Abstract
Primary systemic amyloidosis (AL) is a protein conformation disorder in which monoclonal immunoglobulin light chains produced by clonal plasma cells are deposited as amyloid in the kidneys, heart, liver, or other organs. Why patients with AL present with amyloid disease that displays such organ tropism is unknown. This study tested the hypothesis that both the light-chain variable region (IgVL) germ line genes used by AL clones and the plasma cell burden influenced AL organ tropism. To assess the renal tropism of some light chains, an in vitro renal mesangial cell model of amyloid formation was used. With reverse transcription-polymerase chain reaction, Ig VL genes were sequenced from 60 AL patients whose dominant involved organs were renal (52%), cardiac (25%), hepatic (8%), peripheral nervous system (8%), and soft tissue and other (7%). Patients with clones derived from the 6a VλVI germ line gene were more likely to present with dominant renal involvement, whereas those with clones derived from the 1c, 2a2, and 3r Vλ genes were more likely to present with dominant cardiac and multisystem disease. Patients withVκ clones were more likely to have dominant hepatic involvement and patients who met the Durie criteria for myeloma (38%, 23 of 60) were more likely to present with dominant cardiac involvement independent of germ line gene use. In the in vitro model, unlike all other AL light chains tested, λVI light chains formed amyloid rapidly both with and without amyloid-enhancing factor. These data support the hypothesis that germ line gene use and plasma cell burden influence the organ tropism of AL.
Introduction
Primary systemic amyloidosis (AL) is a rare protein conformation and clonal plasma cell disorder similar to multiple myeloma.1,2 In AL, fibrillar material usually composed of the amino termini of immunoglobulin light chains (often of the λ isotype) is deposited in key viscera by an unknown mechanism.3-10 At diagnosis, AL usually displays a tropism for one organ system or another, resulting in various dominant symptomatic clinical presentations. For example, patients with dominant cardiac involvement may present with right-sided heart dysfunction and may have electrocardiograms showing low precordial voltage and a pattern of myocardial infarction in the absence of coronary artery disease (pseudoinfarct pattern). Patients with renal involvement often present with proteinuria in the nephrotic range, whereas those whose peripheral nervous system is involved present with orthostasis or sensorimotor polyneuropathy.10 These distinctive features—fibrillar deposits, λ predominance, and the tropism of organ involvement—remain unexplained.
During early B-cell development, immunoglobulin germ line genes rearrange with retention of selected genes, endowing each B cell with one heavy-chain and one light-chain variable region germ line gene to encode the hypervariable or complementarity-determining regions (CDRs) of the immunoglobulin protein. In the antigen-dependent stage of B-cell development, the rearranged germ line genes mutate at a much higher rate than somatic genes, a process that results in unique immunoglobulin variable region gene sequences (IgVL;VL = Vλ andVκ). In a clonal B-cell disorder, these sequences provide a signature for the clone. Identification of clonal Ig VL genes with the polymerase chain reaction (PCR) has proven useful in the study of B-cell disorders and has permitted the compilation of comprehensive directories of IgVλ and Vκ germ line genes.11-14 Using these databases, the clonal IgVL genes of patients with B-cell disorders can be assigned germ line genes and assessed for homology to germ line sequences. In addition, investigators have determined the relative contributions of germ line genes to the circulating B-cell pool in healthy individuals (the normal expressed repertoire).15-19 By such approaches, a germ line gene rarely used in the normal repertoire may be shown to be used preferentially in a particular B-cell disorder.20
In this report we identify clonal Ig VLgenes from patients with AL to test the hypothesis that the tropism of organ involvement is a function of Ig VL germ line gene use and plasma cell burden. We used the presence of myeloma as defined by the Durie diagnostic criteria as a surrogate for plasma cell burden and, for the sake of analysis, assumed that patients with myeloma have a greater plasma cell burden and produce more amyloid-forming light chains.21 In addition, we used an in vitro renal mesangial cell model for amyloid formation to assess the relative activities of AL light chains of different isotypes. Our results demonstrate that Ig VL germ line gene use in AL is preferential, involving several genes that make minimal contributions to the normal repertoire, and that the tropism of organ involvement in AL is significantly influenced by IgVL germ line gene use and clonal plasma cell burden.
Patients, materials, and methods
Patients and plasma cell disease
The patients with AL were evaluated for the extent of amyloid-related organ involvement and for dominant organ involvement by standard criteria as previously described.22-24 Briefly, at presentation patients were categorized according to clinical manifestations as having renal, cardiac, hepatic, or neuropathic dominant organ involvement. Patients with more than one of these features were categorized according to the most prominent and symptomatic organ involvement. Other manifestations, such as soft-tissue involvement, were defined based on tissue biopsy results or pathognomonic physical findings (eg, shoulder-pad sign). Plasma cell disease was evaluated as previously described,23 and clonal plasma cell disorders were categorized as meeting or not meeting diagnostic criteria for myeloma using the Durie major and minor criteria as described.21
Renal involvement was defined as proteinuria more than 0.5 g/d and renal failure as dialysis dependence or creatinine clearance less than 10 mL/min. Cardiac involvement was defined as the presence of a mean left ventricular wall thickness on echocardiogram more than 11 mm in the absence of a history of hypertension or valvular heart disease, or as the presence of unexplained low voltage (< 0.5 mV) on the electrocardiogram. Patients who were New York Heart Association (NYHA) class 1 with evidence of cardiac amyloid by echocardiogram or electrocardiogram were categorized as having asymptomatic cardiac involvement. Patients who were NYHA class 2 or higher with evidence of cardiac involvement were categorized as having dominant cardiac involvement. Neuropathic involvement was defined based on clinical history, autonomic dysfunction with orthostasis, gastric atony by gastric emptying scan, and abnormal sensory or motor findings on neurologic examination. Hepatic involvement was defined as hepatomegaly with an alkaline phosphatase level more than 200 U/L.
Specimen preparation and cloning of IgVLgenes
Bone marrow aspirates were obtained from patients with biopsy-proven AL who gave written informed consent under institutional review board–approved protocols. Marrow cells were treated with ammonium chloride to lyse red blood cells, washed, pelleted, and frozen as previously described.25,26For preparation of complementary DNA (cDNA), total RNA was extracted from 107 marrow cells using TRIzol (Gibco-BRL, Gaithersburg, MD). Preparations of cDNA were made with Superscript II reverse transcriptase (Life Technologies, Grand Island, NY) as previously described.25,26 The cDNA was amplified by PCR using 5′ oligonucleotide FR1 primers specific for the expressed Ig VL subtypes with aCκ or pan-Cλprimer.27 Each specimen was subject to multiple amplifications and bands were selected for cloning from 3 separate PCR experiments. The amplified material from the PCR reactions was prepared in a TA Cloning Kit (Invitrogen, San Diego, CA). Twelve colonies representative of the 3 separate reactions were picked for plasmid isolation and screened for appropriately sized inserts byEcoR1 digest and electrophoresis. Five to 10 inserts were sequenced backward and forward at core facilities. A clonal IgVL was identified provided that one gene was distinctly overrepresented in each patient and that the gene was present in a minimum of 3 inserts. Because the FR1 primers routinely introduce minor sequencing errors, primers forVL leader (L) regions were designed and additional PCR amplifications with L-CLprimers were performed to sequence FR1 correctly (Table1, Figure1). Genes with correctly sequencedFR1 regions were analyzed for homology to germ line donor sequence and evidence of somatic hypermutation.
Polymerase chain reaction sense primers for germ line gene leader regions and amplicon lengths when used with 5′ constant region antisense primers
| Germ line donor (subtype) . | Primer sequence (5′-3′) . | Amplicon length (bp) . |
|---|---|---|
| 1c (VλI) | ATG GCC AGC TTC CCT CTC CTC | ∼ 447 |
| 2a2 (VλII) | ATG GCC TGG GCT CTG CTG CTC | " |
| 3r (VλIII) | ATG GCA TGG ATC CCT CTC TTC | " |
| 6a (VλVI) | ATG GCC TGG GCT CCA CTA CTT | ∼ 435 |
| O18-O8 (VKI) | ATG GAC ATG AGG GTC CCT GCT | ∼ 414 |
| LFVK431 (VKI) | ATG GAC ATG AGA GTC CTC GCT | " |
| Germ line donor (subtype) . | Primer sequence (5′-3′) . | Amplicon length (bp) . |
|---|---|---|
| 1c (VλI) | ATG GCC AGC TTC CCT CTC CTC | ∼ 447 |
| 2a2 (VλII) | ATG GCC TGG GCT CTG CTG CTC | " |
| 3r (VλIII) | ATG GCA TGG ATC CCT CTC TTC | " |
| 6a (VλVI) | ATG GCC TGG GCT CCA CTA CTT | ∼ 435 |
| O18-O8 (VKI) | ATG GAC ATG AGG GTC CCT GCT | ∼ 414 |
| LFVK431 (VKI) | ATG GAC ATG AGA GTC CTC GCT | " |
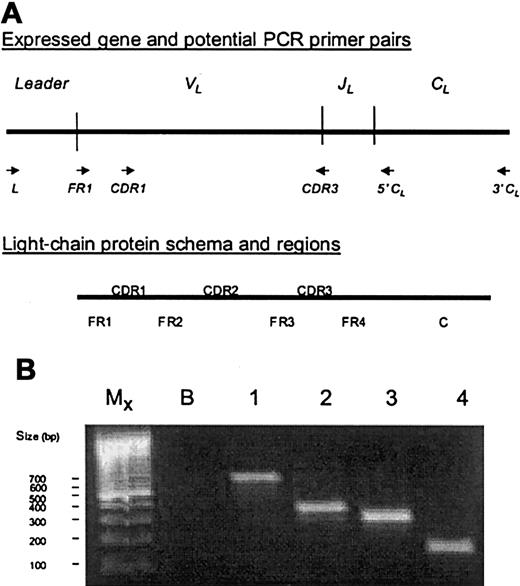
Light-chain variable region gene, protein, and PCR.
(A) The upper sketch shows the Leader,VL (variable), JL(joining), and CL (constant) gene segments as found in an expressed gene, and the various sets of primers that may be designed to amplify the variable region gene or portions of it. The lower sketch shows a schema of the light-chain protein and its regions. Framework regions (FR1-4) provide structure and usually have few amino acid replacements due to somatic mutation. Complementarity-determining regions (CDR1-3) provide sites for antigen binding and have frequent amino acid mutations due to somatic mutation. The constant region (C) also provides structure. Note that theVJ junction comprises the middle of the CDR3 region. (B) A PCR gel is shown of the same VλVI light-chain cDNA amplified with different sets of primers. Lanes 1 through 4 show segments of different length amplified with primers to different portions of the gene: lane 1, L-3′ CLprimers → 708 bp; lane 2, L-5′ CLprimers → 435 bp; lane 3, FR1-5′ CL primers → 378 bp; and lane 4, CDR1-CDR3 primers → 231 bp. PCR with unique CDR1-CDR3 primers is useful for detecting minimal residual disease and clonotypic contamination of stem cell components. MX indicates size markers as shown in legend; B, no cDNA blank; bp, base pairs.
Light-chain variable region gene, protein, and PCR.
(A) The upper sketch shows the Leader,VL (variable), JL(joining), and CL (constant) gene segments as found in an expressed gene, and the various sets of primers that may be designed to amplify the variable region gene or portions of it. The lower sketch shows a schema of the light-chain protein and its regions. Framework regions (FR1-4) provide structure and usually have few amino acid replacements due to somatic mutation. Complementarity-determining regions (CDR1-3) provide sites for antigen binding and have frequent amino acid mutations due to somatic mutation. The constant region (C) also provides structure. Note that theVJ junction comprises the middle of the CDR3 region. (B) A PCR gel is shown of the same VλVI light-chain cDNA amplified with different sets of primers. Lanes 1 through 4 show segments of different length amplified with primers to different portions of the gene: lane 1, L-3′ CLprimers → 708 bp; lane 2, L-5′ CLprimers → 435 bp; lane 3, FR1-5′ CL primers → 378 bp; and lane 4, CDR1-CDR3 primers → 231 bp. PCR with unique CDR1-CDR3 primers is useful for detecting minimal residual disease and clonotypic contamination of stem cell components. MX indicates size markers as shown in legend; B, no cDNA blank; bp, base pairs.
Sequence analysis of VLgenes and proteins
Sequence alignment analyses were performed by GenBank BLAST (Basic Local Alignment Search Tool) search and germ line gene counterparts were assigned by V-BASE (Ig variable region gene database,http://www.mrc-cpe.cam.ac.uk/imt-doc/vbase-home-page.html) sequence directory comparison based on maximum homology of the nucleotide sequences.28,29 Homology to germ line sequence was calculated for complete VL genes excluding nucleotides associated with the VJ junction (codons 95 and 96) and FR4. All AL gene sequences were submitted to GenBank. Where indicated, protein sequences were deduced and analyzed for characteristics of amyloid-associated light chains.8 30
Isolation of urinary immunoglobulin light chains and culture of human mesangial cells
Urinary immunoglobulin light chains from patients with plasma cells diseases were purified and characterized as previously described.30-32 Human mesangial cells were obtained from kidneys procured for transplantation but not used or from normal areas of nephrectomy specimens, after obtaining written informed consent under protocols approved by the institutional review board. The cortex was dissected and glomeruli isolated as previously described.30-32 The pellets containing glomeruli were resuspended for culture in medium containing RPMI 1640 (Life Technologies) buffered with 12.5 mM HEPES (Sigma-Aldrich, St Louis, MO) at pH 7.4 and supplemented with 20% heat-inactivated fetal bovine serum (Hyclone Laboratories, Logan, UT), penicillin/streptomycin, and 5 μg/mL bovine insulin. Cellular outgrowths were observed 3 to 5 days after attachment of glomeruli to culture plates. Once outgrowths were established, cells were trypsinized, passed through a 75-μm sieve to remove whole glomeruli, and replated on 100-mm tissue culture dishes. Mesangial cells overgrew epithelial cells and became confluent 3 to 4 weeks after plating, were maintained in culture, and were analyzed as previously described for muscle-specific actin, vimentin, factor VIII, and keratin.31-33 The presence of the first 2, and absence of the last 2, confirmed that the cells were a homogeneous population of mesangial cells. In addition, ultrastructural evaluation confirmed morphologic findings including myofilaments and attachment plaques, indicative of mesangial cells. Second-passage mesangial cells grown on coverslips were used for these experiments.
Amyloid formation and evaluation
Amyloid-enhancing factor (AEF) was extracted from murine spleens and purified.34 Three days before incubation with purified light chains, the fetal bovine serum concentration of the mesangial cell medium was reduced to 0.5%. Mesangial cell cultures were incubated with purified light chains with and without AEF as indicated for up to 96 hours in triplicate for each experimental situation as previously described.31-33 Then, mesangial cells on coverslips were fixed in 80% ethanol and stained with hematoxylin and eosin and, for amyloid, with Congo red and thioflavin-T. Congo red–stained sections were viewed in polarized light using a BH2 Olympus microscope (C-squared, Tamarac, FL). The sections stained with thioflavin-T were examined under fluorescent light using a BH2 microscope with UV light capabilities, a Schott 4G2 exciter filter, and a simple UV filter passing only visible light as a barrier filter. Presence of amyloid was evaluated on examination of sections stained with Congo red and thioflavin-T by counting the number of apple-green birefringent or strongly fluorescent complexes at × 10 in 10 fields in each coverslip and computing the average number of complexes. The counts were repeated 3 times for each experimental condition and performed by 2 independent observers. Results were graded as 0 to 3+ using the following criteria: 0 = no amyloid complexes, trace = questionable amyloid complexes, 1+ = 1 to 5 complexes, 2+ = 5 to 10 complexes, and 3+ = >10 complexes.
Statistics
Means, SDs, medians, and ranges were calculated, and tests for significance performed, with PRISM (Graph Pad, San Diego, CA).
Results
Patients with AL and plasma cell disorders
Clonal Ig VL germ line genes were identified in 60 patients with AL, representing 72% of cases in which reverse transcription-PCR (RT-PCR) was attempted (60 of 83). In the unsuccessful instances, either the material was too scant or the percent plasma cells too low. The characteristics of these 60 patients and the plasma cell disorders are shown in Table2. Fifty-two percent had dominant renal and 25% dominant cardiac amyloid. Seventy-five percent had λ clones and 38% had clonal plasma cell disorders that met criteria for myeloma.
Characteristics of 60 patients whose amyloid IgVL genes were cloned
| Characteristic . | Frequency . |
|---|---|
| Age, y (median [range]) | 55 (29-75) |
| Gender (Men/Women) | 41/19 |
| Median number of organ systems involved (range)* | 2 (1-4) |
| Dominant organ system involved | n (%) |
| Cardiac (≥ NYHA class 2) | 15 (25) |
| Renal | 31 (52) |
| Neuropathic | 5 (8) |
| Hepatic | 5 (8) |
| Other | 4 (7) |
| Plasma cell disorders | n (%) |
| Criteria for myeloma met | 23 (38) |
| Criteria not met | 37 (62) |
| Clonal BM plasma cells (median [range]) | 5-10% (< 5-30) |
| Serum monoclonal protein present | 36 (60) |
| IgGλ | 17 |
| IgGK | 2 |
| IgAλ | 2 |
| IgMλ | 3 |
| λ light chains only | 8 |
| K light chains only | 4 |
| Urine monoclonal protein present | 56 (93) |
| IgGλ | 3 |
| IgAλ | 1 |
| IgGλ + λ light chains | 7 |
| λ light chains | 33 |
| K light chains | 12 |
| Characteristic . | Frequency . |
|---|---|
| Age, y (median [range]) | 55 (29-75) |
| Gender (Men/Women) | 41/19 |
| Median number of organ systems involved (range)* | 2 (1-4) |
| Dominant organ system involved | n (%) |
| Cardiac (≥ NYHA class 2) | 15 (25) |
| Renal | 31 (52) |
| Neuropathic | 5 (8) |
| Hepatic | 5 (8) |
| Other | 4 (7) |
| Plasma cell disorders | n (%) |
| Criteria for myeloma met | 23 (38) |
| Criteria not met | 37 (62) |
| Clonal BM plasma cells (median [range]) | 5-10% (< 5-30) |
| Serum monoclonal protein present | 36 (60) |
| IgGλ | 17 |
| IgGK | 2 |
| IgAλ | 2 |
| IgMλ | 3 |
| λ light chains only | 8 |
| K light chains only | 4 |
| Urine monoclonal protein present | 56 (93) |
| IgGλ | 3 |
| IgAλ | 1 |
| IgGλ + λ light chains | 7 |
| λ light chains | 33 |
| K light chains | 12 |
Organ systems are kidneys, heart, peripheral nervous system, liver, gastrointestinal tract, soft tissues.
AL Ig VLgerm line genes
In these 60 cases, a median of 5 identical sequences were cloned per gene (range, 3-7 sequences) and in only 3 cases was a second IgVL gene amplified, requiring sequencing of additional inserts to identify the predominant clone (ratios of 8:1, 4:1, and 6:2). The VλI,VλVI , andVκI subtypes provided germ line donors for 75% of the clones, whereas the remainder derived from germ line donors of the VλII andVλIII subtypes (Table3).
Distribution of dominant organ involvement as a function of germ line gene use
| VL subtype . | Features of clonal disease . | Amyloid organ-system involvement . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dominant involved organ . | No. involved . | |||||||||
| Germ line donor . | No. patients . | Myeloma criteria met . | Cardiac . | Renal . | PNS . | Hepatic . | Other . | 1-2 . | ≥ 3 . | |
| VλI | 1c | 8 | 6 | 4 | 0 | 2 | 1 | 1 | 1 | 7 |
| DPL5 | 3 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | |
| 1e | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | |
| 1a | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1b | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| VλII | 2a2 | 6 | 1 | 3 | 2 | 1 | 0 | 0 | 3 | 3 |
| 2c.118D9 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| VλIII | 3r | 7 | 1 | 3 | 2 | 1 | 1 | 0 | 5 | 2 |
| 3p | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| VλVI | 6a | 18 | 5 | 2 | 15 | 0 | 0 | 1 | 16 | 2 |
| VKI | LFVK431 | 4 | 2 | 0 | 2 | 0 | 1 | 1 | 3 | 1 |
| O8-O18 | 7 | 4 | 2 | 3 | 0 | 1 | 1 | 5 | 2 | |
| DPK7 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| TOTALS | 60 | 23 | 15 | 31 | 5 | 5 | 4 | 42 | 18 | |
| VL subtype . | Features of clonal disease . | Amyloid organ-system involvement . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Dominant involved organ . | No. involved . | |||||||||
| Germ line donor . | No. patients . | Myeloma criteria met . | Cardiac . | Renal . | PNS . | Hepatic . | Other . | 1-2 . | ≥ 3 . | |
| VλI | 1c | 8 | 6 | 4 | 0 | 2 | 1 | 1 | 1 | 7 |
| DPL5 | 3 | 1 | 1 | 2 | 0 | 0 | 0 | 3 | 0 | |
| 1e | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | |
| 1a | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| 1b | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| VλII | 2a2 | 6 | 1 | 3 | 2 | 1 | 0 | 0 | 3 | 3 |
| 2c.118D9 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
| VλIII | 3r | 7 | 1 | 3 | 2 | 1 | 1 | 0 | 5 | 2 |
| 3p | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| VλVI | 6a | 18 | 5 | 2 | 15 | 0 | 0 | 1 | 16 | 2 |
| VKI | LFVK431 | 4 | 2 | 0 | 2 | 0 | 1 | 1 | 3 | 1 |
| O8-O18 | 7 | 4 | 2 | 3 | 0 | 1 | 1 | 5 | 2 | |
| DPK7 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | |
| TOTALS | 60 | 23 | 15 | 31 | 5 | 5 | 4 | 42 | 18 | |
In the Vλ cases, there was preferential germ line gene utilization. The 1c gene was used in 8 of 15VλI cases, 2a2 in 6 of 7VλII cases, 3r in 7 of 8VλIII cases, and the 6a gene in all 18 VλVI cases. In the normal expressed repertoire, 7% to 8% of light chains are derived from1c, 20% to 35% from 2a2, 7% to 8% from3r, and < 5% from 6a.17,18 Light chains of the rare VλVI subtype have been found frequently in AL.20,26 In theVκ cases, there was also preferential utilization. The relatively rare LFVK431 germ line gene was used in 4 cases and the more common O18-O8 gene in 7.20 All of the Vκ genes used by the κ clones in this series were members of the VκIsubtype. In normal usage, genes of theVκIII subtype dominate.20
AL organ system tropism
Data on germ line gene use, myeloma, and organ disease are summarized in Table 3. The 1c, 2a2, and3r germ line genes are associated with dominant cardiac and multisystem disease, whereas the 6a gene is associated with dominant renal disease. Indeed, the association between the6a donor and dominant renal involvement is striking and a comparison of the frequency of dominant renal involvement in6a patients versus all others achieves significance (P << .01, χ2 = 12.61, degrees of freedom [df] = 1, relative risk = 2.5, 95% confidence interval [CI], 1.56-4.02). In contrast, a comparison of the frequency of dominant cardiac involvement among 6a patients versus all others does not (P = .19). A comparison of the frequency of dominant cardiac involvement among patients with myeloma (10 of 23) versus those without myeloma (5 of 37) also achieves significance (P < .01, χ2 = 6.79, df = 1, relative risk = 3.22, 95% CI, 1.26-8.23). In addition, 3 of 12 patients withVκ clones had dominant hepatic involvement as compared to only 2 of 48 patients with Vλ clones (P < .05, Fisher exact test, 2-tailed; relative risk = 6.0, 95% CI, 1.13-32.0).
AL VLgene analysis
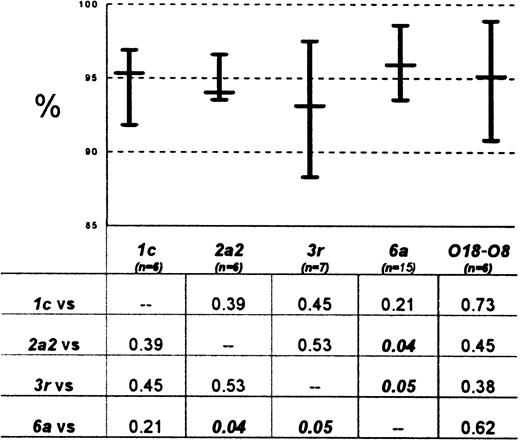
Having identified germ line genes in 60 cases, marrow cDNA was then used in PCR with L-CL primers to amplify and sequence the correct FR1, as depicted in Figure 1. Eighty-five percent (51 of 60) were successfully amplified and sequenced directly from PCR tubes with this technique; the remaining 9 cases gave no identifiable amplicons and further cloning attempts were not pursued (VλI = 3,VλIII = 2,VλVI = 3,VκI = 1). All 51 genes sequenced in this fashion corresponded identically to their previously cloned counterparts except for the differences in FR1 introduced by the FR1 primers (≤ 3 nucleotides). Using these correctly sequenced 51 AL VL genes, we determined the percent homology of AL VL genes with germ line sequences, as an indication of the degree of antigen-driven mutation. All 51 genes showed evidence of likely prior antigenic challenge. The median percent homology to germ line was 95.5% (range, 88.3%-98.9%). Of note, as depicted in Figure 2, theVλVI AL genes were more homologous to germ line than the other AL Vλ genes.
Percent homology to germ line for commonly used AL
IgVL genes. The percent homology (%) is a reflection of the frequency with which individual nucleotides in an Ig gene have been mutated from germ line. Percents were determined for individual AL Ig VL genes correctly sequenced with L-CL primers. Aggregate results are depicted by germ line subgroup; medians and ranges for specific subgroups are shown on the chart and P values of Mann-Whitney comparisons in the table. The 6a genes are the most homologous to germ line and the 3r genes the least. The difference between 6a versus 2a2 or 3r Vλ genes achieves statistical significance. Because the 6a donor is not a common contributor to the normal expressed repertoire, the difference in homology with genes of other Vλ subgroups may reflect a normal process that distinguishes infrequently used germ line genes but may also reflect the propensity of 6a clones to produce amyloid due to germ line-encoded features. It may also reflect a difference in the origins of AL clones. For example, one might in theory see such a contrast if 6a clones were derived from de novo postgerminal center B cells, whereas clones of other Vλdonors were derived from memory B cells subject to additional circuits through the germinal center.
Percent homology to germ line for commonly used AL
IgVL genes. The percent homology (%) is a reflection of the frequency with which individual nucleotides in an Ig gene have been mutated from germ line. Percents were determined for individual AL Ig VL genes correctly sequenced with L-CL primers. Aggregate results are depicted by germ line subgroup; medians and ranges for specific subgroups are shown on the chart and P values of Mann-Whitney comparisons in the table. The 6a genes are the most homologous to germ line and the 3r genes the least. The difference between 6a versus 2a2 or 3r Vλ genes achieves statistical significance. Because the 6a donor is not a common contributor to the normal expressed repertoire, the difference in homology with genes of other Vλ subgroups may reflect a normal process that distinguishes infrequently used germ line genes but may also reflect the propensity of 6a clones to produce amyloid due to germ line-encoded features. It may also reflect a difference in the origins of AL clones. For example, one might in theory see such a contrast if 6a clones were derived from de novo postgerminal center B cells, whereas clones of other Vλdonors were derived from memory B cells subject to additional circuits through the germinal center.
AL VL protein analysis
The protein sequences were deduced for ALVL genes derived from the 1c,2a2, 3r, 6a, O18-O8, andLVFK431 germ line genes, and the deduced sequences were assessed for amino acid replacement mutations that have been associated with amyloid light chains and for mutations that result in the creation of sites for N-glycosylation (N-x-S/T).8,33Amino acid positions are as designated by Kabat-Wu numbering.29 The results of this analysis are shown in Table 4 and the features of the replacement amino acids are indicated. Germ line–encoded residues that may play a role in amyloid formation are not included.8 In these Vκ and Vλ AL light chains, residues with frequent replacement mutations are located along the protein surface in areas involved with the binding of antigen such as positions 30 to 32, 50 to 52, and 93 to 96.
Mutated amino acid residues in deduced amyloid light-chain proteins
| VL subtype . | Germ line donor . | Mutated residue . | No. cases . | Replacement(s) . | Amino acid features . |
|---|---|---|---|---|---|
| VλI | 1c (n = 8) | Gln38 | 6 | His | Neutral to basic |
| Pro40 | 1 | Ser | Both neutral | ||
| Ser50 | 3 | Thr, Iso | All neutral | ||
| Arg | Neutral to basic | ||||
| VλII | 2a2 (n = 6) | Gln38 | 5 | His | Neutral to basic |
| Glu50 | 4 | Asp | Both are acidic | ||
| Ser52 | 4 | Asn (n = 2) | All neutral | ||
| Thr (n = 2) | |||||
| Ser94 | 1 | Asn | N-gly site | ||
| VλIII | 3r (n = 7) | Lys31b | 3 | Glu | Basic to acidic |
| 2 | Tyr, Asn | Basic to neutral | |||
| N-gly site | |||||
| Pro40 | 2 | Ala, Ser | All neutral | ||
| Ser52 | 6 | Thr (n = 3), | All neutral | ||
| Asn (n = 2) | |||||
| Asp | Neutral to acidic | ||||
| Ala90 | 3 | Thr (n = 2), | All neutral | ||
| Ser | |||||
| Ser94 | 4 | Gly (n = 2), Asn, | All neutral | ||
| Arg | Neutral to basic | ||||
| VλVI | 6a (n = 18) | Arg25 | 5 | Gly | Basic to neutral |
| Ser31a | 6 | Ala, Gly, Val, | All neutral | ||
| Asn (n = 2) | |||||
| Asp | Neutral to acidic | ||||
| Asn31b | 5 | Ser, Tyr (n = 2) | All neutral | ||
| Asp | Neutral to acidic | ||||
| Pro40 | 3 | Thr, Gly, Ser | All neutral | ||
| Ser43 | 8 | Ala (n = 8) | All neutral | ||
| Asn52 | 9 | Asp (n = 4) | Neutral to acidic | ||
| Lys | Neutral to basic | ||||
| Ser (n = 3), Thr | All neutral | ||||
| Gln53 | 5 | Val (n = 2) | Both neutral | ||
| Glu (n = 2) | Neutral to acidic | ||||
| Lys | Neutral to basic | ||||
| Ser93 | 12 | Asn (n = 4), Thr (n = 2), | All neutral | ||
| Gly (n = 4) | |||||
| Asp (n = 2) | Neutral to acidic | ||||
| Ser94 | 11 | Asn (n = 9), Gly | All neutral | ||
| Arg | Neutral to basic | ||||
| VKI | O18-O8 (n = 7) | Ser30 | 4 | Ala (n = 2), Thr, Asn | All neutral |
| Asn31 | 1 | Lys | Neutral to basic | ||
| Tyr32 | 3 | Phe (n = 2) | Both neutral | ||
| His | Neutral to basic | ||||
| Glu55 | 3 | Gln (n = 2) | Neutral to acidic | ||
| Lys | Acidic to basic | ||||
| Asp70 | 3 | Asn (n = 2) | Acidic to neutral, | ||
| N-gly sites | |||||
| His | Acidic to basic | ||||
| Thr72 | 2 | Iso | Both neutral | ||
| Asn | Neutral to basic, | ||||
| N-gly site | |||||
| Leu96 | 4 | Tyr, Pro, Thr, Phe | All neutral | ||
| LFVK431 (n = 4) | Gly28 | 3 | Asp (n = 3) | Neutral to acidic | |
| Tyr32 | 3 | Phe (n = 3) | Both neutral | ||
| Ala50 | 2 | Ser (n = 2) | Both neutral | ||
| Lys61 | 3 | Asn (n = 3) | All neutral, | ||
| N-gly sites | |||||
| Asp70 | 2 | His | Acidic to neutral | ||
| Asn | Acidic to neutral, | ||||
| N-gly site | |||||
| Ser93 | 1 | Asn | Both neutral | ||
| Arg96 | 3 | Tyr (n = 2), Pro | Basic to neutral |
| VL subtype . | Germ line donor . | Mutated residue . | No. cases . | Replacement(s) . | Amino acid features . |
|---|---|---|---|---|---|
| VλI | 1c (n = 8) | Gln38 | 6 | His | Neutral to basic |
| Pro40 | 1 | Ser | Both neutral | ||
| Ser50 | 3 | Thr, Iso | All neutral | ||
| Arg | Neutral to basic | ||||
| VλII | 2a2 (n = 6) | Gln38 | 5 | His | Neutral to basic |
| Glu50 | 4 | Asp | Both are acidic | ||
| Ser52 | 4 | Asn (n = 2) | All neutral | ||
| Thr (n = 2) | |||||
| Ser94 | 1 | Asn | N-gly site | ||
| VλIII | 3r (n = 7) | Lys31b | 3 | Glu | Basic to acidic |
| 2 | Tyr, Asn | Basic to neutral | |||
| N-gly site | |||||
| Pro40 | 2 | Ala, Ser | All neutral | ||
| Ser52 | 6 | Thr (n = 3), | All neutral | ||
| Asn (n = 2) | |||||
| Asp | Neutral to acidic | ||||
| Ala90 | 3 | Thr (n = 2), | All neutral | ||
| Ser | |||||
| Ser94 | 4 | Gly (n = 2), Asn, | All neutral | ||
| Arg | Neutral to basic | ||||
| VλVI | 6a (n = 18) | Arg25 | 5 | Gly | Basic to neutral |
| Ser31a | 6 | Ala, Gly, Val, | All neutral | ||
| Asn (n = 2) | |||||
| Asp | Neutral to acidic | ||||
| Asn31b | 5 | Ser, Tyr (n = 2) | All neutral | ||
| Asp | Neutral to acidic | ||||
| Pro40 | 3 | Thr, Gly, Ser | All neutral | ||
| Ser43 | 8 | Ala (n = 8) | All neutral | ||
| Asn52 | 9 | Asp (n = 4) | Neutral to acidic | ||
| Lys | Neutral to basic | ||||
| Ser (n = 3), Thr | All neutral | ||||
| Gln53 | 5 | Val (n = 2) | Both neutral | ||
| Glu (n = 2) | Neutral to acidic | ||||
| Lys | Neutral to basic | ||||
| Ser93 | 12 | Asn (n = 4), Thr (n = 2), | All neutral | ||
| Gly (n = 4) | |||||
| Asp (n = 2) | Neutral to acidic | ||||
| Ser94 | 11 | Asn (n = 9), Gly | All neutral | ||
| Arg | Neutral to basic | ||||
| VKI | O18-O8 (n = 7) | Ser30 | 4 | Ala (n = 2), Thr, Asn | All neutral |
| Asn31 | 1 | Lys | Neutral to basic | ||
| Tyr32 | 3 | Phe (n = 2) | Both neutral | ||
| His | Neutral to basic | ||||
| Glu55 | 3 | Gln (n = 2) | Neutral to acidic | ||
| Lys | Acidic to basic | ||||
| Asp70 | 3 | Asn (n = 2) | Acidic to neutral, | ||
| N-gly sites | |||||
| His | Acidic to basic | ||||
| Thr72 | 2 | Iso | Both neutral | ||
| Asn | Neutral to basic, | ||||
| N-gly site | |||||
| Leu96 | 4 | Tyr, Pro, Thr, Phe | All neutral | ||
| LFVK431 (n = 4) | Gly28 | 3 | Asp (n = 3) | Neutral to acidic | |
| Tyr32 | 3 | Phe (n = 3) | Both neutral | ||
| Ala50 | 2 | Ser (n = 2) | Both neutral | ||
| Lys61 | 3 | Asn (n = 3) | All neutral, | ||
| N-gly sites | |||||
| Asp70 | 2 | His | Acidic to neutral | ||
| Asn | Acidic to neutral, | ||||
| N-gly site | |||||
| Ser93 | 1 | Asn | Both neutral | ||
| Arg96 | 3 | Tyr (n = 2), Pro | Basic to neutral |
These represent the most commonly mutated residues found in our sample with particular focus on amyloid-associated light-chain residues as described in references 8 and 30. Residues in bold are in complementarity-determining regions (CDR). N-gly site indicates the N-glycosylation site.
Amyloid formation in vitro
Ten urinary immunoglobulin light chains were tested in vitro. Seven were from patients with biopsy-proven AL with renal disease including 4 of the 60 whose Ig VL genes were identified (λVI = 3, λII = 2, κI = 1, κII = 1). Three were controls; 2 were light chains from patients with myeloma with nonamyloid renal disease (λI = 1, λII = 1) and one a κI light chain from a patient with light-chain deposition disease. The results of incubation in mesangial cell cultures with and without AEF at a light chain concentration of 10 μg/mL are shown in Table 5. Of note, all of the AL light chains and none of the controls formed amyloid when incubated with AEF. In addition, 2 of the 3 λVI light chains tested in this model system formed more amyloid than all other AL light chains and, unlike all other AL light chains, the λVI light chains formed similar amounts of amyloid with and without AEF. These differences highlight the propensity of mesangial cells to form amyloid without AEF when incubated with λVI but not other types of light chains.
Amyloid formation in an in vitro renal mesangial cell system
| Type of light chains . | No. tested . | Amyloid complexes . | |
|---|---|---|---|
| + AEF . | − AEF . | ||
| Amyloid | 7 | ||
| λII | 2 | 1+, 2+ | 0, 0 |
| λVI | 3 | 3+, 3+, 1+ | 3+, 3+, 1+ |
| KI | 1 | 1+ | 0 |
| KII | 1 | 2+ | 0 |
| Controls | 3 | ||
| λI | 1 | 0 | 0 |
| λII | 1 | 0 | 0 |
| KI | 1 | 0 | 0 |
| Type of light chains . | No. tested . | Amyloid complexes . | |
|---|---|---|---|
| + AEF . | − AEF . | ||
| Amyloid | 7 | ||
| λII | 2 | 1+, 2+ | 0, 0 |
| λVI | 3 | 3+, 3+, 1+ | 3+, 3+, 1+ |
| KI | 1 | 1+ | 0 |
| KII | 1 | 2+ | 0 |
| Controls | 3 | ||
| λI | 1 | 0 | 0 |
| λII | 1 | 0 | 0 |
| KI | 1 | 0 | 0 |
AEF indicates amyloid-enhancing factor.
GenBank accession numbers
Note: Italicized accession numbers relate to sequences previously described by subtype only in reference 26.
Discussion
In this report, we identify the immunoglobulin light-chain variable region (Ig VL) germ line genes used by the plasma cell clones of 60 patients with AL. We assess the plasma cell disorders in these patients as meeting or not meeting the Durie criteria21 for multiple myeloma and analyze the amyloid-related organ disease in these patients as a function of both Ig VL germ line gene use and plasma cell burden (patients with or without myeloma). The hypothesis that germ line gene use and plasma cell burden influence the tropism of AL organ involvement is supported by this analysis. Patients with 6a VλVI clonal disease are more likely to have dominant renal involvement, whereas those with otherVλ clones often have dominant cardiac amyloid and multisystem disease. And patients with Vκ clones are more likely to have dominant hepatic involvement, an association that has been linked by Stevens and others to a possible contribution ofN-glycosylation of kappa light chains to the propensity to form amyloid (Table 4).30 In addition, patients with AL with myeloma are more likely to have dominant cardiac amyloid independent of germ line gene use. The association between increased plasma cell burden and cardiac amyloid is consistent with a recent report in which the degree of plasma cell clonality and marrow plasma cell burden were shown to confer a poor prognosis in AL.35
These results also support the claim of preferential germ line gene use in AL, based both on the well-described preponderance of λ clones and on the frequent use of genes such as 1c, 3r, 6a, andLFVK431 that are uncommon in the normal expressed repertoire. Moreover, given the range of homologies to germ line sequences, these results support the claim that AL clones originate from postgerminal center B cells subject to prior antigenic challenge, as depicted in a recent analysis of the sequences of 14 AL genes.36 Furthermore, the striking association of6a VλVI clonal plasma cell disease and renal amyloidosis is given additional credibility by the results of in vitro testing in a human renal mesangial cell model. There is specificity to this association; it is clearly apprehended but remains unexplained.
It should be emphasized that these data are not free of selection bias. First, the patients tested were seen on referral to tertiary centers, possibly explaining the disproportionate number of male patients. Second, in many instances the patients were referred for consideration of stem cell transplantation and, therefore, represent a possibly younger and healthier segment of the AL patient population.37-39 Third, the patients also represent a portion of the AL population whose clonal immunoglobulin genes could be amplified and identified, for the 60 successful cases represent just over two thirds of the cases in which RT-PCR was attempted. This point is particularly relevant to claims made with respect to plasma cell burden. Fourth, we assume that plasma cell burden can be estimated using criteria designed to distinguish myeloma from monoclonal gammopathy of undetermined significance and that this distinction correlates with light-chain production.21
Nevertheless, concerns of bias duly noted, 2 significant associations between clonal AL VL gene use and dominant organ involvement emerge from this analysis. Indeed, because we evaluated AL organ involvement by standard accepted criteria, it is important to note that the categories used for dominant organ involvement have been shown to possess prognostic significance with respect to survival.10,22 Therefore, the respective associations identified between dominant cardiac involvement and the 1c,2a2, and 3r Vλ genes, and between dominant renal involvement and the 6a gene, also contain prognostic significance. That is, links are likely to exist between germ line gene use and overall survival, as well as between germ line gene use and survival after stem cell transplant, as we have recently suggested.26
With respect to the technique of clonal gene identification, RT-PCR was used to amplify clonal light-chain genes from bone marrow cDNA usingFR1-CL primers and strict rules developed for identification of candidate cloned genes. Clonal ALVL genes were assigned germ line donors and, for purposes of further analysis, 85% of them (51 of 60) were successfully amplified a second time with Leader primers and sequenced directly to identify potential errors in FR1 introduced byFR1 primers. In these instances, this second round of amplification also served to confirm the identity of the cloned genes as clonal genes. In addition, the distribution of germ line genes and the percent homologies to germ line indicate that certain germ line genes likely possess intrinsic features predisposing to amyloid formation.8 40
The critical physicochemical aspects of the proteins encoded by preferentially used light-chain genes remain unexplained. Although the mechanisms underlying organ tropism also remain unexplained, our analysis based on plasma cell burden indicates that light-chain availability or concentration is likely to be an important variable because patients with AL with myeloma are more likely to have dominant cardiac amyloid independent of germ line gene. This conclusion fits with long-standing clinical observations and the results of a similar analysis.10,35 Recent evidence in support of a role for receptor-dependent cell stress in secondary amyloid formation may be relevant in this regard. A multiligand receptor in the immunoglobulin superfamily (RAGE or receptor for advancedglycation end-products) was shown to be up-regulated by amyloid-prone proteins and integral to amyloid deposition.41 It is possible that, in primary amyloidosis, light-chain concentration plays a role in up-regulating RAGE receptors on macrophages and mononuclear phagocytes in different organs and that up-regulation of RAGE receptors in relevant cardiac cells may require a higher concentration of light chains. Of more concrete relevance, however, the amino acid replacements we identified in deduced protein sequences (Table 4) are in positions that may be associated with amyloid formation although specific substitutions that play a causal role or contribute to light-chain instability have not yet been generically identified.8,30 Nevertheless, the presence of these replacements in positions along the protein surface further supports a molecular model of amyloid fibril formation that involves initial dimerization of VL molecules due to interactions among CDR residues.8 A specific role for receptor-ligand interactions involving RAGE or other cell surface receptors and this molecular model are not mutually exclusive in theory.
The AL Ig VL genes we have identified are likely derived from postgerminal center B cells, as is indicated by the assessment of homology to germ line sequences. Furthermore, a difference is seen among the subtypes with respect to homology to germ line, as highlighted by the significant difference between the6a and the 2a2 or 3r clones. It is unknown whether the difference is typical of germ line genes such as6a that are rarely used in the expressed repertoire. Indeed, the development of the expressed repertoire is incompletely documented with respect to marrow plasma cell Ig VL gene expression. It is also possible that the less homologous subtypes represent clones derived from the memory B-cell pool; their emergence may involve antigenic challenge or persistence in ways not well appreciated, and may be related to repeat journeys through the germinal center resulting in several generations of mutations in sequence.42 43 Although the difference in homology may reflect the emergence of AL clones against the backdrop of such hypothetical sources, it more likely represents the inherent tendency of 6a clones to cause amyloid because of germ line–encoded features. Indeed, given the renal tropism of 6a clones and the in vitro data we offer, a specific receptor-ligand interaction is suggested.
In conclusion, we report a series of 60 clonal IgVL gene sequences from patients with AL, the largest series to date. We demonstrate that germ line gene use and plasma cell burden contribute to the tropism of organ involvement, one of the hallmarks of AL. Ig VL germ line donors were associated with dominant hepatic, dominant cardiac and multisystem disease, and dominant renal disease. Patients with AL with myeloma in this series were at increased risk of developing cardiac amyloid independent of germ line donor. Both germ line gene use and clonal plasma cell burden contribute to the tropism of organ involvement observed in AL. Of particular note, the specific association between the 6a VλVI germ line gene and dominant renal disease is supported by data from an in vitro assay using human renal mesangial cells. The specificity of the association justifies our current effort to understand its physicochemical basis in order to develop pharmaceutical approaches that may impede amyloid deposition and the progression of disease.
We thank Dr Stephen D. Nimer for his advice and continued support; and Joanne Santorsa, RN, and Dr Carl O'Hara for assistance obtaining specimens.
Supported by grants from the Food and Drug Administration (FD-R-001346-01), the Arthritis Foundation, and the National Blood Foundation. C.M. was supported by the Program Generalitat de Catalunya-Fulbright.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Raymond L. Comenzo, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail:comenzor@mskcc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal