Hairy cell leukemia (HCL) derives from a mature B cell and expresses markers associated with activation. Analysis of immunoglobulin variable region genes has revealed somatic mutation in most cases, consistent with an origin from a cell that has encountered the germinal center. One unusual feature of hairy cells (HCs) is the frequent expression of multiple immunoglobulin heavy chain isotypes, with dominance of immunoglobulin (Ig)–G3, but only a single light chain type. The origin and clonal relationship of these isotype variants have been unclear. In order to probe the isotype switching status of HCL, RNA transcripts of VHDJH– constant region sequences from 5 cases of typical HCL, all expressing multiple surface immunoglobulin isotypes, were analyzed. Tumor VHDJH-Cμ sequences were identified and found to be somatically mutated (range, 1.4% to 6.5%), with a low level of intraclonal heterogeneity. Additional immunoglobulin isotypes of identical VHDJHsequence were also identified, including IgD (5 of 5), IgG3 (5 of 5), IgG1 (3 of 5), IgG2 (2 of 5), IgA1 (4 of 5), and IgA2 (1 of 5). Derivation of multiple isotypes from individual cells was demonstrated by analyzing transcripts in single sorted cells from one patient, with evidence for coexistence of isotype variants in 10 of 10 cells. These findings indicate that clonally related multiple isotypes coexist in single HCs, with individual isotypes presumably generated via RNA splicing. Production of IgG3 appears common, but IgG1, IgG2, IgA1, and IgA2 also arise, indicating a continuing influence of a directed process on the tumor clone. These HCs appear to be arrested at the point of isotype switch, where RNA processing may precede deletional recombination.
Introduction
Hairy cell leukemia (HCL) is a relatively rare leukemia of B-cell origin, which derives its name from the characteristic cytoplasmic projections of the pathognomonic cell. Clinically, HCL is characterized by pancytopenia with monocytopenia and splenomegaly. Hairy cells (HCs) circulate in the blood and infiltrate the bone marrow. Characteristically, HCs populate the splenic red pulp, and eventually the white pulp atrophies and is replaced, but lymph nodes are generally not involved.1 HCs express FMC7, a normal B-cell activation marker, and commonly express the α chain of the interleukin-2 receptor CD25. Other important diagnostic markers are CD11c, HC2, B-Ly7, and HML-1 (CD103), the latter 2 recognizing the β7 subunit of the same integrin and being the most specific for the diagnosis of HCL.2-4
The status of the immunoglobulin genes provides an indicator of the maturation stage reached by B-cell tumors.5 Naive B cells prior to antigen encounter have unmutated V genes, while B cells that have entered the site of somatic mutation, generally considered to be in the germinal center of the lymph node, are likely to have accumulated somatic mutations.6 For B-cell tumors, the mutational status can have clinical relevance, as found in chronic lymphocytic leukemia (CLL) where the approximately 50% of cases with unmutated VH genes have a significantly worse prognosis.7,8 The majority of B-cell tumors show evidence of somatic mutation, and those located in germinal center sites commonly continue to accumulate mutations after transformation.9-11 However, tumors of more mature B cells, such as multiple myeloma, have VH genes with mutated but homogeneous sequences, indicative of silencing of mutational activity at the postfollicular stage.12 13
Isotype switching events can clearly take place in the germinal center environment,14 but normal IgM+memory cells can also exit from this site, circulate in the blood,15 and move to the marrow.16 In general, B-cell tumors have reflected the known sequence of isotype switching events in normal cells and express IgM(IgD) or, following deletional recombination, a downstream isotype.17 However, immunophenotypic studies have indicated dual isotype expression in some B-cell tumors, and switching events leading to additional isotypes have been confirmed by analysis of RNA transcripts in CLL18-20and in diffuse large B-cell lymphoma (DLBCL).21 In myeloma, where the tumor clone has usually undergone isotype switching, and where there is no phenotypic evidence for IgM+precursor cells, transcripts of CDR3-Cμ have also been detected.22 23 Clearly B-cell tumor clones can include cells at different stages of both somatic mutation and isotype switching.
Analysis of functionally rearranged VH gene segments in HCL has indicated that most sequences contain somatic mutations consistent with a cell of origin that has encountered the germinal center.24 The level of mutation in the functional Vκ genes of 3 cases of HCL appeared low, probably reflecting the generally lower rate of mutation in VL as compared with VH.25,26 There has been a suggestion that a low level of ongoing somatic mutation is occurring in the tumor clone, which could indicate continuing influence of the mutator.27 However, the low level of intraclonal diversity found and the absence of common base changes among different clones have left this question open.27
The most unusual feature of HCs is expression of multiple immunoglobulin isotypes on the tumor cell surface in about 40% of cases.28 Although preferential expression of IgG3 has been documented,29 multiple isotype expression on HCs has been reported previously.30-33 It was initially suggested that this may be the result of passively adsorbed polyclonal immunoglobulin, since HCs have avid Fc receptors,34-36 but rosetting techniques and the use of F(ab′)2reagents have confirmed expression of multiple immunoglobulin isotypes in HCL.37
An analysis of the clonal relatedness of the multiple isotypes expressed was made by generating heterohybridomas from HCs by fusion to a mouse myeloma cell line.38 It was found by Southern blotting that hybridomas contained the tumor VHrearrangement, and that individual hybridomas secreted IgM, IgG, or IgA, indicating that the tumor clone was undergoing isotype switching.38 However, this approach and the possible perturbing effect of heterohybridization did not allow further insight into the production of the multiple isotypes.38 Since conventional isotype switching involves intrachromosomal recombination, with looping out and deletion of intervening constant-region genes, expression of multiple isotypes within a tumor clone is exceptional. Either the clone is heterogeneous, with subsets undergoing a switch to different isotypes, or processing of RNA transcripts to generate different isotypes within the same cell is occurring.39Our analysis of transcripts of 5 cases of HCL in which all cells express multiple immunoglobulin isotypes appears consistent with the latter.
Patients, materials, and methods
Patients
Peripheral blood mononuclear cells (PBMCs) were obtained from 5 patients with diagnosis of HCL referred to the Division of Hematology in Siena, Italy. Three were males and 2 females, with a median age of 55 years (range, 30-59 years). Diagnosis was based on immunohistochemical examination of bone marrow biopsies and morphological and immunological findings of the PBMCs.
Phenotypic analysis
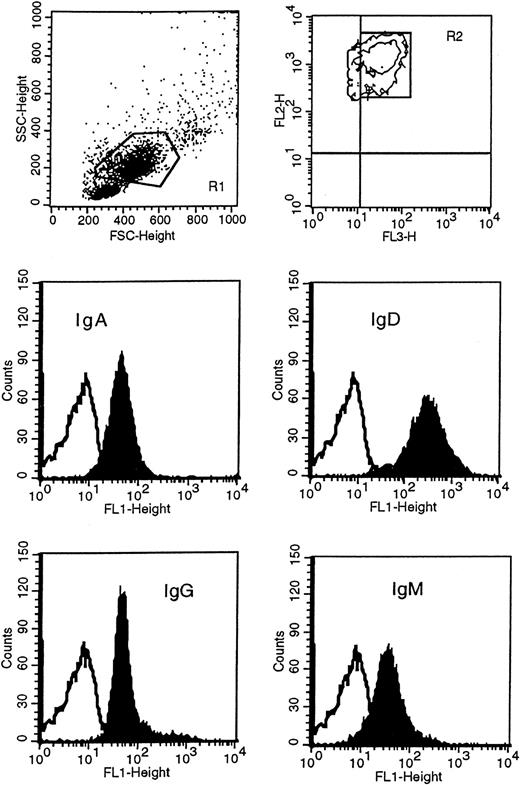
Leukemic cells were obtained following Lymphoprep (Nycomed Pharma, Oslo, Norway) gradient separation from peripheral blood of HCL patients. Immunophenotypic studies were carried out on either fresh or previously cryopreserved leukemic cells, since antigenic expression was not modified by cryopreservation in dimethyl sulfoxide (DMSO) and thawing. Before incubating cells with conjugated monoclonal antibody, mononuclear cells were incubated in RPMI medium with 10% fetal calf serum at 37°C in 5% CO2 overnight to remove polyclonal immunoglobulin absorbed to the class-specific Fc receptors, and then separated again to remove dead cells. Immunophenotypic analysis was performed with direct immunofluorescence technique by dual- or triple-color staining with a large panel of monoclonal antibodies, including anti-CD19, anti-CD11c, anti-CD20, anti-CD23, anti-CD25, anti-CD27, and anti-CD38 (Becton Dickinson, San Diego, CA); anti-CD103, anti-FMC7, anti–κ/λ light chains (Dako, Glostrup, Denmark); and anti-SmIg [surface membrane Ig], anti-IgA, anti-IgD, anti-IgG, and anti-IgM (Caltag Laboratories, Burlingame, CA). In particular, expression of immunoglobulin isotypes on HCs was performed by 3-color staining by means of fluorescein isothiocyanate (FITC)–conjugated F(ab′)2 anti-IgG, anti-IgM, anti-IgD, anti-IgA; peridinin chlorophyll protein (PerCP)-conjugated anti-CD19; and phycoerythrin-conjugated anti-CD11c. HCs were identified as high side scatter (SSC) and high forward scatter (FSC) PBMCs, CD19+/CD11c+ (Figure1). Negative control was performed by incubating the cells with appropriate isotype IgG1- or IgG2-conjugated monoclonal antibodies. Data acquisition and analysis were performed by flow cytometry (FACScan) (Becton Dickinson) with CellQuest software (Becton Dickinson). In all cases 10 000 events were acquired. Expression of antigens was defined from both the percentage of positive cells and the mean fluorescence index (MFI). The MFI represents the ratio of sample mean channel to control mean channel; expression was designated positive when the MFI was greater than 1.
Immunophenotypic analysis of isotype proteins on HCs.
HCs were identified in the R1 region as CD11c+ (FL2) and CD19+ (FL3) and gated in R2. Cells in R1 and R2 were then analyzed for the surface expression of IgM, IgD, IgG, and IgA with FITC(FL1)-conjugated F(ab′)2 monoclonal antibody specific to each class. This example represents patient 163. The unfilled curves in the histogram plots represent the isotype control.
Immunophenotypic analysis of isotype proteins on HCs.
HCs were identified in the R1 region as CD11c+ (FL2) and CD19+ (FL3) and gated in R2. Cells in R1 and R2 were then analyzed for the surface expression of IgM, IgD, IgG, and IgA with FITC(FL1)-conjugated F(ab′)2 monoclonal antibody specific to each class. This example represents patient 163. The unfilled curves in the histogram plots represent the isotype control.
Identification and analysis of the tumor-related VHDJH-Cμ and VHDJH-Cδ isotype transcripts
Total RNA was isolated by means of RNAzol (Cinna Biotecx Labs, Houston, TX) from 5 × 106 mononuclear cell fractions prepared from anticoagulated peripheral blood and cryopreserved in 10% DMSO freezing medium. Reverse transcription to complementary DNA (cDNA) was carried out with Not I-d(T)18 primer and First-Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech, Uppsala, Sweden) in a final volume of 15 μL according to the manufacturer's protocol. One fifth of the cDNA was subjected to amplification.
For identification of tumor VH-Cμtranscripts, a mixture of 5′-primers specific for each of the VH leader sequences of VH1 through VH7 families (VH leader mix)11 was used together with a 3′-primer specific for Cμ(Cμ16) (Table 1). In all cases, polymerase chain reaction (PCR) was carried out in a final volume of 50 μL with 20 pmol each primer, 50 μmol deoxynucleotide triphosphates (dNTPs), and 2.5 U Taq DNA polymerase with reaction buffer (Qiagen, Crawley, United Kingdom). Amplification consisted of an initial denaturation step of 5 minutes at 94°C, followed by 30 cycles of 94°C for 1 minute, 56°C for 1 minute, and 72°C for 1 minute, with a final extension step of 5 minutes at 72°C. In order to prevent contamination, the previously described general procedure were followed, including a control for each PCR with no added template to check for any contaminant.40 For the identification of VH-Cδ transcript, the same conditions were used with either a 5′-primer specific for the tumor-related VH leader sequence or a VH leader mix and 3′-primer specific for Cδ (Cδ1) (Table 1).
Oligonucleotide heavy-chain primers
| Primers . | Location* . | Orientation . | Sequence (5′-3′) . |
|---|---|---|---|
| CH-specific | |||
| Cμ16 | μCH1 (120-115) | Antisense | TTG GGG CGG ATG CAC T |
| Cδ1 | δCH1 (173-164) | Antisense | TTC TCT GGG GCT GTG TC |
| CγFF1 | γCH1 (192-184) | Antisense | GGT CAC CAC GCT GCT GAG GGA GTA GA |
| CαFF1 | αCH1 (187-179) | Antisense | CGT GGT GTA CAG GTC CCC GGA GGC A |
| Cα4 | αCH1 (125-118) | Antisense | GCT CAG CGG GAA GAC CTT GGG GCT |
| VH(patient)-specific | |||
| 163-CDR3 | V1-02 (92-99) | Sense | GTG CGT CGC TCC CAT ATG ATT ACA |
| 163-CDR1 | V1-02 (27-35) | Sense | CAC CTT CAC CGG CTA CTA TAT GCA |
| 103-CDR3 | V4-31 (92-100) | Sense | TGT GCG AGA GAT TTT ACC CTT CCG A |
| 103-CDR1 | V4-34 (32-37) | Sense | GGT GGT TAC TAC TGG AGC TGG G |
| 83-CDR3 | V3-23 (92-99) | Sense | TGT GCG AAA GAT TGG GCT GGG GA |
| 83-CDR1 | V3-23 (26-34) | Sense | GGA TTC ACC TTT AGC AGC TAT GCC A |
| 42-CDR3 | V3-33 (90-99) | Sense | CTA CTG TGC GAG AGA TAT GGG GCT A |
| 42-CDR1 | V3-33 (24-31) | Sense | GCG TCT GGA TTC CCC TTC AGT GA |
| 93-CDR3 | V3-23 (91-99) | Sense | TAT TGT GCG AGA GAT CAA CAC AGT A |
| 93-CDR1 | V3-23 (27-34) | Sense | TTC ATC TTC AGT ACC TAT GGC ATG |
| β-actin RNA | |||
| 5′ H-βA | Exon 2 | Sense | TGA TAT CGC CGC GCT CGT |
| 3′ H-βA | Exon 4 | Antisense | CTC GGC CGT GGT GGT GAA |
| Primers . | Location* . | Orientation . | Sequence (5′-3′) . |
|---|---|---|---|
| CH-specific | |||
| Cμ16 | μCH1 (120-115) | Antisense | TTG GGG CGG ATG CAC T |
| Cδ1 | δCH1 (173-164) | Antisense | TTC TCT GGG GCT GTG TC |
| CγFF1 | γCH1 (192-184) | Antisense | GGT CAC CAC GCT GCT GAG GGA GTA GA |
| CαFF1 | αCH1 (187-179) | Antisense | CGT GGT GTA CAG GTC CCC GGA GGC A |
| Cα4 | αCH1 (125-118) | Antisense | GCT CAG CGG GAA GAC CTT GGG GCT |
| VH(patient)-specific | |||
| 163-CDR3 | V1-02 (92-99) | Sense | GTG CGT CGC TCC CAT ATG ATT ACA |
| 163-CDR1 | V1-02 (27-35) | Sense | CAC CTT CAC CGG CTA CTA TAT GCA |
| 103-CDR3 | V4-31 (92-100) | Sense | TGT GCG AGA GAT TTT ACC CTT CCG A |
| 103-CDR1 | V4-34 (32-37) | Sense | GGT GGT TAC TAC TGG AGC TGG G |
| 83-CDR3 | V3-23 (92-99) | Sense | TGT GCG AAA GAT TGG GCT GGG GA |
| 83-CDR1 | V3-23 (26-34) | Sense | GGA TTC ACC TTT AGC AGC TAT GCC A |
| 42-CDR3 | V3-33 (90-99) | Sense | CTA CTG TGC GAG AGA TAT GGG GCT A |
| 42-CDR1 | V3-33 (24-31) | Sense | GCG TCT GGA TTC CCC TTC AGT GA |
| 93-CDR3 | V3-23 (91-99) | Sense | TAT TGT GCG AGA GAT CAA CAC AGT A |
| 93-CDR1 | V3-23 (27-34) | Sense | TTC ATC TTC AGT ACC TAT GGC ATG |
| β-actin RNA | |||
| 5′ H-βA | Exon 2 | Sense | TGA TAT CGC CGC GCT CGT |
| 3′ H-βA | Exon 4 | Antisense | CTC GGC CGT GGT GGT GAA |
Parentheses represent codons aligned by the primers. Numbering is according to criteria defined by Chothia et al.61
Amplified products were run on 1.5% to 2% agarose gel and purified with the Gene Clean III kit (Bio 101, Vista, CA). DNA was ligated into pGEM-T vector (Promega, Southampton, United Kingdom). Cloning was performed with Escherichia coli strain JM109 (Promega), and sequencing was performed with the Big Dye sequencing reaction kit, a GeneAmp 9600 PCR system, and an ABI Prism 377 DNA sequencer (PerkinElmer, Warrington, United Kingdom). M13 forward and reverse primers or Sp6 primer was used to sequence in both directions. The data were analyzed by means of MacVector 4.5.3 software and aligned to Entrez and V-BASE databases.41 62
Analysis of VH gene usage and mutation pattern was performed as previously described.11 Alternative transcripts were considered tumor derived when they shared the same CDR3 sequence of the tumor VHDJH-Cμ transcript. Intraclonal heterogeneity was assessed in the cloned products and was distinguished from Taq infidelity by an increased frequency as compared with Taq error rate and by the finding of the same mutations in more than one clone and/or in different PCRs.
Qiagen Taq polymerase error rate was assessed by means of the following strategy. A pair of primers, 5′H-βA and 3′H-βA, specific for human β-actin were designed (Table 1) to amplify a band of 568 base pairs (bps). We used cDNA from patient 103 with PCR conditions identical to those used for the amplification of IgM transcripts. The amplified band of the right size was cut, purified, and cloned as described above. Twelve clones were then sequenced in both directions by means of either M13 and 5′H-βA or 3′H-βA primers. The error rate was calculated as [(v × 1000) / (568 × n)] × 10−3bp−1 where v is the number of observed nucleotide variations from the germline sequence and n is the number of clones sequenced.
Identification of VHDJH-Cα/Cγisotype transcripts
After identification of a patient's tumor-related variable heavy chain, CDR3-specific 5′-primers were designed and used together with either Cα- or Cγ-specific 3′-primers. After the CDR3–constant region products were identified by PCR, cloning, and sequencing, then clonal CDR1-specific 5′-primers were designed and used with the same Cα (CαFF1) or Cγ (CγFF1) primers to confirm the presence of the alternative transcripts and to obtain extended VHDJH–constant sequence. This strategy also allowed comparison of the pattern of mutation among different isotypes in the tumor. The positioning of the constant-region primers facilitated discrimination among different subclasses in the PCR product and could provide an analysis of Cγ and Cα subclasses present within the tumor clone. Table 1describes primer sequences and location. In all cases, conventional PCRs, without the need of nested or seminested PCRs, were performed as described above, by means of 30 cycles with 65°C annealing temperature. Amplified products were then cloned, sequenced, and analyzed.
Analysis of tumor-derived transcripts in HCL single cells
Material from patient 103 was available for further analysis. Leukemic cells were preincubated, and labeled with CD11c. HCs were identified as high SSC, high FSC PBMCs, and CD11c+ and sorted on a Becton Dickinson FACS Vantage instrument equipped with an automated cell-deposition unit (Becton Dickinson). Sorting of single events had previously been confirmed by separation of 2.49 μm Nile Red beads (Becton Dickinson). In addition, sorting and V gene analysis of normal CD19+ B cells were found to give single, unrelated sequences (C.I.M., unpublished data, September 2000). Single leukemic cells were sorted into 0.2-mL PCR tubes containing 10 μL of 1 × Superscript II First-Strand Buffer (Gibco BRL, Life Technologies, Paisley, United Kingdom). Sorted cells were snap-frozen on dry ice and stored at −80°C. Reverse transcription to cDNA was carried out in a final volume of 25 μL with random hexamers and Superscript II RNase H− Reverse Transcriptase according to the manufacturer's instructions (Gibco BRL).
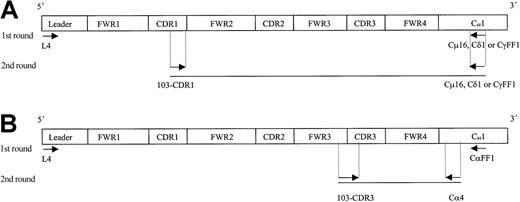
Individual IgM, IgD, and IgG transcripts were identified from single cells by means of a seminested PCR approach, while IgA transcripts were identified by means of a nested PCR (Figure2). In the first round, 3 μL cDNA was amplified by means of VH4 leader primer and a respective downstream Cμ16, Cδ1, CγFF1 or CαFF1 primer. Initial PCR was for 40 cycles with an annealing temperature of 56°C (for IgM) or 65°C (for IgD, IgG, and IgA).
Strategy for the identification of IgM, IgD, IgG, and IgA transcripts from single HCs.
Ten single tumor cells (FSChi/SSChi/CD11c+) from patient 103 were sorted on a FACS Vantage, and cDNA was prepared by means of random hexamers. A seminested (A, for IgM, IgD, IgG) or a nested (B, for IgA) PCR approach was used to identify each specific isotype transcript from individual HCs. Arrows indicate location of the primers used. Sequence and codon location for each primer are described in Table 1 and Thompsett et al11 (family 4–specific leader primer). FWR indicates framework region; CDR, complementary determining region; and C, constant region.
Strategy for the identification of IgM, IgD, IgG, and IgA transcripts from single HCs.
Ten single tumor cells (FSChi/SSChi/CD11c+) from patient 103 were sorted on a FACS Vantage, and cDNA was prepared by means of random hexamers. A seminested (A, for IgM, IgD, IgG) or a nested (B, for IgA) PCR approach was used to identify each specific isotype transcript from individual HCs. Arrows indicate location of the primers used. Sequence and codon location for each primer are described in Table 1 and Thompsett et al11 (family 4–specific leader primer). FWR indicates framework region; CDR, complementary determining region; and C, constant region.
For secondary PCR, 3 μL of the first PCR DNA product was amplified by means of the internal 103-CDR1 primer with individual 3′-CHprimers. For the identification of IgA transcripts, the internal Cα4 primer was used (Table 1). PCR conditions were maintained as in the first round. Amplified DNA products of the expected size were excised from agarose gels, purified, and directly sequenced bidirectionally by means of PCR primers.
Results
Immunophenotype and immunoglobulin isotype surface expression
Table 2 summarizes the clinical and immunophenotypic characteristics of the 5 HCL patients who entered this study. Phenotypic analysis of PBMCs confirmed diagnosis of HCL since all cases were CD19+, CD20+, CD25+, FMC7+, CD103+, CD11c+, CD79b+, with strong expression of SmIg and negative expression of CD23. The leukemic population, identified as CD19+/CD103+ cells, represented 20% to 77% of PBMCs. It is noteworthy that no cases expressed CD38 or the memory B-cell marker CD27.42
Clinical and immunophenotypic characteristics of patients with hairy cell leukemia
| Patient No. . | Age/gender . | Therapy* . | CD19† . | SmIg . | κ/λ . | CD11c . | CD103 . | CD20 . | CD25 . | FMC7 . | CD79β . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 42 | 57/M | No | 28 | 25 | λ | 21 | 20 | 16 | 28 | 21 | 27 |
| 83 | 30/F | No | 63 | 86 | κ | 63 | 77 | 76 | 77 | 67 | 87 |
| 93 | 55/M | IFN-α | 63 | 74 | λ | 63 | 63 | 76 | 63 | 79 | 79 |
| 103 | 59/M | No | 65 | 67 | λ | 65 | 65 | 65 | 68 | 68 | 68 |
| 163 | 45/F | No | 45 | 56 | κ | 44 | 37 | 56 | 54 | 38 | 43 |
| Patient No. . | Age/gender . | Therapy* . | CD19† . | SmIg . | κ/λ . | CD11c . | CD103 . | CD20 . | CD25 . | FMC7 . | CD79β . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 42 | 57/M | No | 28 | 25 | λ | 21 | 20 | 16 | 28 | 21 | 27 |
| 83 | 30/F | No | 63 | 86 | κ | 63 | 77 | 76 | 77 | 67 | 87 |
| 93 | 55/M | IFN-α | 63 | 74 | λ | 63 | 63 | 76 | 63 | 79 | 79 |
| 103 | 59/M | No | 65 | 67 | λ | 65 | 65 | 65 | 68 | 68 | 68 |
| 163 | 45/F | No | 45 | 56 | κ | 44 | 37 | 56 | 54 | 38 | 43 |
SmIg indicates surface membrane Ig; IFN, interferon.
Treatment at the time when material was obtained for phenotypic and molecular immunoglobulin studies.
Percentage of positive cells in the mononuclear cell fraction.
To determine the surface expression of immunoglobulin isotypes on the neoplastic cells of our patients, we used FITC-conjugated F(ab′)2 isotype-specific monoclonal antibodies (Figure 1 illustrates patient 163 as an example). As described in Table3, all patients expressed IgM, IgD, and IgG at varying levels. IgA was expressed in 2 cases (patients 103 and 163) in 94% and 85% of the tumor population, respectively. In patients 83 and 93, we could not detect the clear presence of the IgA protein although a small shift of the sample peak as compared with the isotype control peak was observed. Only patient 42 was phenotypically completely negative for IgA. Although the level of expression of each isotype appeared different within some of the cases, there was no indication of separate populations expressing different immunoglobulin classes.
Immunoglobulin isotype protein expression in hairy cell leukemia
| Ig . | Expression, % (MFI), by patient no.3-150 . | ||||
|---|---|---|---|---|---|
| 42 . | 83 . | 93 . | 103 . | 163 . | |
| IgM | 59 (4.1) | 21 (16.2) | 61 (8.4) | 94 (9.1) | 87 (8) |
| IgD | 93 (12) | 60 (6.4) | 70 (16) | 98 (39) | 95 (55) |
| IgG | 59 (3.0) | 28 (22.1) | 40 (8.3) | 90 (4.1) | 99 (10.7) |
| IgA | NEG | NEG | NEG | 85 (1.9) | 94 (8.6) |
| Ig . | Expression, % (MFI), by patient no.3-150 . | ||||
|---|---|---|---|---|---|
| 42 . | 83 . | 93 . | 103 . | 163 . | |
| IgM | 59 (4.1) | 21 (16.2) | 61 (8.4) | 94 (9.1) | 87 (8) |
| IgD | 93 (12) | 60 (6.4) | 70 (16) | 98 (39) | 95 (55) |
| IgG | 59 (3.0) | 28 (22.1) | 40 (8.3) | 90 (4.1) | 99 (10.7) |
| IgA | NEG | NEG | NEG | 85 (1.9) | 94 (8.6) |
Ig indicates immunoglobulin; MFI, mean fluorescence index; NEG, negative.
Percentage of positive cells and mean fluorescence index (in parentheses) for each isotype were determined on the CD19/CD11c hairy cells.
Analysis of tumor-derived VH genes
Preparations of cDNA from all 5 patients were amplified, cloned, and sequenced initially by means of a VH leader mix and Cμ primers. The identification of tumor-derived VH genes was based on a common CDR3 signature sequence among multiple clones from each patient's amplified cDNA. Table4 shows the number of tumor-derived clones identified and the total number of clones sequenced for the IgM transcript. Sequences not related to the tumor clone differed individually and were most likely to derive from normal B cells. The VH gene used by each tumor, together with deviation in homology from the germline counterpart, is also shown in Table 4. The deduced amino acid sequences are shown in Figure3. Nucleotide sequences have been deposited in the GenBank database (accession numbers: AF302819,AF302820, AF302821, AF302822, AF302823).
VH gene analysis in hairy cell leukemia
| Patient no. . | Ig . | Tumor-derived clones per total no. clones . | VH . | Homology, %4-150 . | JH . | D . |
|---|---|---|---|---|---|---|
| 42 | M | 8/10 | 3 -33 | 93.5 | 4b | 6-13 |
| 83 | M | 8/8 | 3 -23 | 97.6 | 4b | 2-21 |
| 93 | M | 7/8 | 3 -33 | 96.9 | 6b | 4-114-151 4-044-151 |
| 103 | M | 10/11 | 4 -31 | 98.6 | 2 | — |
| 163 | M | 11/12 | 1 -02 | 98.2 | 4b | 3-16 |
| Patient no. . | Ig . | Tumor-derived clones per total no. clones . | VH . | Homology, %4-150 . | JH . | D . |
|---|---|---|---|---|---|---|
| 42 | M | 8/10 | 3 -33 | 93.5 | 4b | 6-13 |
| 83 | M | 8/8 | 3 -23 | 97.6 | 4b | 2-21 |
| 93 | M | 7/8 | 3 -33 | 96.9 | 6b | 4-114-151 4-044-151 |
| 103 | M | 10/11 | 4 -31 | 98.6 | 2 | — |
| 163 | M | 11/12 | 1 -02 | 98.2 | 4b | 3-16 |
Ig indicates immunoglobulin.
Homology to the closest germline VH gene segment.
Case 93 CDR3 tumor sequence was aligned to 2 different D-gene segments of the D4 family with a significant but identical level of confidence.
Deduced amino acid sequences of tumor-derived VH-Cμ transcripts from patients with HCL.
Dashes indicate homology; upper case, replacement mutations; lower case, silent mutations.
Deduced amino acid sequences of tumor-derived VH-Cμ transcripts from patients with HCL.
Dashes indicate homology; upper case, replacement mutations; lower case, silent mutations.
VH gene segments were derived from V3-33in 2 cases, and V3-23, V4-31, and V1-02 in the other 3 cases. CDR3 length varied from 12 to 19 codons (mean, 16 codons). JH4b was used in 3 cases, while JH6b and JH2 were used in the remaining 2 cases. According to the rule proposed by Corbett et al,43 D gene segment could be assigned with confidence in 3 cases (patients 163, 42, and 93), In patient 83, CDR3 alignment did not reach the interval of confidence, while in patient 103 the D segment was not identified. All tumor-related VH genes revealed nucleotide substitutions with the percentage of homology ranging from 93.2% to 98.6%.
Intraclonal heterogeneity
In all our 5 cases, intraclonal heterogeneity exceeding Taq error (0.8 × 10−3 bp−1) was observed. The general level of intraclonal variation was low, ranging from 1.2 × 10−3 (patient 163) to 3.6 × 10−3bp−1 (patient 103) with a median of 2 × 10−3 bp−1 in the IgM transcripts analyzed, and it is difficult to assess the significance of this level. However, the reality of mutations was supported by the presence of repeats among different PCRs. In fact, there was clear evidence of repeated identical intraclonal changes common to at least 2 different isotype transcripts in all cases: in particular, identical nucleotide variations were seen in tumor-derived IgM and IgD clones in patient 163; in IgM and IgG in patient 103; in IgD, IgG, and IgA in patient 42; and in all isotypes in patients 83 and 92. Figure4 illustrates intraclonal heterogeneity in patient 83 as an example.
Analysis of VH–constant region transcripts from case 83.
Alignment is made to the closest germline gene segments V3-23 and JH 4b (bold characters). By means of leader (for IgM and IgD) or VH primers (for IgA and IgG), together with constant region primers for μ, δ, γ, and α, transcripts for IgM, IgD, IgG2, IgG1, IgA1, and IgG3 were identified. All isotype transcripts share an identical mutational grade of homology to the germline sequence. Identical intraclonal variations, like GG → AA substitution (amino acid G → N) in codon 55 or C → G in codon 101, are seen in different isotype transcripts. Letters and numbers above germline sequence indicate amino acid sequence and codon numbers. The number of identical clones are indicated. Sequence identity is shown by dots; mutations are shown by mismatch characters.
Analysis of VH–constant region transcripts from case 83.
Alignment is made to the closest germline gene segments V3-23 and JH 4b (bold characters). By means of leader (for IgM and IgD) or VH primers (for IgA and IgG), together with constant region primers for μ, δ, γ, and α, transcripts for IgM, IgD, IgG2, IgG1, IgA1, and IgG3 were identified. All isotype transcripts share an identical mutational grade of homology to the germline sequence. Identical intraclonal variations, like GG → AA substitution (amino acid G → N) in codon 55 or C → G in codon 101, are seen in different isotype transcripts. Letters and numbers above germline sequence indicate amino acid sequence and codon numbers. The number of identical clones are indicated. Sequence identity is shown by dots; mutations are shown by mismatch characters.
Transcripts for alternative isotypes
After identification of the VHDJH-Cμ transcripts, we sought VHDJH-Cδ, VHDJH-Cγ, and VHDJH-Cα tumor-derived transcripts, with results shown in Table5. For all patients, we found transcripts for IgD, and full VHDJH-Cδ–derived sequences had a mutational pattern identical to the clonal IgM transcript. Tumor-derived VHDJH-Cγ clonal sequences were also identified in 5 of 5 patients, with IgG3 transcripts in all patients, IgG1 in 3 of 5, and IgG2 in 2 of 5. In terms of frequency within the IgG class, IgG3 appeared predominant (Table 5) in all patients except for patient 93, where IgG1 and IgG2 were most evident. IgA-derived clonal sequences were detected in 4 of 5 patients, with predominance of IgA1 (Table 5).
Constant-region transcripts in hairy cell leukemia
| Patient no. . | IgM . | IgD . | IgG . | IgA . |
|---|---|---|---|---|
| 42 | + (8) | + (8) | γ3 + (12) | α1 + (10) |
| γ1 + (4) | ||||
| γ2 + (1) | ||||
| 83 | + (8) | + (10) | γ3 + (20) | α1 + (7) |
| 93 | + (7) | + (2) | γ3 + (3) | α1 + (15) |
| γ1 + (11) | α2 + (1) | |||
| γ2 + (10) | ||||
| 103 | + (10) | + (6) | γ3 + (27) | α1 + (16) |
| 163 | + (11) | + (8) | γ3 + (28) | ND |
| γ1 + (1) |
| Patient no. . | IgM . | IgD . | IgG . | IgA . |
|---|---|---|---|---|
| 42 | + (8) | + (8) | γ3 + (12) | α1 + (10) |
| γ1 + (4) | ||||
| γ2 + (1) | ||||
| 83 | + (8) | + (10) | γ3 + (20) | α1 + (7) |
| 93 | + (7) | + (2) | γ3 + (3) | α1 + (15) |
| γ1 + (11) | α2 + (1) | |||
| γ2 + (10) | ||||
| 103 | + (10) | + (6) | γ3 + (27) | α1 + (16) |
| 163 | + (11) | + (8) | γ3 + (28) | ND |
| γ1 + (1) |
The numbers in parentheses are the number of clones identified for each class (Cμ and Cδ) or subclass (Cγ and Cα). For Cα and Cγ, the number of clones represents the sum of CDR3-CH and CDR1-CH sequences obtained for each subclass.
Ig indicates immunoglobulin; ND, not detected.
Full-length VHDJH sequences linked to Cγ and Cα from all patients had mutational patterns identical to the tumor-related VHDJH-Cμ sequences, with no significant further accumulation of mutations; this is consistent with the silencing of somatic mutation at the IgM stage. An example (patient 83) is shown in Figure 4, where it is clear that, in spite of some intraclonal heterogeneity, the sites and level of mutational change are similar in all isotypes.
Expression of immunoglobulin protein generally correlated with detection of transcripts. All cases expressed both the message and the protein for IgM, IgD, and IgG isotypes. However, for IgA there was some disparity. This was most evident in patients 42 and 163. In patient 42, α1 transcript was detected but no IgA protein was detectable at the cell surface: the reason is unclear but may reflect a difference in sensitivity of the 2 assays. In patient 163, although IgA protein was clearly expressed, no transcript could be detected: this is likely to be due to primer design.
Multiple tumor-derived transcripts in single HCs
Table 6 summarizes the results obtained from 10 individual HCs. PCR controls without template were negative in all cases, excluding possibility of contamination. All individual cells were tumor- derived and, apart from 2 nonidentical base changes in 2 separate cells, probably due to Taq infidelity, shared an identical pattern of VH mutation to the tumor sequence, previously identified by whole population analysis, irrespective of isotype. All cells expressed the IgG3 transcript. In one cell, IgM, D, G3, and A transcripts could be identified. Seven out of 10 cells expressed at least 3 different isotypes (3 IgM/IgD/IgG3; 4 IgD/IgG3/IgA), and the remaining 2 cells expressed 2 different isotypes (1 IgD/IgG3; 1 IgG3/IgA).
Alternative isotype transcripts in single hairy cells from patient 103
| Cell . | IgM . | IgD . | IgG . | IgA . |
|---|---|---|---|---|
| A2 | + | + | + | |
| A3 | + | + | ||
| A4 | + | + | + | |
| A5 | + | + | + | |
| A6 | + | + | + | + |
| A7 | + | + | + | |
| A8 | + | + | + | |
| A9 | + | + | + | |
| A10 | + | + | + | |
| A12 | + | + |
| Cell . | IgM . | IgD . | IgG . | IgA . |
|---|---|---|---|---|
| A2 | + | + | + | |
| A3 | + | + | ||
| A4 | + | + | + | |
| A5 | + | + | + | |
| A6 | + | + | + | + |
| A7 | + | + | + | |
| A8 | + | + | + | |
| A9 | + | + | + | |
| A10 | + | + | + | |
| A12 | + | + |
+ indicates that isotype transcripts were detected; a blank cell indicates that isotype transcripts were not detected.
Discussion
HCL appears to be derived from a mature B cell, and the presence of somatic mutations in VH genes confirms that the cell of origin has encountered a site where mutation is activated. The clear evidence for intraclonal variation also indicates that mutation continued at a low level after transformation, consistent with a possible influence of activated T cells and antigen.44 The most distinctive feature of approximately 40% of cases of HCL, and of all our randomly selected cases, is the expression of multiple immunoglobulin isotypes, apparently by the majority of cells in the clone. In normal B cells, isotype switching generally occurs by recombination between donor and acceptor switch (S) sites located 5′ of each constant-region gene,45-48 with consequent looping out and deletion of the intervening CH genes. The outcome of this deletional mechanism is that a B cell will usually express only a single isotype. The exceptions are IgM and IgD, which are commonly coexpressed owing to the lack of a conventional Sδsite.49,50 Expression of IgM and IgD relies on differential splicing of the long RNA transcript.49 51Since this will have the same VHDJH upstream sequence, the mutational patterns in IgM and IgD should be identical.
The deletional model, however, has to accommodate observations that normal B cells in the mouse can express IgM together with downstream isotypes, and that mouse B cells expressing IgM and IgG1 contain nuclear RNAs that have both Cμ and Cγ1 sequences.52 Similar results were obtained for cells coexpressing IgM and IgG3, which had pre–messenger RNA (mRNA) transcripts containing both Cμ and Cγ3.53 The likely mechanism for dual expression in this case therefore involves RNA processing, rather than translation of long-lived VHDJH-Cμ mRNA.53
Most studies of human B cells have centered on Burkitt lymphoma cell lines, where isotype switching appears to occur mainly by deletional recombination, with 75% of cases deleting Cμ on both allelic chromosomes.54 Normal Epstein-Barr virus–transformed B cells show a similar tendency to undergo switching on both alleles, often to the same constant region.55,56However, the switching process on the nonproductive allele can also be surprisingly variable, with complex deletions and duplications observed.57 In the cases of HCLs that express a single downstream isotype, most appear to have undergone this process, often on both alleles.58
Analysis of the switching process in spontaneous human B-cell tumors that synthesize both IgM and a downstream isotype has been more limited. One difficulty is that such tumors, although clonally related, may be arrested at different points of differentiation. This was evident in our study of a subset of lymphoplasmacytoid leukemia, where the tumor clone can express and secrete IgM and IgG.59Analysis of VH gene sequences in these cases revealed clonal relatedness between the isotypes. However somatic mutational patterns differed in 2 of 5 patients, clearly showing that the tumor population was heterogeneous, with a divergent mutational activity at the IgM and IgG stages.59 Cases with identical mutational patterns were not assessed, since material for single-cell analysis would be required. In our cases of DLBCL, clonally related isotype-switch variants again showed distinct mutational patterns, with clustering into an IgMD/IgG3 set or an IgG1 set.21Evidence for existence of subpopulations in at least some tumors was therefore clear.
The cases of HCL we have studied show no evidence for the existence of subpopulations, either phenotypically or in somatic mutational pattern; this suggests that most cells were synthesizing multiple isotypes. However, because suitable material was available for one case, we were able to take the investigation further by analyzing events in single cells. The finding that 9 of 10 cells contained coexisting transcripts of preswitched (IgM/IgD) and postswitched (IgG/IgA) immunoglobulins allowed us to conclude that individual cells were able to synthesize multiple isotypes. It excluded the alternative possibility that subpopulations were present, each having undergone deletional switch events to different isotypes. It is also in complete accord with the phenotypic data. Strikingly, in one cell, IgM, IgD, IgG, and IgA isotypes were detected. In all cases, transcripts were derived from the same VHDJHrecombination and are therefore from the functionally re-arranged allele. The most likely explanation is that, as for the mouse B cells studied previously, the cells are processing long nuclear RNA transcripts.39,52 At present, it is not known whether this involves the cis or the trans allele. Previous analysis of constant-region genes in cases of HCL expressing multiple isotypes had shown that 2 of 3 cases had abnormalities in the JH-Cμ intronic region28 similar to those identified previously by Laffan and Luzzatto.60The abnormalities arose from the presence of an inverted Cμ-containing sequence 5′ to a normal Cγ3 sequence, presumed to result from an attempt to undergo deletional isotype switch.17 If that kind of process is occurring in our cases, it must be confined to the nonfunctional allele.
It has been suggested that the stage of RNA processing may be passed through by normal B cells, and that trans-spliced chimeric germ line transcripts may serve as “bridging templates” for normal immunoglobulin class-switching recombination.39 It is possible that this subset of HCL is arrested at a stage in which isotype switching is being attempted. The potentially functional transcripts that we have identified from VHDJH–constant region sequences appear in the main to produce immunoglobulin protein. Events in the tumor cells of HCL appear to support the concept that multiple isotype-switch variants are first generated by RNA processing. If so, HCL may provide a useful model to probe these events and to investigate the reason for arrest.
Supported by Associazione Italiana contro le Leucemie (Italy); Multiple Myeloma Research Foundation (United States); and Leukaemia Research Fund (United Kingdom).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Francesco Forconi, Molecular Immunology Group, Tenovus Laboratory, Southampton University Hospitals, Tremona Rd, Southampton S016 6YD, United Kingdom; e-mail: ff1@soton.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal