Abstract
Hepatitis C virus (HCV) infection is associated with extrahepatic B-cell lymphoproliferative disorders. To determine whether a viral antigen drives this B-cell expansion, the B-cell receptors were cloned from HCV-associated lymphomas and were expressed as soluble immunoglobulins. The rescued immunoglobulins were then tested for their ability to bind the HCV-E2 envelope glycoprotein, an antigen that was previously implicated in the pathogenesis of HCV-associated B-cell diseases. One of 2 lymphoma immunoglobulin test cases bound the E2 protein in a manner identical to a bona fide human anti-E2 antibody. Moreover, it bound E2 from multiple viral genotypes, suggesting reactivity with a conserved E2 epitope. These findings support the hypothesis that some HCV-associated lymphomas originate from B cells that were initially activated by the HCV-E2 protein and might explain the association between HCV infection and some B-cell lymphoproliferative disorders.
Introduction
More than 100 million people worldwide are infected by hepatitis C virus (HCV).1 Infection causes prolonged and persistent disease in the majority of carriers, often leading to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (reviewed in Houghton2 and Liang et al3). HCV is also the leading cause of mixed cryoglobulinemia (MC), a systemic immune complex–mediated disorder characterized by polyclonal or monoclonal B-cell proliferation.4,5 MC can evolve into overt B-cell non-Hodgkin lymphoma (NHL),6 7 although the incidence of malignant transformation appears to be restricted to certain areas of the world.
In type II MC, immunoglobulin immune complexes contain both monoclonal immunoglobulin M (IgM) and polyclonal IgG.8 The monoclonal IgM has rheumatoid factor (RF) activity and is encoded mostly by a restricted set of variable (V) region genes, specifically VH1-69 (also known as 51p1) and Vκ3-A27 (also known as κv325).9-12 The same set of V region genes is expressed by the majority of HCV-associated NHL, suggesting a malignant progression from type II MC.13 Immunoglobulin V region genes expressed by these lymphomas exhibit an ongoing process of somatic mutations, indicative of antigen selection.13Moreover, the histologic presentation of many HCV-associated NHLs is typical of germinal center (GC) and post-GC B cells.14Therefore, it is likely that lymphomagenesis occurs when B cells proliferate in response to antigen. To date, the antigen driving these extrahepatic lymphoproliferative diseases is unknown.
Nevertheless, immune responses occurring in HCV patients against one HCV antigen, the E2 envelope glycoprotein (E2), suggest that the restricted V gene usage seen in the lymphoproliferative disorders may be linked to this antigen. V region genes from human anti-E2 antibodies, derived from B cells of HCV-infected individuals, show similar V gene bias to that observed in HCV-associated MC and NHL.15 16 These studies implicate the specific immune response to the E2 antigen in the pathogenesis of B-cell lymphoproliferative diseases and potentially in HCV-associated lymphomas.
A possibility exists that cognate B cells, which bind E2 via their specific B-cell receptor (BCR) could engage 2 signaling complexes simultaneously—the BCR and the CD19/CD21/CD81 complex (CD81 is the putative E2 receptor17). Dual engagement of these 2 signaling complexes has been shown to reduce the threshold of B-cell activation.18 To test the possibility that E2 may be the antigen involved in driving B-cell proliferation, we cloned the V region genes expressed by 2 HCV-associated lymphomas, expressed them as soluble immunoglobulins, and assayed them for binding to E2. The immunoglobulin from one of 2 HCV-associated lymphomas bound the E2 protein in a manner identical to a bona fide human anti-E2 antibody. This result supports the hypothesis that some HCV-associated lymphomas, and by inference, HCV-associated MC, can originate in B cells responding directly to viral antigen stimulation.
Patients, materials, and methods
Patient material
Two Stanford University Medical Center patients with overt B-cell NHL (without MC) and chronic HCV infection were identified. Both patients' sera were positive for anti-HCV antibodies and HCV-RNA at disease diagnosis; in neither case was the viral genotype reported. Patient 1, a 57-year-old man, was diagnosed with stage IV low-grade lymphoma, consistent with extranodal marginal zone lymphoma. Patient 2, a 54-year-old man, had stage IV diffuse large B-cell lymphoma.
Reverse transcriptase–polymerase chain reaction and V gene sequencing
Messenger RNA (mRNA) was isolated from the frozen lymphoma biopsies (Invitrogen, Carlsbad, CA) and reverse transcribed (Promega, Madison, WI). VH genes were amplified by polymerase chain reaction (PCR),19 using 5′ family-specific leader primers and a 3′ antisense joining (J) consensus primer. VL genes were amplified by using 5′ κ or λ leader primers and a 3′ antisense J consensus primer. The PCR products (∼500 base pairs [bp]) were gel purified (Qiaquick; Qiagen, Valencia, CA) and directly sequenced (373; Applied Biosystems, Foster City, CA). Clonal sequences, defined by identical CDR3 sequences from at least 2 independent PCRs, were aligned with germline sequences by using VBASE.20 Somatic mutations were determined by comparison to the most homologous germline gene. The probability (P) of excess or scarcity of replacement (R) mutations in VH complementarity determining region (CDR) and framework (FR) regions was calculated by a multinomial distribution model.21
Expression of NHL immunoglobulins
Clonal VH and VL genes were ligated into the PCR 2.1 cloning vector (Invitrogen), digested withSalI/NheI (VH) orBglII/BsiWI (VL), cloned into an immunoglobulin expression vector,22 and resequenced. Supernatants from transiently transfected COS-7 cells contained 1 to 2 μg lymphoma immunoglobulin/mL. VH and VLgenes were also ligated into pAdXCJL1/CMV adenovirus (AdV) transfer vector.23 Both vectors encode human IgG1/κ constant regions driven by 2 independent cytomegalovirus promoters, enabling expression of soluble IgG1 molecules.
The AdV-transfer vector was recombined with pTG3652 AdV DNA in bacteria, transfected into 293A cells, and plaque purified by cesium chloride centrifugation.24 Supernatants from HeLa cells, infected for 72 hours with purified virus (multiplicity of infection = 50), contained 10 to 100 μg human IgG1/mL as quantified by enzyme-linked immunosorbent assay (ELISA).
Expression of soluble HCV-E2661
A HindIII/XbaI (blunt-ended) fragment encoding amino acids 384 to 661 of E2 (genotype 1a) fused to the tissue plasminogen activator leader peptide derived from p14.tE2.661.hiv25 was ligated intoHindIII/XhoI (blunt-ended)–digested pXCJL2MCS-CMVp/A AdV transfer vector. Purified AdV encoding HCV-E2661 (or LacZ) was prepared as described above, and infected HeLa cell supernatants were used as a source of secreted E2661. Supernatants from AdV-LacZ–infected cells served as mock antigen in subsequent assays.
Expression of intracellular HCV glycoproteins
Plasmids encoding E2661(p14.tE2.661.hiv), full-length E2 (pE2384-746), and E1 plus E2 (pE1E2) were derived from genotype 1a and described elsewhere.25,26 pE1E2p7NS2 was constructed by PCR amplification of E2p7NS2 (encoding amino acids 364 to 1026) with primers (5′-GCGCAAGCTTCCATGGTGGGGAACTGG-3′ and 5′-GAGCTCCTGCAGTCACAGCAACCTCCACCCCTTGG-3′), using the pBRTM/HCV1-3011 (provided by C. M. Rice, Washington University, St Louis, MO) template. The SapI/PstI-digested PCR product was ligated into similarly digested pE1E2. pE1 was constructed by PCR amplification of full-length E1 (encoding amino acids 171-383) with primers (5′-GCGAGCAAGCTTCCATGGGTTGCTCTTTCTCTATC-3′ and 5′-AGTATCATGCATTCACGCGTCGACGCCGGCAAATAG-3′), using template pBRTM/HCV1-3011. The HindIII/NsiI-digested PCR product was ligated with HindIII/PstI digested pCDM8 (-ApaI).26
Plasmids (10 μg) were transfected into 293A cells (GenePORTER 2; Gene Therapy Systems, San Diego, CA), and intracellular glycoproteins were solubilized with lysis buffer (0.1 M Tris-HCl, pH 8.0, 1 mM EDTA, 4% Triton X-100, and protease inhibitors). Lysates from cells transfected with vector alone served as mock antigen in subsequent assays. Q1a, Q1b, Q2a, and Q2b vaccinia virus constructs encode full-length E2 from HCV genotypes 1a, 1b, 2a, and 2b, respectively.27Infection of HeLa cells and preparation of lysates was performed as described.27
Lymphoma immunoglobulin binding to HCV-E2
Sandwich ELISA was performed as described.27Supernatants (50 μL) containing E2661were added to wells precoated with purified Galanthus nivalis lectin (GNA; Sigma, St Louis, MO) and incubated overnight at 4°C. To test for possible cross-reactivity with human immunoglobulin, some of the assays were also performed in the presence of Sepharose-bound human immunoglobulin. Bound lymphoma immunoglobulin or anti-E2 monoclonal antibody (mAb)27 was visualized by using horseradish peroxidase (HRP)–conjugated anti–human IgG (Southern Biotechnology Associates, Birmingham, AL). Purified immunoglobulins from 23 non–HCV-associated NHL rescue hybridomas28 served as controls. Binding of lymphoma immunoglobulins or anti-E2 mAbs to E2 proteins of different genotypes expressed by vaccinia virus was performed as described.27 GNA-captured E2 was detected by the conformation-dependent H53 mAb (mouse IgG1),29 kindly provided by Jean Dubuisson (Institut Pasteur, Lille, France), and HRP-conjugated anti–mouse IgG (Southern Biotechnology Associates).
Immunoprecipitation and Western blot
Human IgG1 antibodies adsorbed to protein A–Sepharose CL-4B beads (Sigma) were incubated overnight at 4°C with supernatants (1 mL) from AdV-infected HeLa cells expressing E2661 or LacZ or with cell lysates containing intracellular E2. Immunoprecipitates were electrophoresed on a nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel,30 transferred to nitrocellulose (Schleicher & Schuell, Keene, NH), and immunoblotted with anti-E2 mAb 3/1125 and HRP-conjugated anti–rat immunoglobulin.
Results
A rescued BCR from an HCV-associated NHL binds soluble E2
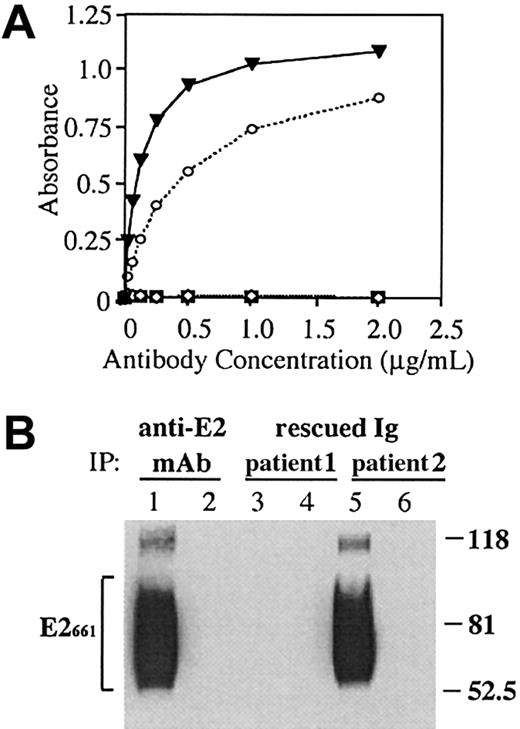
A direct test to determine whether the BCR, expressed on the surface of HCV-associated lymphomas, could bind E2 is not possible. The reason is that E2 also binds CD81, a cell surface–expressed tetraspanin molecule.17 We therefore molecularly rescued the variable heavy (VH) and light (VL) region genes from 2 independent lymphoma biopsies, obtained from patients with chronic HCV infection. The 2 patients did not have a history of MC and both were diagnosed at Stanford University Medical Center. The molecularly rescued V genes were then inserted into an expression vector for the production of soluble immunoglobulins as detailed in “Patients, materials, and methods.” The test antigen was a soluble, secreted form of E2, C-terminally truncated at amino acid 661 (E2661). Truncated E2 has been shown to bind CD81 on liver31 and lymphoid cell lines.17 32
One of the 2 rescued lymphoma immunoglobulins (from patient 2) reacted with this antigen (Figure 1A). The level of reactivity was similar to that of a well-characterized human anti-E2 mAb.27 In contrast, none of the 23 rescued immunoglobulins from non–HCV-associated lymphoma cases bound E2661 (Figure1A). The reactivity of the immunoglobulin from patient 2 with E2 was not reduced by the inclusion of Sepharose-bound human IgG in the assay, indicating that the rescued immunoglobulin was not a RF. Because soluble, recombinant E2 is glycosylated and heterogeneous,33 we compared the pattern of E2 molecules recognized by the rescued lymphoma immunoglobulin to those recognized by the human anti-E2 mAb. HCV-E2661 was immunoprecipitated with the HCV-associated lymphoma immunoglobulins or with the anti-E2 mAb. The precipitated E2 was analyzed by SDS-PAGE, followed by immunoblotting with an independent anti-E2 mAb, which recognizes a linear E2 epitope25 (Figure 1B). The HCV-associated NHL immunoglobulin that bound E2661 by ELISA precipitated E2 molecules with a size distribution that was similar, if not identical, to that immunoprecipitated with the known anti-E2 mAb. Both immunoglobulins bound multiple E2661 glycoforms, seen as a broad banding pattern of heavily glycosylated E2 molecules. Binding was specific for E2 because incubation of the lymphoma immunoglobulins or the anti-E2 mAb with a control supernatant not containing the E2 protein did not precipitate an appropriate size band. Lymphoma immunoglobulins from 2 non–HCV-associated lymphoma cases failed to bind E2 in this assay (data not shown).
A rescued immunoglobulin from an HCV-associated lymphoma binds soluble E2661.
(A) An anti-E2 mAb, CBH-4G (▾),27 HCV-associated immunoglobulin (patient 1, ⋄), HCV-associated immunoglobulin (patient 2, ○) and non-HCV lymphoma immunoglobulin controls (n = 23; ▪) were serially diluted and added to E2661-coated wells, and bound immunoglobulin was determined by ELISA. (B) Supernatants from AdV-infected HeLa cells expressing HCV-E2661 (lanes 1, 3, and 5) or LacZ (lanes 2, 4, and 6) were immunoprecipitated with immunoglobulins derived from an anti-E2 mAb (CBH-4G) and from the 2 HCV-associated lymphoma cases. Proteins were separated by SDS-PAGE, and blots were probed with an antibody against HCV-E2. Sizes (in kd) of protein markers are indicated on the right, and the bracket on the left indicates the precipitated heterogeneously glycosylated E2661 protein.
A rescued immunoglobulin from an HCV-associated lymphoma binds soluble E2661.
(A) An anti-E2 mAb, CBH-4G (▾),27 HCV-associated immunoglobulin (patient 1, ⋄), HCV-associated immunoglobulin (patient 2, ○) and non-HCV lymphoma immunoglobulin controls (n = 23; ▪) were serially diluted and added to E2661-coated wells, and bound immunoglobulin was determined by ELISA. (B) Supernatants from AdV-infected HeLa cells expressing HCV-E2661 (lanes 1, 3, and 5) or LacZ (lanes 2, 4, and 6) were immunoprecipitated with immunoglobulins derived from an anti-E2 mAb (CBH-4G) and from the 2 HCV-associated lymphoma cases. Proteins were separated by SDS-PAGE, and blots were probed with an antibody against HCV-E2. Sizes (in kd) of protein markers are indicated on the right, and the bracket on the left indicates the precipitated heterogeneously glycosylated E2661 protein.
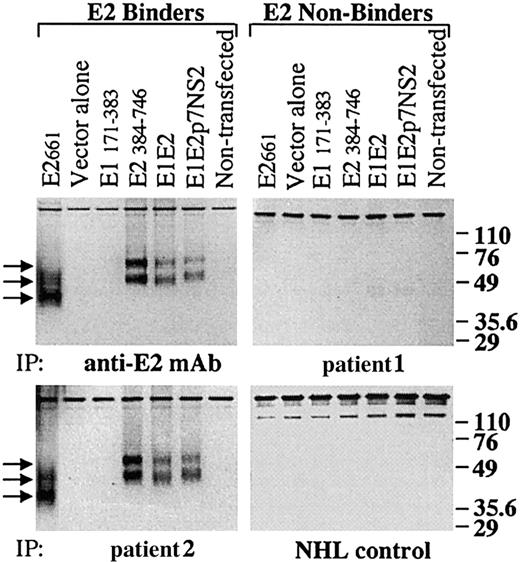
Binding to intracellular forms of E2
Intracellular forms of E2 are believed to adopt a native conformation34,35 and to bind CD81 with greater affinity than the secreted form. Likewise, coexpression of E1 and E2 results in E1E2 heterodimers, resembling native viral particles.33 We therefore tested whether full-length E2 (E2384-746) and E2 expressed in the presence of E1 could bind the rescued lymphoma immunoglobulin. The HCV-associated lymphoma immunoglobulin (patient 2) bound all intracellular forms of E2 (Figure2). Once more, the binding pattern was identical to that of the well-characterized anti-E2 mAb. The lymphoma immunoglobulins did not precipitate the appropriate size band when incubated with nontransfected cell lysates or with cells expressing E1 alone, demonstrating specificity of binding to E2. A NHL immunoglobulin from a non–HCV-associated case did not bind intracellular HCV-E2 (NHL control).
A rescued immunoglobulin from an HCV-associated lymphoma precipitates multiple intracellular forms of the HCV-E2 glycoprotein.
Cell lysates containing each of the HCV glycoproteins were immunoprecipitated with immunoglobulins derived from an anti-E2 mAb (CBH-4G), from the 2 HCV-associated NHL cases, and from a non-HCV NHL control immunoglobulin. Proteins were separated by SDS-PAGE, and blots were probed with an antibody against HCV-E2. Sizes (in kd) of protein markers are indicated on the right, and arrowheads on the left point to the precipitated forms of E2. IP indicates the source of immunoprecipitating immunoglobulin.
A rescued immunoglobulin from an HCV-associated lymphoma precipitates multiple intracellular forms of the HCV-E2 glycoprotein.
Cell lysates containing each of the HCV glycoproteins were immunoprecipitated with immunoglobulins derived from an anti-E2 mAb (CBH-4G), from the 2 HCV-associated NHL cases, and from a non-HCV NHL control immunoglobulin. Proteins were separated by SDS-PAGE, and blots were probed with an antibody against HCV-E2. Sizes (in kd) of protein markers are indicated on the right, and arrowheads on the left point to the precipitated forms of E2. IP indicates the source of immunoprecipitating immunoglobulin.
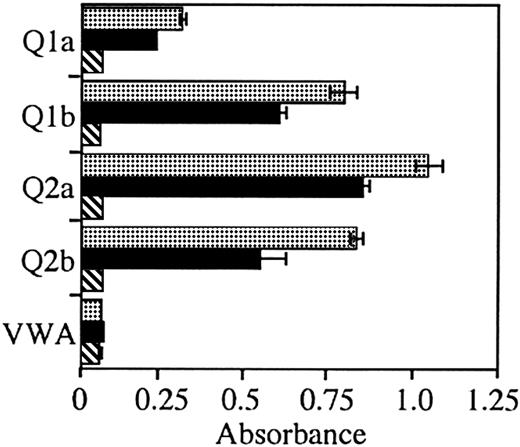
HCV-associated NHL BCR binds E2 of multiple genotypes
To test the breadth of reactivity of the immunoglobulin from patient 2 and to test whether the immunoglobulin from patient 1 could react with E2 from a different HCV genotype, we tested binding of E2 antigens derived from the 1a, 1b, 2a, and 2b genotypes. The lymphoma immunoglobulin from patient 2 bound E2 from all tested genotypes (Figure 3). Again, the level of binding was similar, albeit lower than the control anti-E2 mAb. In contrast, the lymphoma immunoglobulin from patient 1 did not bind E2 from any of the genotypes tested.
A rescued immunoglobulin from an HCV-associated lymphoma reacts with HCV-E2 from multiple genotypes.
Anti-E2 mAb CBH-4G (░), HCV-associated immunoglobulin patient 2 (▪), and HCV-associated immunoglobulin patient 1 (▧) were added to wells coated with E2, obtained from cells infected with recombinant vaccinia virus expressing the recombinant protein from HCV genotypes 1a (Q1a), 1b (Q1b), 2a (Q2a), and 2b (Q2b) or with vaccinia wild-type virus (VWA). Bound immunoglobulin was determined by ELISA. Error bars indicated 1 SD from the mean.
A rescued immunoglobulin from an HCV-associated lymphoma reacts with HCV-E2 from multiple genotypes.
Anti-E2 mAb CBH-4G (░), HCV-associated immunoglobulin patient 2 (▪), and HCV-associated immunoglobulin patient 1 (▧) were added to wells coated with E2, obtained from cells infected with recombinant vaccinia virus expressing the recombinant protein from HCV genotypes 1a (Q1a), 1b (Q1b), 2a (Q2a), and 2b (Q2b) or with vaccinia wild-type virus (VWA). Bound immunoglobulin was determined by ELISA. Error bars indicated 1 SD from the mean.
Gene usage and distribution of mutations in HCV-associated NHL VH and VL immunoglobulin genes
We determined the germline V region gene counterparts of the lymphoma immunoglobulins by searching VBASE.20 They were VH4-34/Vκ3-A27 (patient 1) and VH3-15/Vκ1-O12 (patient 2) (Figure4). None of these CDR3 regions had significant similarity to CDR3 regions of anti-HCV antibodies or to CDR3 sequences derived from RF. The 2 expressed VH and one of the VL genes contained numerous nucleotide differences from their corresponding germline genes, whereas the Vκ1-O12 of patient 2 had only one amino acid difference in framework 3. To address the possible role of antigen selection in the evolution of these HCV-associated immunoglobulins, we analyzed the highly mutated VH region genes for the distribution of R and silent (S) mutations in the CDRs and FRs by using a multinomial distribution model21 (Table 1). Both VH genes contained a significantly lower number of R mutations in FRs than would be expected if mutations had occurred by chance alone, indicating a selective pressure to preserve functional immunoglobulin molecules. However, only 1 of the VH genes (patient 2) showed a significant excess of R mutations in the CDR region (P = .01901), exceeding that expected to occur by chance alone. Thus, both immunoglobulins demonstrated a negative selection pressure against changes in FR, and the E2-binding HCV-associated NHL immunoglobulin (patient 2) showed evidence of positive selection.
Deduced amino acid sequences of HCV-associated lymphoma VHand VL genes.
The amino acid sequences of the most homologous germline genes are shown above the lymphoma sequences (VH, panel A; VL, panel B). Amino acid replacement mutations are indicated by uppercase letters and silent mutations are indicated by lowercase letters.
Deduced amino acid sequences of HCV-associated lymphoma VHand VL genes.
The amino acid sequences of the most homologous germline genes are shown above the lymphoma sequences (VH, panel A; VL, panel B). Amino acid replacement mutations are indicated by uppercase letters and silent mutations are indicated by lowercase letters.
Distribution of VH gene mutations in HCV-associated NHL immunoglobulins
| Patient . | Most similar germline VHgene . | Percentage of VH gene homology . | FR/CDR . | Observed mutations . | ||
|---|---|---|---|---|---|---|
| R . | S . | P* . | ||||
| 1 | VH4-34 | 95.3 | FR | 4 | 4 | .03128† |
| CDR | 2 | 3 | .55921 | |||
| 2 | VH3-15 | 92.6 | FR | 5 | 6 | .00148† |
| CDR | 8 | 2 | .01901† | |||
| Patient . | Most similar germline VHgene . | Percentage of VH gene homology . | FR/CDR . | Observed mutations . | ||
|---|---|---|---|---|---|---|
| R . | S . | P* . | ||||
| 1 | VH4-34 | 95.3 | FR | 4 | 4 | .03128† |
| CDR | 2 | 3 | .55921 | |||
| 2 | VH3-15 | 92.6 | FR | 5 | 6 | .00148† |
| CDR | 8 | 2 | .01901† | |||
The P value is the probability that excess (for CDRs) or scarcity (for FRs) of mutations occurred by chance.
Statistically significant values (P < .05).
Discussion
E2 has been shown to be the primary target of humoral immune responses against HCV.36 Antibodies against E2 were detected in 88% of chronically infected and in 49% of acutely infected HCV patients.37 mAbs have been made from such patients and V region sequences have been determined. Remarkably, these immunoglobulins use a restricted V gene repertoire similar to that used by MC and by HCV-associated NHLs immunoglobulins,9-11namely a strong bias for VH1-69 and Vκ3-A27.15,16 This restricted usage suggests that benign lymphoproliferation in MC and overt B-cell lymphomas may be sequential phases in an antigen-driven process.14 This hypothesis is supported by a recent study in which the same B-cell clone, present in an HCV-infected MC patient early in the course of the disease, was later detected as a NHL.14 The V regions expressed by this B cell showed significant intraclonal diversity and accumulated multiple somatic mutations, indicative of an antigen-driven clonal expansion process.13,14,38 Moreover, the CDR3 regions of this case and of several other lymphoma immunoglobulins showed homology to the CDR3 regions of anti-E2 antibodies.14 38 These data support the hypothesis that the E2 antigen drives B-cell expansion, leading to a population at risk for malignant transformation.
The native structure of E2 is currently unknown. Viral particles have not been isolated nor is a cell culture system available to support efficient HCV replication. Recombinant soluble proteins, truncated to remove the hydrophobic C-terminus at amino acids 661 (E2661) or 715 (E2715), have been used as soluble surrogates for viral particles in immune assays and functional studies. For example, the identification of CD81 as the putative cellular receptor for HCV is based on its binding to a truncated form of the E2 protein.17 Therefore, we tested several different E2 forms in binding studies with the lymphoma-derived immunoglobulins. Intracellular E2661, which lacks many of the complex sugars acquired by secreted E2661 during transit through the host-cell secretory pathway39 has been shown to bind more avidly to cell surface–expressed CD81 than does secreted E2661.34,35 Nevertheless, the rescued lymphoma immunoglobulin from patient 2 bound both the secreted and the intracellular forms of E2661 and also the noncovalently linked E1-E2 heterodimer (Figures 1 and 2), which is believed to be the prebudding form of the HCV envelope glycoprotein complex.40,41 Moreover, the level of binding of the rescued immunoglobulin and the reactivity pattern with multiple E2 glycoforms was comparable to that of a well-characterized anti-E2 mAb (Figures 1 and 2). Interestingly, the characterized mAb expresses the VH1-69 gene,16 whereas neither of the lymphoma cases expressed this VH gene (Table 1). In contrast, both Vκ chains expressed by the lymphoma cases have been frequently reported in immunoglobulin derived from HCV-associated NHLs and in MC. To ascertain whether patient 2 could have evolved from a nonapparent MC with RF activity, we analyzed sequence homologies and found no similarity between the rescued CDR3 regions to those of sequenced RF-encoding immunoglobulins. In addition, the rescued immunoglobulin of patient 2 did not cross-react with human IgG, suggesting that its reactivity is limited only to the viral antigen. Thus, in this case there is no evidence of progression from MC to overt B-cell lymphoma.
Anti-E2 antibodies that block E2 binding to cells have been referred to as neutralization of binding (NOB) antibodies.32 Subsequent studies characterizing 10 human anti-E2 mAbs showed that they could be either NOB-positive or NOB-negative,27 the later do not block E2 binding to CD81. The rescued lymphoma immunoglobulin (patient 2) exhibited properties similar to those of one of the NOB-negative mAbs. The lymphoma-derived immunoglobulin and the anti-E2 mAb recognized a similar spectrum of E2 glycoproteins (Figures 1B and 2) and CD81-bound E2 molecules (data not shown). Both immunoglobulins bound to E2 glycoproteins of multiple viral genotypes (Figure 3), implying reactivity with a conserved E2 epitope. Because of the biased V gene usage in HCV-associated lymphoproliferative disorders, it is likely that a conserved epitope is involved in the process. A benign B-cell proliferation, initiated in direct response to the viral antigen, may render cells at risk for an additional rare event that results in malignant transformation. HCV-infected patients may also develop lymphomas that are either independent of viral infection or are the result of an indirect activation of B cells by the virus. Patient 1 of our study may represent an example of a patient with such lymphoma. It remains to be determined whether reactivity of HCV-associated lymphoma immunoglobulin with the E2 envelope protein is a rare or more prevalent event.
The results of our study are consistent with the receptor-mediated lymphomagenesis hypothesis that was proposed nearly 2 decades ago.42 In that model, chronic antigen stimulation by retroviral env protein was suggested to be both necessary and sufficient to induce virus-specific T-cell proliferation. In the absence of immune regulatory mechanisms, proliferating T-cell clones with high-affinity receptors for env could eventually give rise to frank T-cell lymphomas.42 Our study implicates E2 in receptor-mediated lymphomagenesis leading to HCV-associated NHL. However, our hypothesis of B-cell activation has the additional feature of dual binding of E2 to a cognate BCR and to the CD81 molecule, which is a component of a signaling complex. To the best of our knowledge, this is the first identification of a cognate antigen for a human lymphoma BCR.
We thank J. A. McKeating for the E2 encoding plasmids and for the anti-E2 mAb 3/11, and J. Dubuisson for the anti-E2 mAb H53. We thank Susanne Auffermann-Gretzinger, Wen-Kai Weng, John Timmerman, and Ron Levy for reviewing the manuscript.
GenBank accession numbers for the VH genes areAF372845 and AF372844 (for patients 1 and 2, respectively) and for the VL genes are AF372843 and AF372842 (for patients 1 and 2, respectively).
Supported by grant CA34233 (S.L.) and by grants DA06596 and AI47355 (S.K.H.F.) from the National Institutes of Health, and by grant 2110046 from the Cancer Research Fund, under Interagency Agreement 97-12013 (University of California, Davis contract 98-00924V) with the Department of Health Services, Cancer Research Section (S.L).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Shoshana Levy, Department of Medicine, Division of Oncology, Stanford University Medical Center, 1105a North Wing, CCSR Bldg, 269 Campus Dr, Stanford, CA 94305-5151; e-mail:levy@cmgm.stanford.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal