane antigen, and anaplastic lymphoma kinase (ALK) consistent with a null cell anaplastic large cell lymphoma (ALCL). The malignant cells contained a t(2;19)(p23;p13.1) that interrupted ALK and translocated it to the der(19). Reverse transcriptase-polymerase chain reaction and nucleotide sequence analysis revealed fusion of ALK to tropomyosin 4, an ALK fusion partner not described previously in hematologic malignancies. The clinical presentation and phenotypic features of this malignancy were not typical for ALCL because tumor cells expressed both myeloid (CD13, CD33, HLA-DR) and natural killer (NK) cell antigens. The neoplastic cells most resembled NK cells because in addition to being CD3−/CD56+ with germline TCR genes, these cells were CD25+/CD122+/granzyme B+ and possessed the functional properties of immature NK cells. The unusual clinical presentation, immunophenotype, and functional properties of these neoplastic cells suggest that this malignancy may be derived from the putative myeloid-NK precursor cell. Furthermore co-expression of NK and ALCL features supports the concept that a minority of null-ALCL may be derived from NK cells and expands the spectrum of phenotypes that can be seen in tumors produced by ALK fusion proteins.
Introduction
Anaplastic large cell lymphoma (ALCL) is a heterogeneous subtype of non-Hodgkin lymphomas (NHLs) unified by a characteristic pattern of lymph node invasion by malignant cells that express epithelial membrane antigen (EMA) and CD30.1Because EMA and CD30 can be expressed by a variety of neoplastic cells, co-expression of anaplastic lymphoma kinase (ALK) aids in the diagnosis of ALCL. Aberrant ALK function is thought to play a critical role in the pathogenesis of ALCL, because most ALCLs contain chromosome translocations that create ALK fusion proteins with constitutive kinase activity.1,2 The prototypic ALK fusion protein is nucleophosmin (NPM)-ALK, created by the t(2;5)(p23;q35).3Four variant ALK chimeras have been identified recently in ALCL.4-9 Tumors containing variant ALK chimeras are characterized by cytoplasmic-restricted ALK expression.1
The precise cellular origin of ALCL is unknown. The majority of ALCLs express T-cell–associated antigens and have rearranged TCRgenes.2 The remaining cases express neither T- nor B-cell antigens and are termed null-ALCL. Most null-ALCLs are considered to be of T lineage because they have either α/β or γ/δ TCRrearrangements.10 The derivation of the remaining rare cases that lack expression of T- or B-specific antigens andTCR/Ig rearrangements is unknown. Recently, it has been postulated that ALCLs originate from cells with cytolytic potential, including cytotoxic T cells and possibly natural killer (NK) cells because most ALCLs express cytotoxic proteins, including TIA-1, perforin, and granzyme B, proteins normally expressed only by activated cytotoxic T cells and NK cells.10-12
NK cells are thought to be derived from a bipotential common T cell/NK cell progenitor and express some cell surface antigens associated with T cells; they do not express T-cell–specific antigens, are by definition surface CD3−/CD56+, and have germline TCR and Ig genes.13-15Functionally, NK cells do not require cytokine stimulation or major histocompatibility complex recognition to become cytolytically active and can lyse target cells without major histocompatibility complex restriction or prior sensitization.15 Malignancies arising from cells of NK origin are rare, but they are increasingly recognized as distinct clinicopathologic entities. All neoplasms presumed to be derived from NK cells, including precursor myeloid/NK cell malignancies, express CD56.16,17 However, CD56 expression is not specific for NK malignancies, as some ALCLs, acute lymphoblastic leukemias, acute myeloid leukemias (AMLs), and multiple myelomas also express this antigen.18-22 Recent reports that some NK malignancies express myeloid antigens suggest that the putative T/NK bipotential progenitor may be myeloid-antigen positive.16
In this report, we describe an 18-month-old boy with an unusual malignancy characterized by a capillary leak and hemophagocytic syndrome, hyperleukocytosis, and circulating malignant cells with minimal bone marrow involvement and no identified nodal or extranodal tumor mass. The tumor cells were CD3−/CD56+, lacked TCR gene rearrangements, and had other features seen in the subset of null-ALCLs of unknown derivation, including strong expression of CD30, EMA, and ALK. They also contained a t(2;19)(p23;p13.1) that we determined creates a new variant ALK fusion protein, tropomyosin 4 (TPM4)-ALK. However, the clinical picture was very atypical for an ALCL, and the tumor cells also displayed some features suggestive of a myeloid malignancy (CD13+/CD33+/HLA-DR+) and others classically associated with NK malignancies (granzyme B+with the cytotoxic activity of incompletely differentiated NK cells). This unique case further expands the spectrum of hematologic malignancies associated with ALK fusion proteins.
Patient and methods
Case history
A previously healthy 18-month-old boy presented with fever, hepatosplenomegaly, peripheral edema, and symptomatic pleural effusions. Pleurocentesis revealed a sterile effusion with nonmalignant-appearing lymphocytosis. Complete blood count showed hemoglobin of 100 g/L (10.0 g/dL), platelet count of 281 × 109L (281 000/μL), and a white blood cell (WBC) count of 11 × 109L (11 000/μL) with 81% neutrophils, 5% bands, and 6% lymphocytes. The patient was diagnosed with pneumonia and treated with antibiotics. Over the next several days, he became critically ill with high fever, disseminated intravascular coagulation, and generalized edema with ascites. His pleural effusions worsened and he required intubation and aggressive ventilatory support. No extranodal masses or adenopathy were identified on computed tomography scans of head, abdomen, and thorax, but massive hepatosplenomegaly was present. Hemophagocytosis was noted on bone marrow and liver biopsies, and a serum interleukin-2 receptor level was markedly elevated at 42 300 IU/dL (normal, < 1000 IU/dL). A diagnosis of infectious-associated hemophagocytic syndrome was considered. Three separate bone marrow aspirates and biopsies obtained from multiple sites over 6 days were consistently moderately hypercellular with trilineage hematopoietic elements and contained no identifiable malignant cells. Cytogenetic studies of liver and bone marrow specimens revealed a normal male karyotype.
The patient was treated initially with high-dose intravenous immunoglobulin and methylprednisolone. An extensive investigation for viral infections, including Epstein-Barr virus and herpesviruses such as human herpesvirus 8, was negative. Because of the patient's deteriorating clinical status, tentative diagnosis of infectious-associated hemophagocytic syndrome, and symptoms suggestive of “cytokine storm,” Cyclosporin A (CsA) was added to the treatment regimen. Within 24 hours of starting CsA and after 1 week of methylprednisolone, the patient's WBC count abruptly increased from 5.5 ×109L (5500/μL) to 39.4 × 109L (39 400/μL) over a 36-hour period, and malignant cells were noted in the peripheral blood for the first time. The day before the development of hyperleukocytosis, a bone marrow was obtained that showed no malignant cells by microscopic examination, but retrospective cytogenetic and fluorescence in situ hybridization (FISH) studies revealed presence of the pseudodiploid clone with an ALKtranslocation. The WBC count eventually peaked at 94 × 109L (94 000/μL) with 66% circulating malignant cells.
The neoplastic cells were homogeneous and monocytoid appearing. Some cells contained numerous cytoplasmic vacuoles and exhibited cytoplasmic buddings. Initial analysis indicated that the malignant cells expressed no T, B, or histiocytic cell-associated antigens but strongly expressed CD13 and CD30 and were negative for Tdt and cytochemical myeloperoxidase (MPO) staining. Cytogenetic studies of peripheral blood and bone marrow specimens detected abnormalities in the short arms of chromosomes 2, 5, and 19. FISH studies showed rearrangement ofALK with translocation to the der(19). Reverse transcriptase-polymerase chain reaction (RT-PCR) forNPM-ALK fusion transcripts was negative. Because of the unusual clinical presentation and evidence of a variantALK translocation in malignant cells that co-expressed CD30 and EMA, the patient was felt to have an atypical ALCL.
On the basis of this diagnosis, CsA was discontinued and plasmapheresis and leukopheresis were performed daily for 5 days with significant improvement in the patient's clinical condition. Induction chemotherapy based on the Children's Cancer Group regimens used to treat pediatric ALCL and other childhood NHLs was initiated. The patient achieved a complete remission within 4 weeks and was treated for a total of 8 months with a regimen that included Adriamycin, cyclophosphamide, cytosine arabinoside, daunorubicin, etoposide, high-dose methotrexate, vincristine, and intrathecal cytosine arabinoside but excluded steroids. The patient is currently in remission 14 months after completion of therapy.
Specimens
Cytogenetic and FISH studies were performed at the Colorado Genetics Laboratory. Malignant cells obtained by leukopheresis were used for cytochemical stains, immunohistochemistry, flow cytometric analysis, and functional killing assays. Cryopreserved samples of the patient's neoplastic cells were obtained for further studies from the leukemia cell bank of The Children's Hospital and analyzed under research protocol approved by the Institutional Review Board.
Cytogenetics and FISH
Giemsa-banded cytogenetic studies were performed from unstimulated cultures of bone marrow and peripheral blood. Samples were prepared by using a direct technique and overnight culture methods as described previously.23,24 Cytogenetic results are described by using the International System for Cytogenetic Nomenclature.25
FISH studies were performed by using a commercial ALK probe (Vysis, Downers Grove, IL) that was directly labeled with Spectrum Green and Spectrum Orange and hybridized to both interphase and metaphase targets following the manufacturer's protocol. In the use of this probe, cells with an intact ALK locus have a fused red/green signal on chromosome 2. If an ALK translocation has occurred, then the probe is split. The proximal Spectrum Green–labeled probe remains on the der(2) and the distal Spectrum Orange–labeled probe moves to the partner derivative chromosome. Additional FISH studies were performed with chromosome 2 and 19 whole chromosome paint probes and with probes for 5p-, 19p-, and 19q-specific subtelomeric sequences under conditions recommended by the manufacturer (Vysis).
Fluorescence-activated cell sorter analysis
Immunophenotyping of malignant cells was performed on the leukopheresis product by using directly conjugated fluorescent monoclonal or polyclonal antibodies to CD3, CD4, CD5, CD8, CD14, CD16/56 (combined), CD19, CD45, CD45RO, HLA-DR, TCR-αβ, and TCR-γδ (Becton Dickinson, Mountain View, CA); CD10, CD20 (Coulter, Miami, FL); CD13, CD61 (DAKO, Carpinteria, CA); CD2 (Immunotech, Miami, FL); and CD7, CD25, CD30, CD33, CD34, CD95, CD95L, and CD122 (Pharmingen, San Diego, CA). Cells were also separately stained with isotype controls for each antibody according to manufacturer's protocol. Cells were analyzed by flow cytometry within 24 hours of preparation at the University of Colorado Cancer Center Flow Cytometry Core or at The Children's Hospital by using Coulter EpicsXL flow cytometers and software.
Immunohistochemistry
Immunohistochemical staining was performed by using the heat-induced antigen retrieval technique on paraffin-embedded cellblock sections made from peripheral blood leukocytes obtained from leukopheresis. The following monoclonal antibodies were used according to protocols recommended by the manufacturers: Granzyme B (Chemicon; CA); CD57 (Becton Dickinson); CD3, CD15, CD20, CD68, ALK-1, EMA, MPO, S-100 (DAKO, Carpinteria, CA); and CD30 (Immunotech). Standard cytochemical stains for MPO and nonspecific esterase (NSE) were performed on bone marrow aspirate smears.
NK cell cytotoxicity assays
A standard short-term (4-hour) 51Cr-release NK cell functional assay using NK-sensitive K562 and NK-insensitive Daudi tumor cells as targets was performed.26 Peripheral blood mononuclear cells (PBMCs) were isolated from a healthy volunteer (control) or from the patient's leukopheresis product by using Histo-paque 1077 (Sigma, St. Louis, MO) gradient separation. PBMCs served as effectors in the 51Cr-release assay. Targets were labeled with 100 μCi 51Cr as sodium chromate (ICN Pharmaceuticals, Irvine, CA) in standard media for 1 to 2 hours at 37°C. Radiolabeled target cells (5000) in 100 μL per well were plated in 96-well tissue culture plates. Patient or control PBMCs in a volume of 100 μL were added at ratios of 50:1; 25:1; 12:1; and 6:1 to the target cells. Plates were pelleted by centrifugation and incubated for 4 hours at 37°C. Following incubation, 100 μL cell-free supernatant was transferred to separate tubes and radioactivity was measured by using a gamma counter. Percentage of specific lysis was calculated by using the following formula: (e − s/m − s) × 100, where e, s, and m equal the amount of radioactivity released from PBMCs incubated with effector cells (experimental lysis), with 100 μL media instead of effector cells (spontaneous lysis), or with 100 μL 1% Triton X-100 (maximum lysis), respectively. Experimental, spontaneous, and maximum lysis samples were plated in triplicate. Percentage of specific lysis was calculated as previously described. Fas/FasL killing assays were performed in an analogous manner as previously described.27
Molecular analyses
RNA extraction and RT-PCR analyses were performed as described previously.28 The sequences of PCR oligomers are provided in Table 1. Nucleotide sequencing of cloned PCR products and sequence analysis was performed as described previously.29
Sequence of oligomers used for reverse transcriptase–polymerase chain reaction
| PCR target . | Primer . | Sequence . | Reference . |
|---|---|---|---|
| NPM-ALK | NPM forward | 5-TCCCTTGGGGGCTTTGAAATAACACC | Morris et al3 |
| ALK reverse | 5-CGAGGTGCGGAGCTTGCTCAGC | ||
| TPM3-ALK | C-TPM1 (outer) | 5-CGAGAAGTTGAGGGAGAAAGG | Lamant et al5 |
| ALK-1 (outer) | 5-GCCAGCAAAGCAGTAGTTGGGGTTG | ||
| C-TPM3 (inner) | 5-CTGGCAGAGTCCCGTTGCC | ||
| ALK-2 (inner) | 5-GTCGAGGTGCGGAGCTTGCTCAGC | ||
| ATIC-ALK | ATIC (outer) | 5-CGCTCGCCCTGAACCCAGTG | Ma et al6 |
| ATIC (inner) | 5-GTGTCCACGGAGATGCAGAG | ||
| ALK (outer and inner) | 5-CGAGGTGCGGAGCTTGCTCAGC | ||
| TPM4-ALK | TPM4Start | 5-CGCGCCATGGCCGGCCTCAAC | This report |
| ALKSM3R | 5-AGCACACTTCAGGCAGCGTCTTC | ||
| ABL | ABLX3 | 5-TTTCTCCAGACTGTTGACTGG | Hunger et al48 |
| ABLX2 | 5-CCTTCAGCGGCCAGTAGCAT |
| PCR target . | Primer . | Sequence . | Reference . |
|---|---|---|---|
| NPM-ALK | NPM forward | 5-TCCCTTGGGGGCTTTGAAATAACACC | Morris et al3 |
| ALK reverse | 5-CGAGGTGCGGAGCTTGCTCAGC | ||
| TPM3-ALK | C-TPM1 (outer) | 5-CGAGAAGTTGAGGGAGAAAGG | Lamant et al5 |
| ALK-1 (outer) | 5-GCCAGCAAAGCAGTAGTTGGGGTTG | ||
| C-TPM3 (inner) | 5-CTGGCAGAGTCCCGTTGCC | ||
| ALK-2 (inner) | 5-GTCGAGGTGCGGAGCTTGCTCAGC | ||
| ATIC-ALK | ATIC (outer) | 5-CGCTCGCCCTGAACCCAGTG | Ma et al6 |
| ATIC (inner) | 5-GTGTCCACGGAGATGCAGAG | ||
| ALK (outer and inner) | 5-CGAGGTGCGGAGCTTGCTCAGC | ||
| TPM4-ALK | TPM4Start | 5-CGCGCCATGGCCGGCCTCAAC | This report |
| ALKSM3R | 5-AGCACACTTCAGGCAGCGTCTTC | ||
| ABL | ABLX3 | 5-TTTCTCCAGACTGTTGACTGG | Hunger et al48 |
| ABLX2 | 5-CCTTCAGCGGCCAGTAGCAT |
PCR indicates polymerase chain reaction; NPM, nucleophosmin; ALK, anaplastic lymphoma kinase; TPM, tropomyosin; ATIC, 5-aminoimadazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase; ABL, Abelson leukemia gene.
Results
Morphologic and cytochemical features of the malignant cells
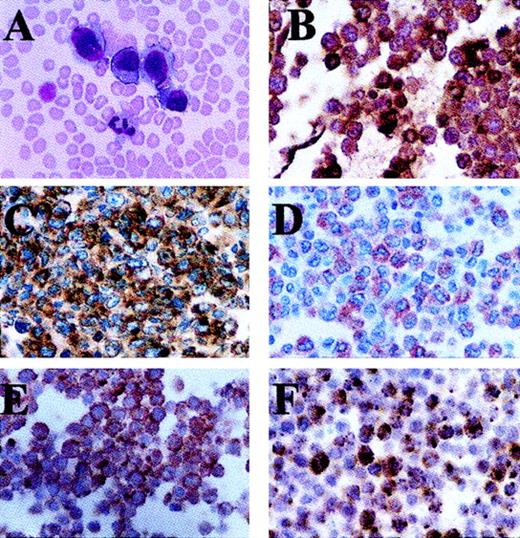
The malignant cells present in Wright-stained peripheral blood were large and homogenous, and they contained large round to oval nuclei, slightly dense chromatin, inconspicuous nucleoli, and abundant blue cytoplasm. Large cytoplasmic blebs were also noted in some cells (Figure 1A). Many tumor cells contained multiple clear cytoplasmic vacuoles, but no cytoplasmic granules or Auer rods were seen. Malignant cells were negative for Sudan black B, NSE, and MPO cytochemical reactivity, although high expression of cytoplasmic MPO protein was detected by immunohistochemistry (Figure 1B).
The malignant blasts have the morphology and immunohistochemical features of ALCL cells.
(A) Wright stain of peripheral blood smear. Sections created from cell blocks of the leukopheresis product stain positive with monoclonal antibodies directed against (B) MPO, (C) CD30, (D) EMA, (E) ALK, and (F) granzyme B. Note that ALK staining is present only in the cytoplasm of the malignant cells (E).
The malignant blasts have the morphology and immunohistochemical features of ALCL cells.
(A) Wright stain of peripheral blood smear. Sections created from cell blocks of the leukopheresis product stain positive with monoclonal antibodies directed against (B) MPO, (C) CD30, (D) EMA, (E) ALK, and (F) granzyme B. Note that ALK staining is present only in the cytoplasm of the malignant cells (E).
Immunophenotypic properties of malignant cells
A fluorescence-activated cell sorter (FACS) light-scatter profile of cells freshly obtained by leukopheresis showed 2 populations of cells expressing CD45 (Figure 2A). The malignant cells comprised the vast majority of CD45+ cells and showed a side-scatter profile characteristic of monocytes (region I). A much smaller population of cells with the side-scatter profile representing normal lymphocytes was also present (region II). FACS analysis gated on the malignant cells (region I) revealed an unusual immunophenotype. All the malignant cells strongly expressed CD13, CD25, CD30, CD45, CD45RO, HLA-DR, CD95 (Fas), and CD122. On dual-color FACS analysis, 68% of malignant cells expressed CD16/CD56 (combined antibody reagent) but lacked CD3 expression consistent with the antigen profile of NK cells (Figure 2B). With the use of single-color analysis, 56% of malignant cells expressed CD56. Approximately 20% of malignant cells expressed CD3dim, CD7dim, CD33bright, and CD14bright; however, it was not possible to determine whether the same population of cells simultaneously expressed a combination of these antigens or whether CD16/56+ cells also co-expressed these antigens because single-color rather than multicolor FACS was performed for these antigens. Less than 10% of the malignant cells expressed T-cell–associated antigens CD2, CD4, CD5, and CD8 or B-cell–associated antigens CD19 and CD20. Tumor cells also did not express CD10, CD34, CD61, Fas ligand, or surface TCR α, β, γ, or δ chains.
Malignant cells display phenotypic and functional properties characteristic of natural killer cells.
(A) FACS light scatter profile and CD45 expression of freshly isolated cells from the patient's peripheral blood leukopheresis product. Cells located in region I represent malignant leukocytes. Cells located in region II represent normal lymphocytes. (B) FACS analysis of cells in region I demonstrates that a majority of neoplastic cells express CD16/56 but lack CD3 surface expression consistent with the phenotype of a NK cell. (C) Percentage of specific lysis of NK-sensitive K562 cells by unstimulated patient malignant cells (▪) versus control peripheral blood mononuclear cells (■) at various effector-to-target ratios as determined by a standard 4-hour chromium release NK cell assay.
Malignant cells display phenotypic and functional properties characteristic of natural killer cells.
(A) FACS light scatter profile and CD45 expression of freshly isolated cells from the patient's peripheral blood leukopheresis product. Cells located in region I represent malignant leukocytes. Cells located in region II represent normal lymphocytes. (B) FACS analysis of cells in region I demonstrates that a majority of neoplastic cells express CD16/56 but lack CD3 surface expression consistent with the phenotype of a NK cell. (C) Percentage of specific lysis of NK-sensitive K562 cells by unstimulated patient malignant cells (▪) versus control peripheral blood mononuclear cells (■) at various effector-to-target ratios as determined by a standard 4-hour chromium release NK cell assay.
On cellblock sections made from peripheral blood, malignant cells were readily identified and appeared similar to neoplastic cells identified in Wright-stained peripheral blood (Figure 1A). Immunohistochemical staining demonstrated that the tumor cells did not express cytoplasmic CD3, CD15, CD57 (mature NK antigen), CD68, or S100 (histiocyte-associated antigens). All the malignant cells stained strongly with antibodies to CD30 (Figure 1C). The majority of neoplastic cells stained positive with antibodies to EMA and ALK (Figure 1D,E). ALK-1 staining was confined to the cytoplasm of the malignant cells. Nearly all neoplastic cells were positive for the cytotoxic protein granzyme B, although only a small percentage of cells stained strongly positive (Figure 1F). TCRα, β, γ, and δ genes remained in germline configuration by Southern blot analysis (data not shown).
Malignant cells display cytotoxic properties of NK cells
NK cells have the capacity to lyse tumor cells by 2 pathways. The predominant pathway involves release of cytotoxic granules containing perforin and granzyme B to target cells. A second pathway, primarily found in fully differentiated/activated NK cells, uses Fas ligand to kill Fas+ targets. In short-term (4-hour)51Cr-release assays, unstimulated NK cells are defined by their ability to lyse NK-sensitive (K562) but not NK-insensitive (Daudi) target cells.
Because the neoplastic cells expressed antigens associated with NK cells, we performed 51Cr-release assays on freshly isolated, unstimulated tumor cells to determine if they possessed NK cytotoxic properties. PBMCs isolated from a normal control contained 12% NK cells as determined by FACS (data not shown). Isolated PBMCs from the patient contained less than 1% normal lymphocytes, and no cells expressing an NK cell phenotype (CD56+CD3−) could be detected within the normal lymphocyte population. Therefore, the vast majority of cells within the patient PBMC isolate were malignant and expressed the NK antigen profile CD56+CD3−. Similar to control PBMCs, unstimulated patient PBMCs were highly cytotoxic in a dose-dependent manner toward NK-sensitive K562 (Figure 2C). In contrast, neither patient nor control PBMCs significantly lysed NK-resistant Daudi cell targets (data not shown). Furthermore, neither control nor patient PBMCs were able to lyse Fas+ targets, indicating that these cells did not express functional Fas ligand (data not shown). This finding was consistent with FACS analysis showing no Fas ligand on the malignant cells. Therefore, the tumor cells express the functional property of immature NK cells, as they can lyse K562 but not Fas+ targets.
Identification of a novel ALK translocation
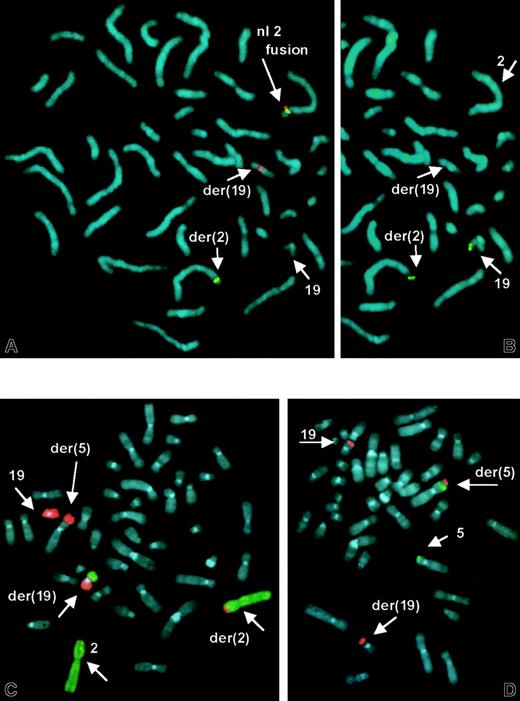
Cytogenetic studies of unstimulated peripheral blood and bone marrow specimens revealed structural abnormalities of chromosomes 2, 5, and 19 (Figure 3). FISH using a 2-color split signal ALK probe cocktail revealed a rearrangement in interphase nuclei. Metaphase FISH demonstrated that the proximalALK sequences remained on the der(2), whereas distalALK sequences were translocated to the der(19) (Figure4A). This was not a cryptic t(2;5) because RT-PCR for NPM-ALK fusion transcripts was negative (data not shown). Additional FISH studies identified a reciprocal t(2;19) (Figure 4B,C) and showed that the der(5) results from an unbalanced translocation of 19q to distal 5p, leaving 5p subtelomeric sequences intact (Figure 4D). These detailed FISH studies established the karyotype of the main clone to be 46, XY, t(2;19)(p23;p13.1), der(5)t(5;19)(p15.3;q13.1).
G-banded karyotype from leukemic clone demonstrates structural abnormalities of chromosome homologues 2, 5, and 19.
Arrows denote the abnormal chromosome 2, 5, and 19 homologs. FISH (see Figure 4) resolved the karyotype as 46, XY, t(2;19)(p23;p13.1), der(5)t(5;19)(p15.3;q13.1).
G-banded karyotype from leukemic clone demonstrates structural abnormalities of chromosome homologues 2, 5, and 19.
Arrows denote the abnormal chromosome 2, 5, and 19 homologs. FISH (see Figure 4) resolved the karyotype as 46, XY, t(2;19)(p23;p13.1), der(5)t(5;19)(p15.3;q13.1).
FISH demonstrates
ALK rearrangement, t(2;19), and der(5)t(5p;19q).(A) ALK split-apart FISH probe shows normal fused orange/green signal on normal No. 2 homologue, and split green signal on der(2) and orange signal on der(19). (B) Same metaphase rehybridized with 19p subtelomeric sequences, showing translocation of 19ptel to der(2). (C) FISH with dual-color whole chromosome paint probes shows No. 19 sequences (orange) translocated to der(2) and der(5) and No. 2 sequences (green) on der(19). (D) Subtelomeric sequences for 5p (green) and 19q (orange) show retention of 5ptel on der(5), and 19qtel translocated distal to 5ptel.
FISH demonstrates
ALK rearrangement, t(2;19), and der(5)t(5p;19q).(A) ALK split-apart FISH probe shows normal fused orange/green signal on normal No. 2 homologue, and split green signal on der(2) and orange signal on der(19). (B) Same metaphase rehybridized with 19p subtelomeric sequences, showing translocation of 19ptel to der(2). (C) FISH with dual-color whole chromosome paint probes shows No. 19 sequences (orange) translocated to der(2) and der(5) and No. 2 sequences (green) on der(19). (D) Subtelomeric sequences for 5p (green) and 19q (orange) show retention of 5ptel on der(5), and 19qtel translocated distal to 5ptel.
TPM4 is a new ALK fusion partner
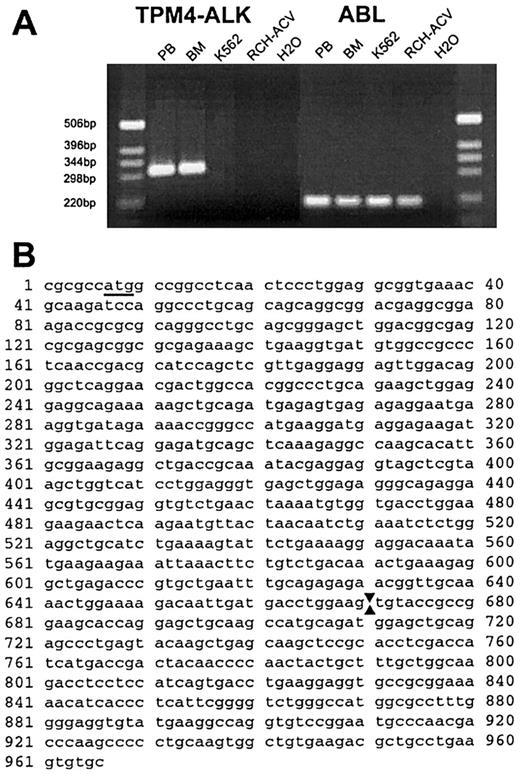
On the basis of the FISH and RT-PCR results, we concluded thatALK was fused to a new 19p13.1 partner gene in this neoplasm. Because the rearrangements were complicated, we undertook studies to exclude the possibility of cryptic fusion of ALKto one of its other known partner genes and performed RT-PCR by using primers designed to amplify ATIC-ALK andTPM3-ALK fusion transcripts. Although theATIC-ALK PCR was negative, we observed robust amplification of an approximate 300–base-pair (bp) product (and weaker amplification of larger products) with theTPM3-ALK primers from both peripheral blood and bone marrow specimens of the patient with the t(2;19) but not from samples lacking ALK translocations (Figure5A). We cloned the PCR product and determined the nucleotide sequences of 2 independent clones with inserts of 308 and 426 bp (GenBank accession No. AF362886 andAF362887). The last 83 nucleotides of both clones were derived fromALK with the site of fusion identical to that seen in otherALK chimeric complementary DNAs (nucleotide 40843). The 5′ portion of both clones shared homology withTPM3, but they were more homologous to alternatively spliced products of TPM4. The longer clone started with the outerTPM3 oligomer and the shorter clone started with the innerTPM3 oligomer. The clones also showed differential splicing within the region homologous to TPM4, but both showed fusion between TPM4 position 714 (numbering as per GenBank accession No. NM_00329030) and ALK 4084.TPM4 has been mapped to chromosome 19p13.1,31indicating that the complex t(2;19) in this case fused ALKto a second member of the tropomyosin gene family. Amplification with primers designed to amplify TPM3-ALK transcripts occurred fortuitously because the TPM3 primers we used were homologous to both TPM3 and TPM4. To confirm theTPM4-ALK fusion, we designed aTPM4-specific oligomer that spanned the start codon. PCR using this and an oligomer located more 3′ in ALK gave an approximate 950-bp product that was cloned and sequenced. This product containedTPM4 nucleotides 45-714 (100% match to NM_003290) fused toALK starting at position 4084 (Figure 5B; GenBank accession No. AF310722).
The t(2;19)(p23;p13.1) fuses
TPM4 to ALK. (A) Photograph of ethidium bromide–stained gel of RT-PCR products. An approximately 300-bp product was amplified with primers for TPM3 andALK from patient peripheral blood (PB) and bone marrow (BM), whereas no product was amplified from control cell lines K562 and RCH-ACV, or from a water-negative control. The results of control RT-PCR analyses for a portion of the ABL complementary DNA, verifying the integrity of isolated RNA, are shown at right. (B) Nucleotide sequence of amplified portion ofTPM4-ALK complementary DNA obtained usingTPM4-specific and ALK primers. The initiation codon in TPM4 is underlined and arrows denote the point of fusion between TPM4 (nucleotides 1 > 670) andALK sequences (671 > 966).
The t(2;19)(p23;p13.1) fuses
TPM4 to ALK. (A) Photograph of ethidium bromide–stained gel of RT-PCR products. An approximately 300-bp product was amplified with primers for TPM3 andALK from patient peripheral blood (PB) and bone marrow (BM), whereas no product was amplified from control cell lines K562 and RCH-ACV, or from a water-negative control. The results of control RT-PCR analyses for a portion of the ABL complementary DNA, verifying the integrity of isolated RNA, are shown at right. (B) Nucleotide sequence of amplified portion ofTPM4-ALK complementary DNA obtained usingTPM4-specific and ALK primers. The initiation codon in TPM4 is underlined and arrows denote the point of fusion between TPM4 (nucleotides 1 > 670) andALK sequences (671 > 966).
Discussion
We describe a leukemic presentation of an unusual extramedullary hematologic malignancy in a young child with fulminant clinical symptoms. This case presented a diagnostic challenge because no solid tumor mass could be identified, the circulating tumor cells did not appear to arise from the bone marrow, and they did not express an antigen profile typical of any subtype of leukemia commonly observed in children. Because the clinical presentation of childhood ALCL is diverse, is often associated with cytokine storm–like symptoms, and can rarely be associated with circulating tumor cells,32-37 the malignant cells were analyzed with this diagnosis in mind. Consistent with a null-ALCL, tumor cells were CD3−/CD56+; had germline TCR genes; strongly expressed CD30, EMA, and ALK; and contained a novelALK translocation. Similar to all other variant ALK fusion proteins described to date, ALK staining was confined to the cytoplasm. On the basis of these features a clinical diagnosis of atypical ALCL was made, and the patient was treated with and responded well to therapy (excluding steroids) used to treat ALCLs and certain other subtypes of childhood NHL.
This case contained a t(2;19)(p23;p13.1), which we found created a new variant ALK fusion protein, TPM4-ALK. Chimeric ALK proteins are present in most childhood ALCLs, which are considered to comprise a distinct histopathologic entity unified by their expression of ALK fusion proteins.2 About 70% of these cases contain a t(2;5)(p23;q35) that produces NPM-ALK, a constitutively active tyrosine kinase that has oncogenic properties in a variety of experimental systems.3,38-41 The other 30% contain variant ALK fusion proteins created by translocations joining 2p23 to other regions of the genome. To date, 4 different ALK fusion variants have been identified in ALCL: t(1;2)(q25;p23) and TPM3-ALK, inv(2)(p23;q35) and ATIC-ALK, t(2;3)(p23;p21) and TFG-ALK, and t(2;22)(p23;q11.2) and CLTCL-ALK.4-9 At the time this patient presented,ALK translocations had only been described in ALCL. After we identified TPM4-ALK fusion in this case, Lawrence et al42 reported that TPM4-ALK andTPM3-ALK fusion genes also occur in a nonhematalogic malignancy termed inflammatory myofibroblastic tumor (IMT).42 Thus, ALK translocations can no longer be considered pathognomonic of ALCL.
We identified 2 species of TPM4-ALK transcripts fortuitously when we cloned and sequenced RT-PCR products obtained withTPM3 and ALK primers. The 3′ portions of the major 308-bp and minor 426-bp products we cloned match the type I and II TPM4-ALK transcripts identified in IMT, whereas the first 81 (major) and 103 (minor) bp are more divergent. GenBank searches performed recently (Hunger SP, unpublished data, 2001) show that these 5′ portions are highly homologous to recently described alternatively spliced TPM4 isoforms. We later amplified and cloned fusion transcripts containing the complete portion ofTPM4 fused to ALK (Figure 5B). These transcripts are identical to type II TPM4-ALK transcripts identified in IMT.42 The relevance of different species ofTPM4-ALK fusion transcripts and proteins is unknown at this time and merits additional study.
Even in the presence of an ALK translocation, the clinical features present in this patient and the phenotype of the malignant cells were very atypical for an ALCL and prompted us to perform additional investigations. These studies demonstrated that the tumor cells also had features typically associated with both myeloid and NK malignancies. Although the profile of CD3−/CD56+ and germline TCR genes is consistent with a null-ALCL, this phenotype can also be present in myeloid and NK tumors. This case did not meet accepted criteria for an AML, as cytochemical stains for MPO were negative and there was no significant marrow involvement. However, the malignant cells did express cytoplasmic MPO by immunostaining and expressed several cell surface antigens typically associated with AML (CD13, CD33, and HLA-DR). Evidence of NK cell derivation was more compelling. Tumor cells expressed granzyme B, which is predominantly restricted to NK cells, cytotoxic T cells, and malignancies derived from these cell types. Granzyme B has also been found in normal CD34+hematopoietic progenitor cells mobilized by chemotherapy combined with granulocyte colony-stimulating factor and in AML cell lines exposed in vitro to genotoxic agents, but granzyme B expression has not been described in myeloid leukemias to our knowledge.43 44 The malignant cells also possessed functional properties of incompletely differentiated NK cells because freshly isolated nonstimulated tumor cells were able to lyse NK-sensitive but not NK-insensitive or Fas-positive cell lines in short-term cytotoxicity assays. Taken together, these findings suggest that this neoplasm may be related to a spectrum of malignancies recently categorized as being derived from the putative myeloid/NK progenitor cell.
The classification of NK cell malignancies is evolving and becoming increasingly complex. Recently proposed classification schemes differentiate immature (precursor) from mature NK cell malignancies and from NK-like T-cell malignancies.16,17 Precursor NK cell disorders resemble acute leukemia and include myeloid/NK cell precursor leukemia, blastic NK cell leukemia/lymphoma, and possibly myeloid/NK cell acute leukemia.16,45,46 Neoplasms derived from mature NK cells include extranodal NK/T-cell lymphoma (nasal type) and aggressive NK-cell leukemia/lymphoma.47 Table2 compares the features of these closely related entities with each other and with the malignancy that we characterized.
Comparison of malignancies with a CD3−/CD56+/CD57− phenotype and leukemic presentation
| . | Patient's tumor . | Myeloid/NK cell acute leukemia . | Myeloid/NK cell precursor acute leukemia . | Blastic NK cell lymphoma/leukemia . | Aggressive NK cell leukemia . |
|---|---|---|---|---|---|
| Phenotype | CD7−, CD13+, CD30+, CD15−, CD33+/−, CD34−, HLA-DR+ | CD33+, CD16−, CD34+/−, HLA-DR− | CD7+, CD33 or CD13+; CD34+; CD16−; CD15+/−; HLA-DR+/− | CD4+/−, CD16−, CD33−, TdT(IHC)+/−, HLA-DR+ | CD2+, CD5−, CD16+/−, CD33−, HLA-DR+ |
| Cytochemical MPO reactivity | Negative | Positive | Negative | Negative | Negative |
| Cytoplasmic expression of MPO by IHC | Positive | Unknown | Frequently positive | Negative | Negative |
| Morphology | Monocytoid | Promyelocytoid | Immature blastoid | Lymphoblastoid | Large granular lymphocytes |
| Granzyme B | Positive | Negative | Negative | Negative | Positive |
| Functional NK cytotoxicity | Present | Present only following IL-2 stimulation | Absent | Absent | Present |
| IL-2R (CD25) | Positive | Unknown | Negative | Negative | Positive |
| Bone marrow involvement | Secondarily involved | Always | Occasional | Rare | Common |
| Extramedullary disease | Present | Uncommon | Common | Occasional | Common |
| Lymph node involvement | Not detected | Common | Common | Occasional | Common |
| TCR rearrangement | Germline | Primarily germline | Primarily germline | Primarily germline | Germline |
| . | Patient's tumor . | Myeloid/NK cell acute leukemia . | Myeloid/NK cell precursor acute leukemia . | Blastic NK cell lymphoma/leukemia . | Aggressive NK cell leukemia . |
|---|---|---|---|---|---|
| Phenotype | CD7−, CD13+, CD30+, CD15−, CD33+/−, CD34−, HLA-DR+ | CD33+, CD16−, CD34+/−, HLA-DR− | CD7+, CD33 or CD13+; CD34+; CD16−; CD15+/−; HLA-DR+/− | CD4+/−, CD16−, CD33−, TdT(IHC)+/−, HLA-DR+ | CD2+, CD5−, CD16+/−, CD33−, HLA-DR+ |
| Cytochemical MPO reactivity | Negative | Positive | Negative | Negative | Negative |
| Cytoplasmic expression of MPO by IHC | Positive | Unknown | Frequently positive | Negative | Negative |
| Morphology | Monocytoid | Promyelocytoid | Immature blastoid | Lymphoblastoid | Large granular lymphocytes |
| Granzyme B | Positive | Negative | Negative | Negative | Positive |
| Functional NK cytotoxicity | Present | Present only following IL-2 stimulation | Absent | Absent | Present |
| IL-2R (CD25) | Positive | Unknown | Negative | Negative | Positive |
| Bone marrow involvement | Secondarily involved | Always | Occasional | Rare | Common |
| Extramedullary disease | Present | Uncommon | Common | Occasional | Common |
| Lymph node involvement | Not detected | Common | Common | Occasional | Common |
| TCR rearrangement | Germline | Primarily germline | Primarily germline | Primarily germline | Germline |
NK, natural killer; IHC, immunohistochemistry; MPO, myeloperoxidase; IL-2, interleukin 2; IL-2R, interleukin-2 receptor; TCR, T-cell receptor.
The clinicopathologic features of the patient we describe do not correspond with any previously described tumor of which we are aware. Rather, this case has some features suggestive of an ALCL-like malignancy (CD30+/EMA+/ALK+ with anALK translocation), some suggestive of a myeloid malignancy (CD13+/CD33+/HLA-DR+ and expressing cytoplasmic MPO but lacking MPO activity) and some features (granzyme B+/CD25+/CD122+ with the cytotoxic activity of incompletely differentiated NK cells but CD2−/FasL− and lacking association with Epstein-Barr virus) that are intermediate to immature and mature tumors within the NK tumor spectrum. This constellation of characteristics may be due to derivation of this malignancy from the putative myeloid-NK precursor cell. Co-expression of NK cell and ALCL-related features by the same tumor provides some support to the theory that ALCL may arise from cells containing cytolytic potential and that a subset of null-ALCL truly may be derived from NK cells or their precursors. However, it is critical to distinguish this subset from ALCLs that simply express CD56 but do not have other NK cell–associated features.22 In the future, it will be important to evaluate null-ALCLs more comprehensively by using CD56 and other NK-associated antigen expression and functional studies to determine whether other cases might also have features of NK-derived malignancies. Similarly, further studies are necessary to clarify whether some myeloid/NK-cell precursor malignancies also express the ALCL triad of CD30+/EMA+/ALK+seen in this tumor. Finally, this case exemplifies issues raised by evolving changes in classification of hematologic malignancies that raise the question of whether tumors should be classified and treatment selected on the basis of clinicopathologic characteristics or sentinel molecular lesions.
We thank Stephan Morris for generously sharing reagents, data on ATIC-ALK before publication, and thoughtful advice. We also thank Mitchell Bitter for his expert review of the pathology of this case and for review of this manuscript, Karen Helms and her colleagues in the flow cytometry core lab for expert assistance, Bette Jamieson for help with morphology studies, and Billie Carstens for expert figure layout and design. In particular, we thank reviewer B for a very detailed and useful critique of this manuscript.
Supported by grants from the Loewenstern Family Foundation (to S.P.H.), Cure For Lymphoma Research Foundation (to S.J.M.), Lymphoma Research Fund at The Children's Hospital Cancer Center and National Cancer Institute Cancer Center Core Grant CA 46934. S.P.H. is a Translational Research Grant Awardee of the Leukemia and Lymphoma Society of America.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stephen P. Hunger, UCHSC Campus Box C229, 4200 East Ninth Ave, Denver, CO 80262; e-mail: stephen.hunger@uchsc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal