The development of gastric mucosa-associated lymphoid tissue (MALT) lymphoma is a multistep process and can be clinico-pathologically divided into Helicobacter pylori-associated gastritis, low-grade tumors, and high-grade tumors. The molecular events underlying this progression are largely unknown. However, identification of the genes involved in MALT lymphoma-specific t(11;18)(q21;q21) and t(1;14)(p22;q32) has provided fresh insights into the pathogenesis of this disease. T(11;18)(q21;q21) results in a chimeric transcript between the API2 and theMALT1 genes, whereas t(1;14) (p22;q32) causes aberrant nuclear BCL10 expression. Significantly, nuclear BCL10 expression also occurs frequently in MALT lymphomas without t(1;14)(p22;q32), suggesting an important role for BCL10 in lymphoma development. Thirty-three cases of H pylori gastritis, 72 MALT lymphomas, and 11 mucosal diffuse large B-cell lymphomas (DLBCL) were screened for t(11;18)(q21;q21) by reverse transcription–polymerase chain reaction followed by sequencing. BCL10 expression in lymphoma cases was examined by immunohistochemistry. The API2–MALT1 fusion transcript was not detected in H pylorigastritis and mucosal DLBCL but was found in 25 of 72 (35%) MALT lymphomas of various sites. Nuclear BCL10 expression was seen in 28 of 53 (53%) of MALT lymphomas. Of the gastric cases, the largest group studied, the frequency of both t(11;18)(q21;q21) and nuclear BCL10 expression was significantly higher in tumors that showed dissemination to local lymph nodes or distal sites (14 of 18 = 78% and 14 of 15 = 93%, respectively) than those confined to the stomach (3 of 29 = 10% and 10 of 26 = 38%). Furthermore, t(11;18)(q21;q21) closely correlated with BCL10 nuclear expression. These results indicate that both t(11;18)(q21;q21) and BCL10 nuclear expression are associated with advanced MALT lymphoma and that their oncogenic activities may be related to each other.
Introduction
The development of mucosa-associated lymphoid tissue (MALT) lymphoma is a multistage process.1 This is best understood in gastric MALT lymphoma, the most common form. Typically, low-grade gastric MALT lymphoma arises from mucosal lymphoid tissue that is acquired usually as a reaction to Helicobacter pylori infection.2,3 Low-grade MALT lymphoma is initially confined to the gastric mucosa, and its growth depends critically on the contact help of H pylori–specific intratumoral T cells; therefore, it responds favorably to H pylori eradication therapy.4-6 However, when the lymphoma invades the deep layers of the gastric wall and disseminates to local lymph nodes and distal sites, the tumor loses its dependence on H pylori-specific T cells and is no longer sensitive toH pylori eradication therapy.7-9 Finally, low-grade gastric MALT lymphoma may transform into a more aggressive diffuse large B-cell lymphoma (DLBCL).10 11
Direct12-14 and indirect antigen stimulation4,5 and several genetic factors, including genetic instability,15 trisomy 3,16 p53 mutation/LOH,17 p16 deletion,18t(1;14)(p22;q32),19 and t(11;18)(q21;q21),20,21 are implicated in MALT lymphoma development. However, the molecular events underlying the multistep progression of the tumor remain largely unknown. Identification of the genes involved in MALT lymphoma-specific t(1;14)(p22;q32)22,23 and t(11;18)(q21;q21)24-26 has provided fresh insights into the pathogenesis of this disease.
T(1;14)(p22;q32) causes overexpression of BCL10, an apoptosis regulatory molecule.22,23 In contrast to its expected oncogenic role, wild-type BCL10 has been shown to be proapoptotic and to behave as a tumor suppressor in cell transformation assays.22,23,27-32 Truncated BCL10 mutants have been shown to gain transforming ability22; however, BCL10gene mutation is not a feature of MALT lymphoma with t(1;14)(p22;q32).33 The mechanism underlying the oncogenic role of t(1;14)(p22;q32) remains unclear; nevertheless, study of BCL10 protein expression pattern provides fresh clues to explain the above paradoxical findings.34 In contrast to normal B cells that express BCL10 in the cytoplasm, MALT lymphoma cells with t(1;14)(p22;q32) express the protein predominantly in the nucleus.34 Interestingly, up to 50% of MALT lymphomas without the translocation also express BCL10 mainly in the nucleus, albeit at a lower level.34 These results suggest that nuclear BCL10 expression is associated with MALT lymphoma development and may confer oncogenic activity.
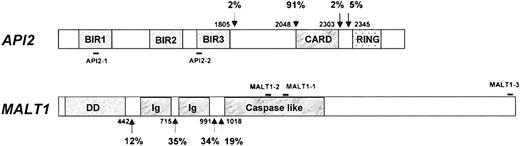
T(11;18)(q21;q21) results in the expression of a chimeric transcript between the API2 and MALT1 genes.24The API2 gene contains 3 N-terminal baculovirus IAP repeats (BIR), a middle caspase recruitment domain (CARD), and a C-terminal zinc-binding RING finger domain.35 Full-length API2 has been shown to inhibit the biologic activity of caspases 3, 7, and 9 and is, therefore, believed to be an apoptosis inhibitor.35,36The MALT1 gene, a paracaspase, comprises an N-terminal death domain (DD), followed by 2 immunoglobulinlike C2-type domains and a caspaselike domain, and its function is unknown.24,37 By reverse transcription–polymerase chain reaction (RT-PCR) of the API2-MALT1 fusion transcript, Southern blot analysis, and interphase fluorescence in situ hybridization for t(11;18)(q21;q21), the translocation has been found in 30% to 50% MALT lymphoma of various sites but not in nodal or splenic marginal zone B-cell lymphoma.38-42 However, it remains to be determined how this translocation contributes to the progression of MALT lymphoma.
Interestingly, there appears to be no difference in histology, immunophenotype, and clinical behavior between MALT lymphomas with t(1;14)(p22;q32) and t(11;18)(q21;q21). It is thus possible that both translocations exert their oncogenic activities through a similar pathway. To understand the role of t(11;18)(q21;q21) and BCL10 nuclear expression during MALT lymphoma development, we screened 33 cases ofH pylori gastritis, 72 MALT lymphomas, and 11 mucosal DLBCL for t(11;18)(q21;q21) and examined patients with MALT lymphoma for BCL10 expression. T(11;18)(q21;q21) and BCL10 expression pattern were correlated with the clinical staging of lymphoma and with each other.
Materials and methods
Materials
Fresh-frozen gastric biopsies from 39 patients with gastritis were collected from the Hospital of Ancona (Italy). The diagnosis of gastritis was made after histologic examination; lymphoid infiltration was mild in 25 patients and severe with aggregated follicles in 14 patients. Gastric ulcer was seen in 3 patients, including 2 with mild and 1 with severe gastritis. H pylori was identified in 33 of 39 gastric biopsy specimens by Warthin Starry staining and histology.
Frozen tissue samples from 72 low-grade MALT lymphomas, including a case with t(1;14)(p22;q32), and 11 mucosal DLBCL were retrieved from the surgical files of the Department of Histopathology, Royal Free and University College Medical School. Three low-grade MALT lymphomas contained a small large-cell component within the B-cell follicles, and no mucosal DLBCL showed any low-grade MALT lymphoma lesion. Fifty-six MALT lymphomas were from the stomach, and the anatomic origins of the remaining cases are summarized in Table1. Nine gastric cases with disseminated disease were accompanied by additional tissue from lymph node, spleen, or small intestine. Clinical staging was available in 15 cases, whereas the extent of tumor spread was estimated in 26 gastric cases in which sufficient surgical material was available for histological examination.
Frequency of t(11;18)(q21;q21) and BCL10 nuclear expression in gastritis, mucosa-associated lymphoid tissue lymphoma, and mucosal diffuse large B-cell lymphomas
| . | Total no. cases . | No. t(11;18)-positive cases . | No. nuclear BCL10-positive cases/no. cases examined . |
|---|---|---|---|
| Gastritis | 39 | 0 | ND |
| MALT lymphoma | |||
| Stomach | 56 | 19 (34%) | 22/41 (54%) |
| Small intestine | 1 | 1 | 1/1 |
| Lung | 7 | 4 (57%) | 4/5 (80%) |
| Salivary gland | 6 | 1 (17%) | 1/5 (20%) |
| Conjunctiva | 1 | 0 | ND |
| Thyroid | 1 | 0 | 0/1 |
| Subtotal | 72 | 25 (35%) | 28/53 (53%) |
| Mucosal DLBCL | |||
| Stomach | 9 | 0 | 0/7 |
| Lung | 1 | 0 | ND |
| Colon | 1 | 0 | 0/1 |
| Subtotal | 11 | 0 | 0/8 |
| . | Total no. cases . | No. t(11;18)-positive cases . | No. nuclear BCL10-positive cases/no. cases examined . |
|---|---|---|---|
| Gastritis | 39 | 0 | ND |
| MALT lymphoma | |||
| Stomach | 56 | 19 (34%) | 22/41 (54%) |
| Small intestine | 1 | 1 | 1/1 |
| Lung | 7 | 4 (57%) | 4/5 (80%) |
| Salivary gland | 6 | 1 (17%) | 1/5 (20%) |
| Conjunctiva | 1 | 0 | ND |
| Thyroid | 1 | 0 | 0/1 |
| Subtotal | 72 | 25 (35%) | 28/53 (53%) |
| Mucosal DLBCL | |||
| Stomach | 9 | 0 | 0/7 |
| Lung | 1 | 0 | ND |
| Colon | 1 | 0 | 0/1 |
| Subtotal | 11 | 0 | 0/8 |
MALT indicates mucosa-associated lymphoid tissue; DLBCL, diffuse large B-cell lymphomas.
RNA extraction and cDNA synthesis
Total RNA was extracted from up to 10 mg frozen tissue using an RNeasy Mini Kit (Qiagen, West Sussex, United Kingdom). Up to 2 μg total RNA was reverse-transcribed into cDNA in a 20 μL volume using SuperScript Preamplification System (Life Technologies, Paisley, United Kingdom) and oligo(dT) primer. If the amount of total RNA was below the measurable level, such as from biopsy tissue samples, a maximum volume of RNA preparation was used for cDNA synthesis.
Amplification and sequencing of the API2-MALT1 fusion transcript
One microliter sample cDNA was added to a 25 μL reaction volume containing 0.2 mM dNTP, 2 mM MgCl2, 0.2 μM sense and antisense primers each, and 1 U Platinum Taq DNA polymerase (Life Technologies) and amplified on a Phoenix thermal cycler (Helena BioSciences, Sunderland, United Kingdom). The PCR was performed using a so-called touchdown program at 94°C for 4 minutes followed by 1 cycle of denaturing at 94°C for 1 minute, annealing at 65°C (1°C down each cycle until 59°C) for 1 minute and extension at 72°C for 1.5 minutes, and then 35 cycles at 94°C for 1 minute, 58°C for 1 minute, and 72°C for 1.5 minutes. A final extension at 72°C for 10 minutes concluded the reaction. Two sets of primers were used to cover all the known breakpoints in both theAPI2 and the MALT1 genes38-41(Figure 1) and were used for PCR of all samples unless otherwise indicated. The first set of primers consisted of sense primer (API2-1, 5′-CTG GTG TGA ATG ACA AGG TC-3′, nucleic acids 897-916, GenBank accession no. NM_001165) and antisense primer (MALT1-1, 5′-TAG TCA ATT CGT ACA CAT CC-3′, nucleic acids 1124-1143, AB026118).39 Primary PCR products were further amplified using nested primers: API2-2 (5′-ACA TTC TTT AAC TGG CCC TC-3′, nucleic acids 1505-1524) and MALT1-2 (5′-CAA AGG CTG GTC AGT TGT TT-3′, nucleic acids 1030-1049).39 Amplified PCR products were analyzed by electrophoresis on 0.9% agarose gels and ethidium bromide staining. All PCR reactions were performed at least in duplicate.
Schematic presentation of
API2 and MALT1 gene structure and primer position. Known breakpoints are indicated by arrows, and nucleic acids are numbered according to cDNA sequence of theAPI2 (GenBank, NM_001165) and MALT1 genes (AB026118). The frequency of individual known breakpoints was given.38-42 Solid bar indicates the position of primers used.
Schematic presentation of
API2 and MALT1 gene structure and primer position. Known breakpoints are indicated by arrows, and nucleic acids are numbered according to cDNA sequence of theAPI2 (GenBank, NM_001165) and MALT1 genes (AB026118). The frequency of individual known breakpoints was given.38-42 Solid bar indicates the position of primers used.
A 256-bp fragment of the glucose-6-phosphate dehydrogenase (G6PD) gene was amplified in parallel as a control to verify RNA quality and RT-PCR efficiency for each sample, using sense primer 5′-GAG GCC GTG TAC ACC AAG ATG AT-3′ and antisense primer 5′-AAT ATA GGG GAT GGG CTT GG-3′. Primers were chosen to flank a region containing introns 10 (104 bp) and 11 (105 bp) (GenBank X55448.1, M12996) so that the amplified product of contaminating DNA was distinguishable from that of cDNA.
To assess the sensitivity of RT-PCR for detection of the API2-MALT1 fusion transcript, cell suspensions of 2 gastric MALT lymphomas with different t(11;18)(q21;q21) were serially diluted with tonsillar lymphocytes. Cell mixtures, each containing a total of 2 × 106 cells but a variable amount of tumor cells, were subjected to RNA isolation and RT-PCR for the G6PD and API2-MALT1 fusion transcript.
PCR products were purified using a Concert Rapid PCR Purification System (Life Technologies) and sequenced in both directions with nested primers using dRhodamine dye terminators and AmpliTaq DNA polymerase, FS, on an ABI Prism 377 sequencer (PE Applied Biosystems, Foster City, CA).
BCL10 immunohistochemistry
BCL10 was immunostained using a mouse monoclonal antibody (clone 151) on formalin-fixed and paraffin-embedded tissues as described previously.34 Briefly, 4 μm tissue sections were heat-retrieved for antigen in target retrieval solution pH 6.0 (Dako, Glostrup, Denmark) in a microwave oven for 25 to 35 minutes, depending on the size of the section. Sections were then incubated with anti-BCL10 antibody at 1:60 dilution for 1 hour followed by biotinylated rabbit antimouse antibody and peroxidase-conjugated ExtroAvidin (Sigma Chemical, St Louis, MO). Finally, the staining was visualized with 3, 3-diaminobenzidine tetrahydrochloride (Kem-En-Tec A/S, Denmark) in H2O2.
Statistical analysis
Fisher exact and χ2 tests were used to analyze the correlation between the clinical staging of lymphoma with t(11;18)(q21;q21) or BCL10 expression pattern and the association between t(11;18)(q21;q21) and BCl10 expression pattern.
Results
T(11;18)(q21;q21) is absent in H pylorigastritis
RT-PCR used for the detection of the API2-MALT1 fusion transcript was highly sensitive. In each of the 2 dilution experiments with MALT lymphomas harboring different t(11;18)(q21;q21), the API2-MALT1 fusion transcript was detectable when the t(11;18)(q21;q21)-positive cells were diluted down to a concentration of 1 in 106 tonsillar cells using a single set of PCR primers (Figure2A).
Detection of t(11;18)(q21;q21) by RT-PCR of the API2-MALT1 fusion transcript.
(A) Sensitivity of RT-PCR for detection of the API2-MALT1 fusion transcript. Tumor cells harboring t(11;18)(q21;q21) were serially diluted with tonsillar cells and were then subjected to RNA extraction and RT-PCR. Using the first set of PCR primers (API2-1 and MALT1), the API2-MALT1 fusion transcript was detectable when the t(11;18)(q21;q21)-positive cells were diluted down to a concentration of 1 in 106 tonsillar cells. M, molecular weight marker; Ton, tonsillar cell; 10−6 to 10−1, various tumor cell concentrations; T, undiluted tumor cells; −, negative control. (B) T(11;18)(q21;q21)-positive MALT lymphoma. The number corresponds to the case number in Table 2. −, negative control. (C) Case 16 shows an identically sized API2-MALT1 product between the primary gastric MALT lymphoma and the tumor-involved lymph node, spleen, and small intestine.
Detection of t(11;18)(q21;q21) by RT-PCR of the API2-MALT1 fusion transcript.
(A) Sensitivity of RT-PCR for detection of the API2-MALT1 fusion transcript. Tumor cells harboring t(11;18)(q21;q21) were serially diluted with tonsillar cells and were then subjected to RNA extraction and RT-PCR. Using the first set of PCR primers (API2-1 and MALT1), the API2-MALT1 fusion transcript was detectable when the t(11;18)(q21;q21)-positive cells were diluted down to a concentration of 1 in 106 tonsillar cells. M, molecular weight marker; Ton, tonsillar cell; 10−6 to 10−1, various tumor cell concentrations; T, undiluted tumor cells; −, negative control. (B) T(11;18)(q21;q21)-positive MALT lymphoma. The number corresponds to the case number in Table 2. −, negative control. (C) Case 16 shows an identically sized API2-MALT1 product between the primary gastric MALT lymphoma and the tumor-involved lymph node, spleen, and small intestine.
RT-PCR of the reference gene G6PD was successful in all 39 patients with gastritis, confirming adequate RNA extraction and cDNA synthesis. Despite this and the high sensitivity of RT-PCR for detection of the API2-MALT1 fusion transcript, none of the patients with gastritis were positive for t(11;18)(q21;q21).
T(11;18)(q21;q21) is associated with more advanced MALT lymphomas but is absent in mucosal DLBCL
As with the gastritis samples, RT-PCR of the G6PD gene was successful in all lymphoma tissues examined. The API2-MALT1 fusion transcript was detected in 25 of 72 (35%) low-grade MALT lymphomas (Figure 2B, Table 2) but in none of the 11 mucosal diffuse large B-cell lymphomas. All these positive cases were detected by the primary PCR. Secondary nested PCR did not increase the sensitivity for detection of t(11;18)(q21;q21).
Summary of clinicopathologic and molecular data of t(11;18)(q21;q21)-positive mucosa-associated lymphoid tissue lymphomas
| Patients . | Age . | Sex . | Origin . | Lymphoma staging* . | BCL10 nuclear expression . | Breaking point‡ . | |
|---|---|---|---|---|---|---|---|
| API2 . | MALT1 . | ||||||
| 1 | 41 | M | Stomach | III (spleen, lymph node)† | + | 2048 | 715 |
| 2 | 49 | F | Stomach | IIE1 (lymph node) | NA | 2048 | 715 |
| 3 | 42 | F | Stomach | IIE1 (lymph node) | + | 2048 | 715 |
| 4 | 44 | M | Stomach | IIE1 | + | 2048 | 715 |
| 5 | 47 | M | Stomach | IIE | + | 2048 | 715 |
| 6 | 51 | M | Stomach | IE | + | 2048 | 715 |
| 7 | 54 | M | Stomach | IIE | NA | 2048 | 715 |
| 8 | 61 | M | Stomach | IIE2 | + | 2048 | 715 |
| 9 | 62 | M | Stomach | IE | + | 2048 | 715 |
| 10 | 65 | M | Stomach | IIE1 | + | 2048 | 715 |
| 11 | 72 | M | Stomach | III (spleen) | NA | 2048 | 715 |
| 12 | NA | M | Stomach | III | + | 2048 | 991 |
| 13 | 39 | M | Stomach | III (spleen, lymph node) | + | 2048 | 991 |
| 14 | 44 | M | Stomach | IV | + | 2048 | 991 |
| 15 | 49 | M | Stomach | IE | + | 2048 | 991 |
| 16 | 60 | F | Stomach | IV (small intestine, spleen, lymph node) | + | 2048 | 991 |
| 17 | 60 | M | Stomach | IE | + | 2048 | 991 |
| 18 | 44 | F | Stomach | IE | + | 2048 | 1018 |
| 19 | 63 | M | Stomach | III | + | 2048 | 1018 |
| 20 | 49 | M | Lung | NA | NA | 2048 | 715 |
| 21 | 66 | F | Lung | NA | + | 2048 | 715 |
| 22 | 49 | F | Lung | NA | + | 2048 | 715 |
| 23 | 46 | M | Lung | NA | NA | 2048 | 991 |
| 24 | 68 | M | Small intestine | IE | + | 2048 | 1018 |
| 25 | 53 | M | Salivary gland | NA | + | 2048 | 715 |
| Patients . | Age . | Sex . | Origin . | Lymphoma staging* . | BCL10 nuclear expression . | Breaking point‡ . | |
|---|---|---|---|---|---|---|---|
| API2 . | MALT1 . | ||||||
| 1 | 41 | M | Stomach | III (spleen, lymph node)† | + | 2048 | 715 |
| 2 | 49 | F | Stomach | IIE1 (lymph node) | NA | 2048 | 715 |
| 3 | 42 | F | Stomach | IIE1 (lymph node) | + | 2048 | 715 |
| 4 | 44 | M | Stomach | IIE1 | + | 2048 | 715 |
| 5 | 47 | M | Stomach | IIE | + | 2048 | 715 |
| 6 | 51 | M | Stomach | IE | + | 2048 | 715 |
| 7 | 54 | M | Stomach | IIE | NA | 2048 | 715 |
| 8 | 61 | M | Stomach | IIE2 | + | 2048 | 715 |
| 9 | 62 | M | Stomach | IE | + | 2048 | 715 |
| 10 | 65 | M | Stomach | IIE1 | + | 2048 | 715 |
| 11 | 72 | M | Stomach | III (spleen) | NA | 2048 | 715 |
| 12 | NA | M | Stomach | III | + | 2048 | 991 |
| 13 | 39 | M | Stomach | III (spleen, lymph node) | + | 2048 | 991 |
| 14 | 44 | M | Stomach | IV | + | 2048 | 991 |
| 15 | 49 | M | Stomach | IE | + | 2048 | 991 |
| 16 | 60 | F | Stomach | IV (small intestine, spleen, lymph node) | + | 2048 | 991 |
| 17 | 60 | M | Stomach | IE | + | 2048 | 991 |
| 18 | 44 | F | Stomach | IE | + | 2048 | 1018 |
| 19 | 63 | M | Stomach | III | + | 2048 | 1018 |
| 20 | 49 | M | Lung | NA | NA | 2048 | 715 |
| 21 | 66 | F | Lung | NA | + | 2048 | 715 |
| 22 | 49 | F | Lung | NA | + | 2048 | 715 |
| 23 | 46 | M | Lung | NA | NA | 2048 | 991 |
| 24 | 68 | M | Small intestine | IE | + | 2048 | 1018 |
| 25 | 53 | M | Salivary gland | NA | + | 2048 | 715 |
Other abbreviations are explained in Table 1.
Patients 11 and 18 had a large cell component within the B-cell follicle.
NA, not applicable.
Ann Arbor-Musshoff staging system for extranodal lymphoma.
Tissues available for RT-PCR and showing fusion transcripts identical to their corresponding primary tumors are indicated in parentheses.
Nucleic acids denote breakpoints according to GenBank NM_001165 for API2 and AB026118 for MALT1.
The frequency of t(11;18)(q21;q21) in MALT lymphoma of various sites is shown in Table 1. Of those arising in the stomach, the largest group examined, the API2-MALT1 fusion transcript, was found in 19 of 56 (34%) samples. Among t(11;18)(q21;q21)-positive gastric MALT lymphomas, frozen tissues from the lymphoma involving lymph nodes, spleen, or small intestine were available in 6 cases. In each case, RT-PCR of these disseminated lesions showed an API2-MALT1 fusion product identically sized to that of their corresponding gastric lymphoma (Figure 2C, Table 2). Of the 3 cases with a large cell component within the B-cell follicle, 2 were t(11;18)(q21;q21) positive. However, the available frozen tumor tissues in these cases did not contain a prominent large cell component to allow examination of whether t(11;18)(q21;q21) was present in the transformed cells.
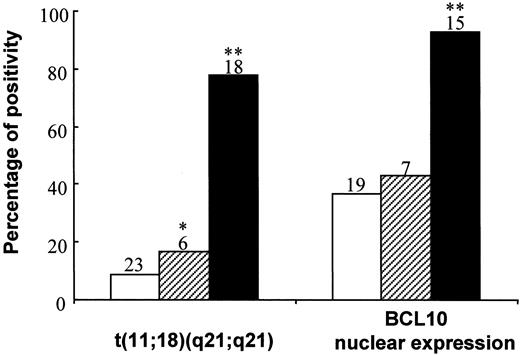
Of 56 low-grade gastric MALT lymphomas, the extent of tumor spread was assessed in 47 cases. As shown in Figure3, t(11;18)(q21;q21) was associated with more advanced gastric MALT lymphomas: 2 of 23 (9%) tumors were confined to the gastric mucosa or submucosa, but 1 of 6 (17%) (P < .01; Fisher exact test) tumors invaded the muscular layer or serosa and 14 of 18 (78%) (P < .0001; χ2 test) tumors disseminated beyond the stomach.
Correlation between clinical staging of gastric MALT lymphoma and t(11;18)(q21;q21) and BCL10 nuclear expression.
The number of cases in individual subgroups is indicated at the top of the corresponding histogram. ■, submucosa; ▨, muscle and serosa; ▪, spread beyond stomach;*P < .01; **P < .005.
Correlation between clinical staging of gastric MALT lymphoma and t(11;18)(q21;q21) and BCL10 nuclear expression.
The number of cases in individual subgroups is indicated at the top of the corresponding histogram. ■, submucosa; ▨, muscle and serosa; ▪, spread beyond stomach;*P < .01; **P < .005.
Sequencing analysis of the RT-PCR products confirmed the presence of an API2-MALT1 fusion transcript in all positive cases. The breakpoint was invariable at nucleotide 2048 (intron 7) on the API2 gene (NM_001165) but varied at nucleotide 715 (intron 4, 15 of 25, 60%), 991 (intron 7, 7 of 25, 28%) or 1018 (intron 8, 3 of 25, 12%) on theMALT1 gene (AB026118),43 fusing the N-terminal of the API2 gene, which contains 3 intact BIR domains, to variable carboxyl terminal regions of the MALT1 gene (Table2).
BCL10 nuclear expression is associated with advanced MALT lymphomas
BCL10 protein expression was investigated in 53 MALT lymphomas. Twenty-eight cases showed BCL10 expression predominantly in the nuclei of most tumor cells, and the remaining cases showed cytoplasmic BCL10 expression. As with t(11;18)(q21;q21), BCL10 nuclear expression was also associated with advanced MALT lymphomas. Of 41 gastric MALT lymphomas in which the lymphoma staging was available, nuclear BCL10 expression was found in 7 of 19 (37%) tumors confined to the mucosa or the submucosa, 3 of 7 (43%) cases in which tumor invaded to muscular layer or serosa, and 14 of 15 (93%) cases in which tumor disseminated beyond the stomach (P < .005; χ2 test) (Figure 3). All 8 mucosal DLBCLs examined showed cytoplasmic but not nuclear BCL10 expression.
MALT lymphoma with t(11;18)(q21;q21) expresses nuclear BCL10
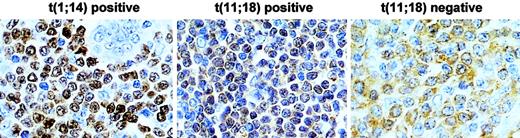
In total, 50 cases were examined for both t(11;18)(q21;q21) and BCL10 expression pattern. BCL10 nuclear expression was significantly higher in t(11;18)(q21;q21)-positive (20 of 20, 100%) than in t(11;18)(q21;q21)-negative MALT lymphomas (7 of 30, 23%) (Fisher exact test, P = .426 × 10−8) (Figure 4).
Comparison of BCL10 protein expression between MALT lymphomas with and without t(11;18)(q21;q21).
T(11;18)(q21;q21)-positive case shows BCL10 (brown) nuclear expression in most of tumor cells, whereas the negative case displays cytoplasmic BCL10 expression. The level of BCL10 nuclear expression in t(11;18)(q21;q21)-positive cells is much lower than that in t(1;14)(p22;q32)-positive cells. Original magnification, × 650. Sections were counterstained with hematoxylin.
Comparison of BCL10 protein expression between MALT lymphomas with and without t(11;18)(q21;q21).
T(11;18)(q21;q21)-positive case shows BCL10 (brown) nuclear expression in most of tumor cells, whereas the negative case displays cytoplasmic BCL10 expression. The level of BCL10 nuclear expression in t(11;18)(q21;q21)-positive cells is much lower than that in t(1;14)(p22;q32)-positive cells. Original magnification, × 650. Sections were counterstained with hematoxylin.
In view of the significant association between t(11;18)(q21;q21) and nuclear BCL10 expression, we re-examined the 7 cases showing nuclear BCL10 expression but not t(11;18)(q21;q21) to ascertain whether the absence of the translocation in these cases was due to a failure of detection by the primer set used. A new MALT primer (MALT1-3, 5′-TTT TTC AGA AAT TCT GAG CCT G-3), which targets the 3′ end of its coding region, and API2-1 primer were used for PCR (Figure 1). All 7 cases consistently showed absence of the API2-MALT1 fusion transcript despite successful amplification of t(11;18)(q21;q21)-positive controls.
Of these 7 cases, 2 showed high BCL10 nuclear expression similar to that seen in t(1;14)(p22;q32)-positive MALT lymphomas.34 However, insufficient frozen tumor tissue was available for Southern blot analysis to test whether these cases harbored t(1;14)(p22;q32).
Discussion
Chromosomal translocations associated with B-cell lymphomas commonly involve the immunoglobulin locus, most likely occur during the VDJ recombination process, and are believed to be the primary events in lymphomagenesis.44 At least some of these translocations are known to occur in prelymphomatous lesions. For example, t(14;18)(q32;q21), which is associated with up to 90% of follicular lymphomas, has also been found in approximately 50% of lymphoid hyperplasias and peripheral blood lymphocytes from healthy controls.45-47 To examine whether t(11;18)(q21;q21), a frequent translocation in MALT lymphoma,38-41 is present in prelymphomatous lesions, we examined H pylori-associated gastritis from cases in Italy, where the frequency of H pylori-associated gastric MALT lymphoma is high.48 Our results indicate that t(11;18)(q21;q21) is absent or at least not a frequent event in H pylori-associated gastritis.
The oncogenic role of t(11;18)(q21;q21) in MALT lymphoma development is further underpinned by the finding that the translocation is associated with advanced gastric tumors, particularly those showing tumor spread beyond the stomach. These results are well in agreement with the observation of a high frequency (75%) of t(11;18)(q21;q21) in gastric MALT lymphomas that were resistant to H pylori eradication therapy but not in those that responded to the therapy.49,50 It has been shown that gastric MALT lymphomas not responsive to H pylori therapy are those with invasion of deeper layers of the gastric wall and involvement of regional lymph nodes, whereas responsive tumors are generally those confined to the gastric mucosa.8 9 Thus, t(11;18)(q21;q21) is a molecular marker for aggressive gastric MALT lymphomas and those not responsive to H pylori therapy.
In view of the significant role of t(11;18)(q21;q21) during MALT lymphoma progression, it is intriguing that the translocation has not been found in transformed MALT lymphoma.38-41 Although mucosal DLBCL may arise de novo, at least a proportion of mucosal DLBCLs are transformed from low-grade MALT lymphomas.10,11It is unlikely that such an important translocation is lost during high-grade transformation. Alternatively, t(11;18)(q21;q21)-positive MALT lymphoma is a distinct subgroup of MALT lymphoma that does not undergo or rarely undergoes high-grade transformation. There are clear differences in gross chromosomal abnormalities between t(11;18)(q21;q21)-positive and -negative MALT lymphomas: the former do not usually show any chromosomal aberrations other than t(11;18)(q21;q21), whereas the latter, including lymphomas with t(1;14)(p22;q32), display various abnormalities including recurrent and rare aberrations.19-21 It remains to be determined whether further differences in histology, immunophenotype, and genetic abnormalities such as genetic instability exist between the 2 groups.
The other significant finding of the present study is the strong correlation between t(11;18)(q21;q21) and BCL10 nuclear expression. It is possible that this correlation was simply the result of their independent association with advanced MALT lymphomas. Alternatively, the oncogenic activity of t(11;18)(q21;q21) may be associated with nuclear BCL10 expression. This is indirectly supported by the low frequency of nuclear BCL10 expression in follicular lymphoma and DLBCL,34 in which t(11;18)(q21;q21) is absent.38-42 More important, both BCL10 and the API2-MALT1 fusion product activate NFκB, and BCL10 binds MALT1 and some of the API2-MALT1 products.22 37 However, the precise mechanisms underlying the molecular interaction between BCL10 and API2-MALT1 and the functional consequence of such an interaction remain to be investigated.
It is interesting to note that 7 t(11;18)(q21;q21)-negative cases also expressed nuclear BCL10. Nuclear BCL10 expression in 2 cases could be attributed to the presence of t(1;14)(p22;q32) because the BCL10 staining pattern resembled that seen in MALT lymphomas with the translocation.34 The factors underlying nuclear BCL10 expression in the remaining cases are not clear. However, involvement of 11q21 by translocations other than t(11;18) (q21;q21) has been reported in lymphoid malignancies.54 Studies are in progress to examine whether these cases harbor API2 rearrangement.
Despite strong evidence of an important role for t(11;18)(q21;q21) in MALT lymphoma progression, the molecular mechanisms underlying its oncogenic activity remain elusive. All the breakpoints in theAPI2 gene occur between the third BIR domain and the C-terminal RING, with 91% occurring just upstream of the CARD.38-41 In contrast, the breakpoints in the MALT1gene are more variable.38-41 Thus, the resultant API2-MALT1 fusion transcripts always comprise a truncated API2 with 3 intact BIR domains but without the C-terminal RING or without both the middle CARD and C-terminal RING in most. The characteristic disruption of the API2 gene by the translocation strongly suggests that truncated API2 with retention of the BIR domain, a critical element for antiapoptotic activity, is required for oncogenic activity. This is supported by the finding that the RING domain of cIAP1 and API2 negatively regulates the antiapoptotic activity of their BIR domains.51 The negative effect of the RING finger on BIR function may be associated with its ability to promote auto-ubiquitination and degradation.51-53 Replacement of the C-terminal of API2 with the C-terminal of MALT1 by the fusion product would release the intrinsic antiapoptotic activity of the BIR domain and, therefore, make the new molecule antiapoptotic. The incoming MALT1 C-terminal may further enhance the antiapoptotic activity of the BIR domain by an unknown mechanism. It has been demonstrated that the API2-MALT1 fusion product, but not API2 or MALT1 alone, was capable of activating NFκB,37 further suggesting the importance of the C-terminal MALT1 in the oncogenic activity of the fusion product.
We thank Dr Tim C. Diss for critical reading of the manuscript.
H.L. and H.Y. contributed equally to this work.
Supported by the Cancer Research Campaign and the Leukaemia Research Fund, United Kingdom.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ming-Qing Du, Department of Histopathology, Royal Free and University College Medical School, Rockefeller Building, University Street, London WC1E 6JJ, United Kingdom; e-mail:m.du@ucl.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal