Chronic granulomatous disease (CGD) is an inherited disorder of host defense against microbial infections caused by defective activity of the phagocyte NADPH oxidase. Based on an increase of neutrophil superoxide-generating ability in response to interferon γ (IFN-γ) in a single patient with CGD, multicentered group studies demonstrated a beneficial effect of prophylactic IFN-γ. However, no apparent increase of the phagocyte superoxide generation was found in patients enrolled in these studies. The present report offers an additional kindred in whom an IFN-γ–dependent increase in neutrophil superoxide production was observed in 3 affected patients. The defect in the CYBB gene for gp91-phox was identified as an otherwise silent mutation adjacent to the third intron of theCYBB gene that alters messenger RNA splicing. By molecular analysis, significant differences were found in the splicing pattern ofCYBB gene transcripts in patient neutrophils between 1 and 25 days after administration of IFN-γ. Furthermore, a complete transcript containing the missing exons could be detected in all specimens after the treatment. The changes in the splicing pattern of the transcripts and the prolonged effect on superoxide-generating ability of patient neutrophils indicate that IFN-γ induced a partial correction of the abnormal splicing of CYBB gene transcripts in myeloid progenitor cells.
Introduction
Chronic granulomatous disease (CGD) is an inherited disorder of host defense against bacterial and fungal infections; affected patients suffer from severe recurrent and intractable infections beginning in early childhood.1 Phagocytes from patients with CGD show impaired microbicidal activity due to defects in the superoxide-generating phagocyte oxidase.1 It is known that the mutations responsible for CGD reside within the genes for 4 essential components of the oxidase designated as gp91-phox(phagocyte oxidase), p22-phox, p47-phox and p67-phox.2,3 The gp91-phox forms membrane cytochrome b558 together with p22-phox and plays an essential role in the transfer of electrons following assembly of the active oxidase with the cytoplasmic p47- and p67-phox components. Patients with CGD with gp91-phox defects account for the majority of cases, and in most instances the cytochrome is reduced or absent in phagocytes and B lymphocytes. The CYBB gene that encodes gp91-phoxis localized to Xp21.1 and encompasses 13 exons spanning approximately 30 kilobases (kb). Mutations in the CYBB gene are heterogenous, and include missense, nonsense, deletion, insertion, and splicing defects.1,4,5 Various cytokines have been studied for enhancement of the neutrophil superoxide generation, showing that IFN-γ may be an effective agent.6,7 Ezekowitz and coworkers first demonstrated that phagocytes of a patient with X-linked CGD were responsive to interferon-γ (IFN-γ), which almost completely corrected the oxidase defect in vitro and in vivo.8,9 These findings motivated multicenter groups to perform double-blinded clinical studies of IFN-γ as a prophylactic agent in CGD, which demonstrated its clinical benefit in the majority of patients with CGD.10,11 In these group studies, however, no apparent increases in phagocyte superoxide generation were observed. The patient studied by Ezekowitz and colleagues, therefore, has been considered to be an exceptional case. Recently, the mutation in this patient has been identified as a single base substitution at the sixth position of the first intron in the CYBBgene.4 Based on additional studies, Condino-Neto and Newburger proposed that IFN-γ improved the splicing efficiency ofCYBB gene transcripts in the patient and corrected a nuclear processing defect due to the intronic mutation by augmenting nuclear export of normal transcripts.12 In this report, we show an IFN-γ–dependent increase of superoxide production associated with a change in the messenger RNA (mRNA) splicing pattern of CYBBgene transcripts in neutrophils from 3 patients in one family, who have a silent mutation adjacent to intron 3 of CYBBgene.
Patients, materials, and methods
Patients and their family
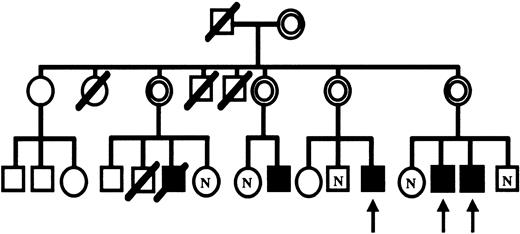
Among the patients enrolled in our INF-γ study, 3, patient 1 (13 years old), patient 2 (17 years old), and patient 3 (16 years old), were studied. These patients are members of the same kindred. The patients have been generally well, though they occasionally suffered from pneumonia, lymphadenitis, and cystitis. Patient 3 also suffered from liver abscess at 12 years of age, but has otherwise had no other severe infections. Figure1 shows the pedigree of the patients' family where the patients studied are marked by arrows. In this family, 5 healthy carriers and 5 X-CGD patients were identified by flow cytometric analysis. Among the descendents, 3 had died of severe bacterial infection in their early life, suggesting that they had CGD. Informed consents were obtained prior to the studies for IFN-γ. In this study, IFN-γ was administered subcutaneously once at a dosage of 50 μg/m2 (SHIONOGI, Osaka, Japan). Peripheral blood samples were drawn and subjected to the studies outlined below. Patient 2 developed a high-grade fever on the first day of the treatment. An intractable acne vulgaris on the face of patients 1 and 2 disappeared after the treatment.
Pedigree of CGD patients and their family.
CGD patients, ▪; carriers, ⌾. Symbols marked as N indicate normal; arrows, the patients studied.
Pedigree of CGD patients and their family.
CGD patients, ▪; carriers, ⌾. Symbols marked as N indicate normal; arrows, the patients studied.
Isolation of neutrophils
Neutrophils from heparinized venous blood of the patients were prepared by dextran sedimentation followed by Ficoll-Paque (Pharmacia Fine Chemicals, Tokyo, Japan) gradient centrifugation and hypotonic lysis to remove contaminating erythrocytes, as described.13
Isolation of genomic DNA and amplification of genomic DNA by polymerase chain reaction
DNA was isolated from neutrophils using PUREGENE DNA Isolation Kit (Gentra Systems, Minneapolis, MN). For mutation analysis, each exon of gp91-phox was amplified by a pair of primers (1 pmol each) complementary to the flanking introns as described previously.14 The reaction mixture (25 μL) contained 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, each dNTP and Ampli Taq Gold (Perkin Elmer Japan, Chiba, Japan). The following conditions were used for 40 cycles of amplification by polymerase chain reaction (PCR) for gp91-phox genomic DNA, except for gp91-phox exon13: the first denaturation was at 95°C for 9 minutes followed in subsequent cycles, denaturation at 95°C for 30 seconds, annealing and extension at 60°C for 90 seconds and final extension at 60°C for 3 minutes. Exon 13 was amplified by using Taq DNA polymerase (Boehringer Mannheim, Tokyo, Japan) under the following conditions: initial denaturation at 94°C for 2 minutes, followed by 10 cycles of DNA denaturation at 94°C for 30 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 1 minute, and then 35 cycles of denaturation at 94°C for 10 seconds, annealing at 50°C for 15 seconds, and extension at 72°C for 1 minute.
Sequencing
Using the cycle sequence method with the Taq Dye Deoxy Terminator Sequencing Kit (Perkin Elmer Japan) with 70 to 80 ng PCR product as a template, sequencing was performed with an ABI model 373A or 310 (Perkin Elmer Japan). Both strands were sequenced.
Neutrophil superoxide production
Neutrophil active oxygen production as a result of superoxide generation was assayed according to the method described by Vowells and coworkers using dihydroxyrhodamine 123 (DHR-123) as a fluorescence indicator and phorbol myristate acetate (PMA) as a stimulus.15 Neutrophils (1 × 106) were suspended in 100 μL phosphate-buffered saline (PBS) containing 5 mM glucose, 0.1% bovine serum albumin (BSA), and 10 mM NaN3. After incubation at 37°C for 5 minutes, the cell suspension was added to 10 μL of 333 μM DHR-123, which was incubated for 5 additional minutes at 37°C. The cells were then stimulated by 1 μL PMA (100 ng/μL) at 37°C for 30 minutes. The tubes containing stimulated or unstimulated cells were chilled on ice. All samples were analyzed with a FACScan using CellQuest software (Becton Dickinson Immunocytometry Systems, San Jose, CA). Neutrophils were gated on the basis of forward and side scatter. Fluorescence signals at 585 nm were measured for 10 000 events and presented as logarithmically amplified signals. The relative amount of active oxygen was estimated on the basis of the increase in green fluorescence intensity of stimulated neutrophils in comparison to resting or unstimulated cells. To compare active oxygen-generating abilities of patients' neutrophils after administration of IFN-γ, we calculated the percentage of increase in mean fluorescence as follows: (mean value of stimulated patient cells − mean value of resting patient cells) × 100/(mean value of stimulated control cells − mean value of resting control cells)
Flow cytometric analysis of cytochromeb558on cell surface of neutrophils
Neutrophils were stained with the anticytochromeb558 monoclonal antibody, 7D5,16 or control IgG1 followed by staining with phycoerythrin-conjugated antimouse antibody. The stained neutrophils were analyzed by flow cytometry (FACScan, Becton Dickinson Immunocytometry Systems) using CellQuest software (Becton Dickinson Immunocytometry Systems).
Extraction of total RNA and reverse transcription-PCR
Total RNA was extracted from neutrophils by the acid guanidinium thiocyanate-phenol-chloroform method.17 Single-strand complementary DNA (cDNA) was synthesized from 1 μg RNA using a First Strand cDNA Synthesis Kit according to manufacturer's instructions (TaKaRa Biomedicals, Kyoto, Japan). A portion of the gp91-phox cDNA was amplified with PCR using a forward primer (f1) on exon 1 and either a reverse primer (r4) on exon 4, namely 5′ AGAGTGAAGTGCAATCATCCATGCCACC 3′ or a reverse primer (r6) on exon 6 as described by Dinauer and coworkers18 under the conditions used for the first round amplification. Amplified products were separated on a 3% agarose gel and stained with ethidium bromide. The ratios of products were analyzed using a Densitometer (ATTO Densitograph software library, Tokyo, Japan).
Sequence of the band from the agarose electrophoresis
The cDNA band separated from the agarose gel was extracted using EASYTRAP Ver. 2 kit (TaKaRa Biomedicals) and sequenced directly.
Results
All 3 patients studied were members of the same kindred as shown in Figure 1. IFN-γ was administered subcutaneously once at a dosage of 50 μg/m2. Peripheral blood samples were drawn from each patient before and after the treatment and subjected to the following studies.
Molecular analysis of gp91-phox gene in the patients
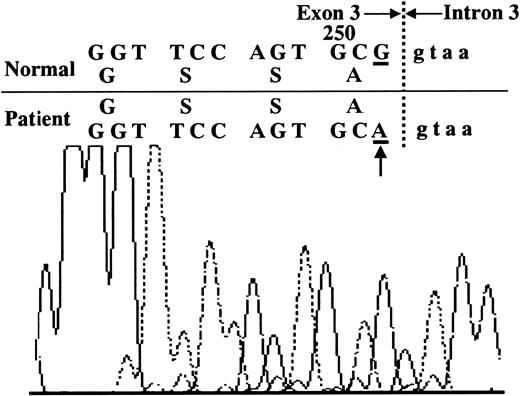
DNA was purified from the peripheral blood neutrophils obtained from each patient prior to IFN-γ treatment, amplified using 13 pairs of primers for the CYBB gene, and analyzed.14It was found that guanosine at residue 252 in the splice donor site of exon 3 was substituted with adenosine (Figure2). This is the sole mutation found in all 3 patients studied and does not result in an amino acid transition, thereby, making it a silent mutation. However, the mutant residue is adjacent to intron 3 and this substitution significantly reduces the matching score from 86.1 to 73.7 for splice region sequence to the consensus, resulting in exon 3 skipping together with exon 2 in most of the transcripts (Figure3).19 The ratio between the transcripts containing and lacking exon 3 was varied among the patients.5
Mutation in the
CYBB gene of the CGD patients. A single substitution of adenosine at residue 252 to guanosine was found. This residue is at the 5′ splice site of the third exon/intron boundary (arrow).
Mutation in the
CYBB gene of the CGD patients. A single substitution of adenosine at residue 252 to guanosine was found. This residue is at the 5′ splice site of the third exon/intron boundary (arrow).
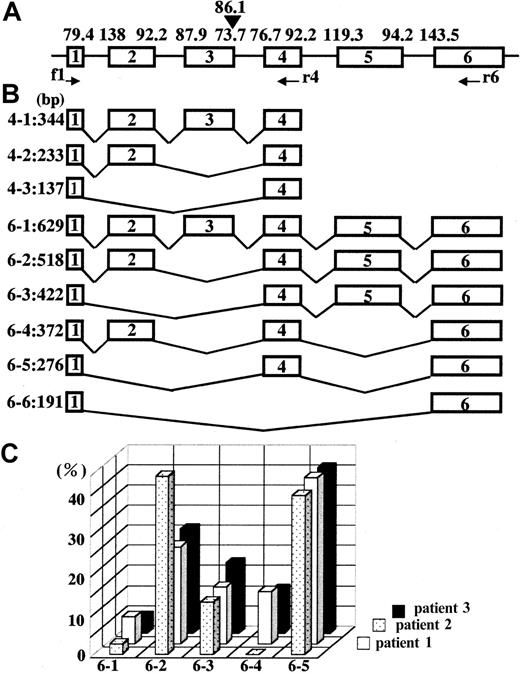
Estimation and analysis of alternatively spliced products.
Alignment of exons 1 to 6 and the matching scores (A), molecular sizes of different construction of exons 1 to 6 (B), and splicing pattern change after IFN-γ treatment (C). In panel C, each spliced band in Figure 6C was quantified using a densitometer, and each relative value is presented as a column.
Estimation and analysis of alternatively spliced products.
Alignment of exons 1 to 6 and the matching scores (A), molecular sizes of different construction of exons 1 to 6 (B), and splicing pattern change after IFN-γ treatment (C). In panel C, each spliced band in Figure 6C was quantified using a densitometer, and each relative value is presented as a column.
Analysis of the superoxide-generating ability of neutrophils before and after IFN-γ treatment
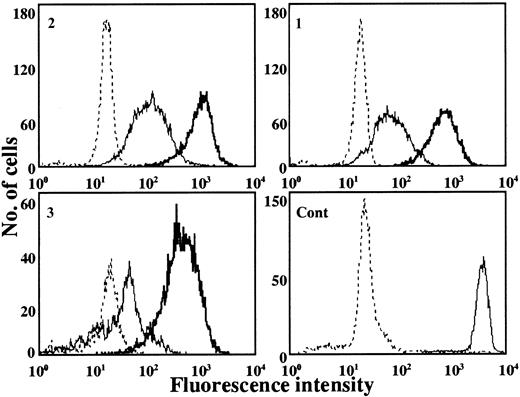
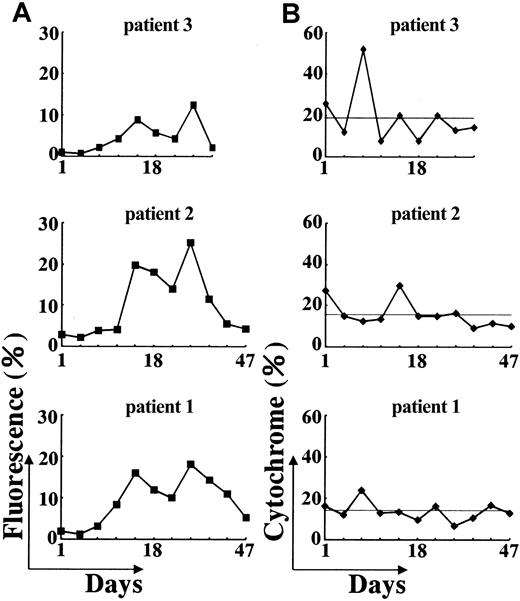
The superoxide-generating ability of neutrophils from the patients was evaluated based on flow cytometry histograms using DHR-123 as a fluorescence probe to detect reactive oxygen species. In these experiments, isolated neutrophils were stimulated with PMA, histograms of DHR fluorescence were obtained, and relative values of fluorescence were calculated and compared. Superoxide-generating activities of neutrophils from all 3 patients before IFN-γ treatment were only 1% to 3% of control neutrophils (data not shown). However, 11 days after a single dose of IFN-γ, a marked increase was observed in neutrophils from all 3 patients, reaching maximum after 25 days (Figure4). As shown in Figure5, the higher activities persisted for 2 to 4 weeks, with neutrophil oxidant production in patients 1, 2, and 3, respectively, being 18.4%, 25.2%, and 12.5% of control neutrophils after 25 days. The activity then decreased gradually and declined to almost pretreatment levels after 47 days.
Flow cytometric analysis of active oxygen generation in neutrophils.
Histograms of DHR-123 fluorescence as indicative of active oxygen generation in neutrophils are shown. Neutrophils were obtained from patients 1, 2, and 3 or from control (Cont) before (solid and dotted line) or 25 days after (bold solid line) IFN-γ administration. Profiles of the cells stimulated with PMA are shown with a solid or bold solid line.
Flow cytometric analysis of active oxygen generation in neutrophils.
Histograms of DHR-123 fluorescence as indicative of active oxygen generation in neutrophils are shown. Neutrophils were obtained from patients 1, 2, and 3 or from control (Cont) before (solid and dotted line) or 25 days after (bold solid line) IFN-γ administration. Profiles of the cells stimulated with PMA are shown with a solid or bold solid line.
Change of active oxygen-generating ability in patients' neutrophils and surface cytochrome
b558 expression after IFN-γ treatment. Active oxygen production in neutrophils obtained from the patients at the indicated days after IFN-γ treatment was analyzed by flow cytometry using DHR-123 as a fluorescence probe and PMA as a stimulus. (A) Increase in fluorescence intensity in comparison to control cells was estimated and expressed as percent increase in mean fluorescence. (B) Surface expression of cytochromeb558 in neutrophils at the indicated days were analyzed by flow cytometry after staining with anticytochromeb558 monoclonal antibody 7D5. The lines indicate the mean values.
Change of active oxygen-generating ability in patients' neutrophils and surface cytochrome
b558 expression after IFN-γ treatment. Active oxygen production in neutrophils obtained from the patients at the indicated days after IFN-γ treatment was analyzed by flow cytometry using DHR-123 as a fluorescence probe and PMA as a stimulus. (A) Increase in fluorescence intensity in comparison to control cells was estimated and expressed as percent increase in mean fluorescence. (B) Surface expression of cytochromeb558 in neutrophils at the indicated days were analyzed by flow cytometry after staining with anticytochromeb558 monoclonal antibody 7D5. The lines indicate the mean values.
Flow cytometric analysis of cell surface cytochrome b558in patients' neutrophils
Isolated intact neutrophils from patients were stained with monoclonal antibody 7D5 and flow cytometric analysis was performed. Before IFN-γ treatment, neutrophils from each patient expressed 16% to 28% cytochrome b558 on their surface compared to that of control neutrophils and no dramatic change was observed after IFN-γ (Figure 5). The results suggest that these patients have a nonfunctional form of cytochrome present on cell surface before IFN-γ treatment.
Changes of splicing pattern of CYBB gene transcripts after IFN-γ treatment
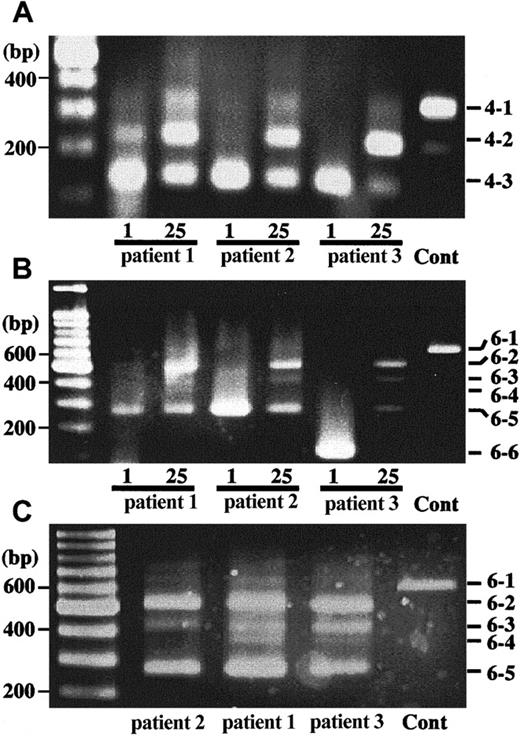
Total RNA was extracted from neutrophils, from which single-strand cDNA was synthesized. To analyze for a change in the pattern of splicing for CYBB gene transcripts, we used 2 sets of primers for PCR that flanked exon 3, namely a forward primer on exon 1 and a reverse primer either on exon 4 or on exon 6. Figure6A shows amplified fragments obtained using primers for exons 1 and 4, which were also extracted and sequenced. One day after the IFN-γ treatment, almost all amplified transcripts in neutrophils from patients 2 and 3 exhibited a 137-bp band that lacked exon 2 and 3 (Figure 6A, 4-3), and for patient 1, 72% were transcripts of this type. At 25 days after IFN-γ treatment, total mRNA for gp91-phox increased (data not shown) and the splicing patterns of gp91-phox transcripts in neutrophils were altered significantly. Although most transcripts still lacked exons 2 and 3, or exon 3 and were amplified as a 137- or 233-bp band, respectively (Figure 6A, 4-3 or 4-2), a significant amount of a 344-bp product that contained all 4 exons was newly detected (Figure 6A, 4-1). The changes in splicing after IFN-γ treatment were confirmed using primers for exons 1 to 6. At day 1, after IFN-γ administration, nearly all of the transcripts in neutrophils from patient 3 were of a size suggesting they contain only exons 1 and 6 (Figure 6B, 6-6), whereas a majority of transcripts for patients 1 and 2 appeared to contain exons 1, 4, and 6 (Figure 6B, 6-5). At 25 days after IFN-γ treatment, a marked change in the pattern of gp91-phox mRNA splicing was noted in neutrophils from all 3 patients (Figure 6B,C). Furthermore, amplified products that contained all 6 exons (Figure 6C, 6-1) were found in neutrophils from all 3 patients, at a relative percentage of 6.8%, 2.8%, and 4.3% in the specimens from patients 1, 2, and 3, respectively. In addition to the transcripts containing all 6 exons and the transcripts that lacked only exon 3 (24.3%, 44.3%, and 25.9% for those from patients 1, 2, and 3, respectively), there were also transcripts that lacked exon 2 and/or exon 5 together with exon 3. It is not clear at the moment why such alternative splicing takes place. The lower matching score of the splicing sequence to the consensus in the 3′ end of exon 3 and the 5′ end of exon 4 may influence such a splicing pattern. Alternatively, several purine-rich sequences known to promote splicing in exon 1 (2 sites), 4, 5 (3 sites each), and 6 (2 sites) may result in such variety of splicing patterns.20
Splicing pattern change of
CYBB gene transcripts after IFN-γ treatment.From neutrophil RNA of the patients who received IFN-γ 26 days before, single-stranded cDNAs were synthesized. PCR products were obtained from the cDNAs by using 2 sets of primers, a forward primer on exon 1 and a reverse primer on exon 4 (A) or a forward primer on exon 1 and a reverse primer on exon 6 (B,C). The products were separated on a 3% agarose gel and stained with ethidium bromide. In panels A and B, 40-cycle and in panel C, 50-cycle amplifications were performed. DNA size markers are shown on left and numbers shown on right indicate differently spliced products illustrated in Figure 3.
Splicing pattern change of
CYBB gene transcripts after IFN-γ treatment.From neutrophil RNA of the patients who received IFN-γ 26 days before, single-stranded cDNAs were synthesized. PCR products were obtained from the cDNAs by using 2 sets of primers, a forward primer on exon 1 and a reverse primer on exon 4 (A) or a forward primer on exon 1 and a reverse primer on exon 6 (B,C). The products were separated on a 3% agarose gel and stained with ethidium bromide. In panels A and B, 40-cycle and in panel C, 50-cycle amplifications were performed. DNA size markers are shown on left and numbers shown on right indicate differently spliced products illustrated in Figure 3.
Discussion
In this study, we report that neutrophils from 3 patients in one family exhibited greatly increased superoxide-generating ability after a single IFN-γ treatment. The mutation in the affected patients was identified as a single base substitution of guanosine to adenosine in the CYBB gene at residue 252 in exon 3 adjacent the splice junction. This mutation does not result in an amino acid transition but most gp91-phox transcripts analyzed showed skipping of exon 2 and 3. The improvement of superoxide-generating ability in neutrophils following IFN-γ was also observed in another patient who has exactly the same mutation but is in a different kindred than for the patients mentioned above (F.I. and H.N., unpublished results, June 2000). Eight other patients with CGD with the same mutation have been registered in the X-CGD database,1 4 and we speculate, they would receive similar clinical benefit of IFN-γ treatment with improved neutrophil superoxide-generating ability.
The results presented here indicate that for some splice junction region mutations in the CYBB gene, IFN-γ treatment at least partially corrects mRNA splicing and associated oxidase defects in vivo. Ezekowitz and colleagues have also reported an X-CGD patient in whom greatly increased phagocyte superoxide release was observed following IFN-γ treatment.8,9 The mutation in this patient was identified recently as a single base substitution in the first intron of the CYBB gene.4 In subsequent analysis of the molecular basis of the IFN-γ effect in this latter patient, Condino-Neto and Newburger found an increased ratio of spliced to unspliced CYBB mRNA in nuclei. They proposed that augmentation of nuclear export of normal transcripts and improvement in the fidelity of splicing at the first intron was the most likely mechanism of the IFN-γ–induced correction.12 In contrast, we report in this study dramatic IFN-γ–induced changes in transcript splicing in a kindred who otherwise had exon skipping due to a silent mutation in CYBB adjacent to intron 3. IFN-γ treatment resulted in increases in full-length transcripts, associated with increased O-generating activity.
Interferon-γ is known to alternate splicing pattern in certain RNA transcripts including tryptophanyl-tRNA,21 nitric oxide synthase mRNA,22 and HLA class I mRNA.23DNA-binding proteins involved in an alternative splicing mechanism such as SF 2/ASF,24 hnRNP A1,25RNPS1,26 SRp40, and SRp5527 are known. SRp40 and SRp55 bind to the splicing enhancer region of cardiac troponin T pre-mRNA and promote splicing of exon 527, and hnRNPA/B binds to the exonic splicing silencer region of human immunodeficiency virus 1 pre-mRNA and inhibits splicing of exon 2.28Further studies are needed to elucidate whether any of such proteins are involved in the splicing pattern change of CYBB gene transcripts described in this paper.
In the present investigation, markedly increased superoxide-generating activity in neutrophils was observed 11 days after a single administration of IFN-γ, and the increased activity was maintained for 2 to 4 weeks. The similar prolonged effect of IFN-γ was reported for the CGD patient described above who has a single base substitution in the first intron.9 It was shown further that IFN-γ–induced restoration of phagocyte function occurred at the level of myeloid progenitor cells.29 Caux and coworkers reported that IFN-γ stimulated the early stage of myelopoiesis by enhancing the frequency of growth factor–responding cells.30 Shiohara and colleagues showed that multipotential progenitors in murine bone marrow were supported by stem cell factors and IFN-γ, where the former was required in the early stage of development and the latter for subsequent growth.31
These studies and the prolonged effect on superoxide-generating ability in the present cases suggest that IFN-γ affected splicing of transcripts at the granulocyte progenitor cell level. This conjecture is further supported by the fact that changes in transcript splicing were not observed in B-cell lines established from the patients whose cells were then cultured in vitro with IFN-γ for 30 minutes to 2 weeks (F.I. and H.N., unpublished result, June 2000).32,33 Enhancement of phagocyte superoxide-generating ability and increased gene expression of individual oxidase components after administration of IFN-γ have been previously reported.34-36 Ahlin and associates reported a 3% to 5% increase of the neutrophil superoxide-generating ability in 2 X-linked CGD patients and one CGD patient with p67-phoxdeficiency.37 The gp91-phox promoter elements required for IFN-γ induction like IFN-γ stimulated response elements (ISRE) may be involved in such an increase.38 In the present investigation, a slight increase in CYBB total mRNA in neutrophils was observed after IFN-γ treatment. Cultivation of the B-cell lines from these patients in vitro with IFN-γ also increased total CYBB RNA (F.I. and H.N., unpublished observation, June 2000). However, changes in splicing of gp91-phox transcripts were not observed with the patients' B-cell lines cultured with IFN-γ as mentioned above. Therefore, increase of total RNA itself is independent of the appearance of correcting spliced CYBB gene transcripts, at least in B-cell lines.
Although there were changes in the superoxide-generating ability and splice pattern of gp91-phox mRNA in the patients' neutrophils in this study, enhanced expression of gp91-phoxon neutrophils was not observed as measured by cell surface staining with the cytochrome-specific monoclonal antibody 7D5. The epitope of the antibody was recently determined to be peptide coded between exon 5 and 6 of CYBB gene.39 Our results may indicate that nonfunctional cytochrome b558 detected prior to IFN-γ treatment included a form of gp91-phoxcontaining exon 5 and 6.
Finally, a significant number of CGD patients with splice site mutations have been registered in the United States, Europe, and Japan. It will be informative to study whether IFN-γ affects the splicing pattern for CYBB gene transcripts in any of such patients and whether this is associated with improved neutrophil superoxide-generating ability.
We are grateful to the patients and their families for their participation in this study. We thank Dr M. Takii (Kitakyushu Municipal Medical Center) and Dr K. Kawakami (Kagoshima University School of Medicine) for providing us useful information on CGD patients, and Dr M. R. Mercado (Kumamoto University School of Medicine) for technical assistance. We thank Dr M. C. Dinauer (Indiana University School of Medicine) for her critical reading of the manuscript. We also appreciate former professor I. Matsuda (Director of Ezuko Institution for Developmental Disabilities) for his support of this work.
Supported by grants from the Ministry of Education, Science, Sports and Culture, Japan, and the Ministry of Health and Welfare, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hiroyuki Nunoi, Department of Pediatrics, Miyazaki Medical College, 5200 Kihara, Kiyotake, Miyazaki 889-1692, Japan; e-mail: h-nunoi@fc.miyazaki-med.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal