Abstract
Fanconi anemia (FA) is a human autosomal recessive cancer susceptibility disorder characterized by cellular sensitivity to mitomycin C and ionizing radiation. Six FA genes (corresponding to subtypes A, C, D2, E, F, and G) have been cloned, and the encoded FA proteins interact in a common cellular pathway. To further understand the in vivo role of one of these human genes (FANCG), we generated a targeted disruption of murine Fancg and bred mice homozygous for the targeted allele. Similar to the phenotype of the previously described Fancc−/− andFanca−/− mice, theFancg−/− mice had normal viability and no gross developmental abnormalities. Primary splenic lymphocytes, bone marrow progenitor cells, and murine embryo fibroblasts from theFancg−/− mice demonstrated spontaneous chromosome breakage and increased sensitivity to mitomycin C and, to a lesser extent, ionizing radiation. Fancg−/−lymphocytes had a defect in the FA pathway, based on their failure to activate the monoubiquitination of the downstream Fancd2protein in response to IR. Finally,Fancg−/− mice had decreased fertility and abnormal gonadal histology. In conclusion, disruption of theFancg gene confirms the role of Fancg in the FA pathway. The Fancg−/− mouse may be useful as an animal model for future gene therapy and cancer susceptibility studies.
Introduction
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome characterized by multiple congenital anomalies and progressive bone marrow failure.1,2 FA patients develop several types of cancers, including acute myeloid leukemias and squamous cell carcinomas of the head and neck.3 FA cells are sensitive to DNA cross-linking agents, such as mitomycin C (MMC) and, to a lesser extent, ionizing radiation (IR).4,5 Based on somatic cell fusion studies, FA is composed of 8 distinct complementation groups.6,7 Six of the human FA genes, including the genes for FANCA,8,9FANCC,10 FANCD2,11 FANCE,12FANCF,13 and FANCG,14 have been cloned.
Recent studies have demonstrated that the 6 cloned FA proteins interact in a common cellular pathway.15 The FANCA, FANCC, FANCE, FANCF, and FANCG proteins assemble in a multisubunit nuclear complex.16-21 The FA protein complex regulates the monoubiquitination of the downstream FANCD2 protein, suggesting that the complex is a multisubunit monoubiquitin ligase or regulates a ligase activity. When normal (non-FA) cells are exposed to DNA-damaging agents, such as IR, MMC, or UV light, FANCD2 is monoubiquitinated and targeted to nuclear foci containing the BRCA1 protein.15Disruption of this pathway leads to the characteristic cellular and clinical abnormalities observed in FA.
In an attempt to understand the in vivo function of FA genes, targeted disruptions of FA genes have been generated. Two murine models, containing disruptions of the murine homolog of FANCC, have been developed. Chen et al22 generated a disruption of exon 8 of Fancc, while Whitney et al23 used homologous recombination to create a disruption of exon 9. In both models, spontaneous chromosome breakage was observed. In addition, an increase in chromosome breaks in splenic lymphocytes in response to bifunctional alkylating agents was observed. In both models,Fancc−/− mice had germ cell defects and decreased fertility. Unlike human FA patients, theFancc−/− mice had no obvious gross developmental abnormalities or cancer susceptibility. To date, theFancc murine knockout model has been useful in examining (1) the role of the Fancc gene in the physiologic response of hematopoietic cells to DNA damage,24,25 (2) the in vivo effects of inhibitory cytokines on FA marrow cells,23,26,27 and (3) the efficacy of gene therapy.28-30
In principle, targeted disruption of other murine FA genes, such as theFancg/xrcc9 gene, may provide additional insight to in vivo function. First, additional FA gene knockout models will allow a side-by-side comparison of disease severity in different FA subtypes. Recent studies have suggested that the severity of FA, in terms of developmental abnormalities and hematologic defects, is dependent on FA subtype.31 For instance, FA-C and FA-G patients exhibit more severe disease, while FA-A patients exhibit more mild disease, with later onset of bone marrow failure and hematologic malignancy. Whether mice with targeted disruptions of different FA genes vary in disease severity remains untested. Second, conflicting studies suggest that some FA patients and cell lines are sensitive not only to mitomycin C (MMC) and diepoxy butane (DEB) but also to IR. Accordingly, murine knockout models may allow the systematic evaluation of differential drug and IR sensitivity. Third, the availability of other FA gene disruptions will allow the generation and characterizaton of mice with multiple FA gene knockouts. For instance, if 2 FA genes function exclusively in the same cellular pathway, a double knockout should have the same phenotype as the single FA gene knockout. In the current study, we have used homologous recombination to disrupt the murine Fancg gene and have analyzed the primary cells and tissues derived from Fancg−/− mice.
Materials and methods
Generation of Fancg-deficient mice and genotyping
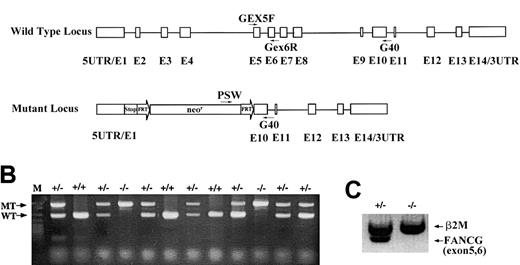
The murine Fancg gene was disrupted by replacing exons 2 to 9 with an FRT-flanked neomycin cassette via homologous recombination in 129/SvJae embryonic stem cells. Mice homozygous for the Fancg mutation within a mixed genetic background of 129/Sv and C57BL were generated following standard protocols. Mouse tail genomic DNA was prepared as previously described32 and used as a template for polymerase chain reaction (PCR) genotyping. PCR reactions were assembled according to the manufacturer's protocols (Roche, Indianapolis, IN) with primers Gex5F, Gex6R, Psv, and G40 (Figure1) in the same reaction tube, at a final concentration of 1μM each. The cycling conditions were 1 cycle of 94°C for 3 minutes; then 30 cycles of 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 45 seconds; followed by 1 cycle of 72°C for 5 minutes. Mouse testis RNA was extracted using the SV Total RNA Isolation System (Promega, Madison, WI). Fancg messenger RNA (mRNA) and β2-microglobulin mRNA were simultaneously amplified from 1 μg total RNA by reverse transcriptase (RT)-PCR using the OneStep RT-PCR kit (Qiagen, Valencia, CA). The amplification conditions were 50°C for 30 minutes for 1 cycle; 95°C for 15 minutes for 1 cycle; 94°C for 45 seconds, 60°C for 45 seconds, and 72°C for 1 minute for 30 cycles; and 72°C for 10 minutes for 1 cycle. Oligonucleotides (Figure 1A) used for PCR and RT-PCR were as follows:
Targeted disruption of the murine
Fancg gene. (A) Schematic representation of the murine Fancg gene showing 14 exons. The targeted allele has exons 2 to 9 replaced by the FRT-flanked neomycin cassette. There are translational stop codons in 3 reading frames 5′ to the neomycin cassette to terminate any potential upstream translation. Primers used for genotype and RT-PCR are indicated. (B) PCR genotype of mouse tail DNA using primers indicated in panel A. Primers Gex5F and Gex6R amplify a 285 base pair (bp) product specifically from the wild-type allele (WT), whereas primers Psv and G40 amplify a 580 bp product specifically from the mutant allele (MT). (C) RT-PCR of the wild-typeFancg mRNA using primers Gex5F and Gex6R fromFancg+/− testes but not fromFancg−/− testes. A 360 bp RT-PCR product of β2-microglobulin was used as an internal control (β2M).
Targeted disruption of the murine
Fancg gene. (A) Schematic representation of the murine Fancg gene showing 14 exons. The targeted allele has exons 2 to 9 replaced by the FRT-flanked neomycin cassette. There are translational stop codons in 3 reading frames 5′ to the neomycin cassette to terminate any potential upstream translation. Primers used for genotype and RT-PCR are indicated. (B) PCR genotype of mouse tail DNA using primers indicated in panel A. Primers Gex5F and Gex6R amplify a 285 base pair (bp) product specifically from the wild-type allele (WT), whereas primers Psv and G40 amplify a 580 bp product specifically from the mutant allele (MT). (C) RT-PCR of the wild-typeFancg mRNA using primers Gex5F and Gex6R fromFancg+/− testes but not fromFancg−/− testes. A 360 bp RT-PCR product of β2-microglobulin was used as an internal control (β2M).
Gex5F: 5′CCTCTGAGGATCTGCTACTACTGC3′.
Gex6R: 5′GTGTACACCTGGACTAACACGGAC3′.
G40: 5′TGGCTAAATTCACTAAGTG3′.
Psv: 5′AAGGTTGGGCTTCGGAATCG3′.
M2F: 5′GTATGCTATCCAGAAAACCCCTC3′.
M2R: 5′CATGTCTCGATCCCAGTAGACGG3′.
Chromosome breakage assay for murine splenocytes
Splenocytes were prepared from 6-week-old mice of knownFancg genotype. Briefly, the spleen was dissected, crushed in RPMI medium into a single-cell suspension, and filtered through a 70 μm filter. Red cells were lysed in hypotonic ammonium chloride. The remaining splenic lymphocytes were washed in phosphate-buffered saline and resuspended in RPMI/10% fetal bovine serum plus phytohemagglutinin. Cells were tested for viability by the trypan blue exclusion assay. Cells were cultured for 24 hours in media and exposed to MMC or DEB for an additional 48 hours. Alternatively, cells were cultured for 60 hours, exposed to IR (2 or 4 Gy, as indicated), and allowed to recover for 12 hours before chromosome breakage or trypan blue exclusion (viability) analysis.
Murine hematopoietic progenitor assay
Mononuclear cells were isolated from the femurs and tibiae of 4- to 6-week-old Fancg+/− orFancg−/− mice, as previously described. A total of 2 × 104 cells were cultured in 1 mL of MethoCult M3434 media (StemCell Technologies, Vancouver, BC, Canada) with or without MMC treatment. Colonies were scored at day 7, when most of the colonies belong to the granulocyte-macrophage colony-forming unit or erythroid burst-forming unit lineages. Each number was averaged from duplicate plates, and the data were derived from 2 independent experiments.
Immunoblotting of the murine Fancd2 protein
Primary splenic lymphocytes, prepared as above, were cultured in RPMI medium plus 10% heat-inactivated fetal bovine serum for 72 hours. Cells were untreated or irradiated with IR (2, 4, 10, and 20 Gy). Cells were lysed, and cellular proteins were electrophoresed, transferred to nitrocellulose, and immunoblotted with a polyclonal antibody raised against human FANCD2.15 This antiserum cross-reacts with the Fancd2 protein in murine cells.
Flow cytometry
Lymphocytes isolated from thymus, spleen, and peripheral lymph nodes were stained for T- or B-lymphocyte surface molecules with fluorescein isothiocyanate–conjugated anti-CD3, anti-CD4, and anti-CD19 and phycoerythrin-conjugated anti-CD8, anti-CD44, anti-CD45RB, immunoglubulin M, and anti-B220 (BD PharMingen, San Diego, CA). Stained cells were analyzed on a Coulter Epics XL flow cytometry system.
Histology
Mice ovaries and testes were isolated and fixed in 4% paraformaldehyde and further processed by the core facility of the Department of Pathology at Massachusetts General Hospital. The specimens were photographed on a Zeiss Axiplot.
Results
Gene targeting and gross phenotype ofFancg−/−mice
We have disrupted the murine Fancg gene by replacing exons 2 to 9 with an FRT-flanked neomycin resistance cassette through homologous recombination in murine embryonic stem cells (Figure1A). Two independent embryonic stem cell clones with the desired mutation were injected into C57BL/6 blastocysts, and one germ-line transmission competent male chimera was obtained and bred with C57Bl/6 females to generate heterozygous (Fancg+/−) mice. Approximately 25% of the offspring of heterozygous breeders were homozygous (Fancg−/−) as determined by genomic PCR (Figure 1B), indicating that there was no embryonic lethality associated with biallelic Fancg mutations. Moreover, no wild-type Fancg mRNA transcripts were detected in the testis of the Fancg−/− mice by RT-PCR using primers amplifying exons 5 and 6 (Figure 1C). This result demonstrates that these mice are null mutants for the Fancg gene. Mutant animals weighed the same as littermate controls, and no macroscopic developmental abnormalities of the limbs or other organ systems were detected (data not shown).
Increased chromosome breakage and decreased survival ofFancg−/−cells in MMC
Cytogenetic analysis demonstrated that primary cells (splenic lymphocytes) from the Fancg−/− mice had significantly higher DEB-induced and MMC-induced chromosomal aberrations, particularly radial forms, compared with control mice (Figure 2A). Spontaneous aberrations were also increased in Fancg−/− splenocytes. In addition, we established primary skin fibroblast cultures (MEF cultures) from day 18 mutant and control embryos (data not shown). Treatment of the mutant fibroblasts with MMC and DEB revealed chromosome breakage and other cytogenetic abnormalities, analogous to primary skin fibroblasts from human FA patients (Figure 2B).
Primary splenic lymphocytes from
Fancg−/− mice have increased DEB- and MMC-induced chromosome breakage. (A) Comparison of the number of aberrations per cell after treatment with the indicated amount of DEB or MMC. (B) Metaphase spreads of splenocytes fromFancg+/− and Fancg−/−mice after 72 hours of in vitro growth. Cells were exposed to either no drug (+0), MMC (20 ng/mL), or IR (4 Gy). Arrows indicate radial forms or breaks.
Primary splenic lymphocytes from
Fancg−/− mice have increased DEB- and MMC-induced chromosome breakage. (A) Comparison of the number of aberrations per cell after treatment with the indicated amount of DEB or MMC. (B) Metaphase spreads of splenocytes fromFancg+/− and Fancg−/−mice after 72 hours of in vitro growth. Cells were exposed to either no drug (+0), MMC (20 ng/mL), or IR (4 Gy). Arrows indicate radial forms or breaks.
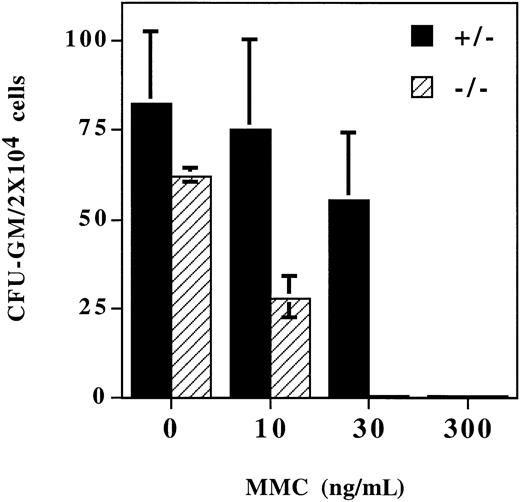
The bone marrow of the Fancg−/− mutant mice was grossly normal compared with the bone marrow of normal littermate controls (data not shown). Also, the peripheral blood count and blood cell morphology of the Fancg−/− mutant mice were grossly normal. There were no significant differences for the hematocrits of Fancg+/+ (47.6% ± 0.2%),Fancg+/− (48.5% ± 3.7%), andFancg−/− (52.3% ± 0.2%) mice. However, when bone marrow progenitor cells were grown in methylcellulose culture in the presence of MMC, the Fancg−/−progenitor cells had a decreased survival rate compared with normal littermate (+/−) control marrow (Figure3). For a MMC concentration of 30 ng/mL, no Fancg−/− progenitor colonies were observed.
Bone marrow progenitor cells from
Fancg−/− mice have increased sensitivity to MMC. Mononuclear cells were isolated from bone marrow fromFancg+/− mice orFancg−/− mice. Cells were plated in methylcellulose in the presence of the indicated concentration of MMC. Colonies were enumerated at day 7.
Bone marrow progenitor cells from
Fancg−/− mice have increased sensitivity to MMC. Mononuclear cells were isolated from bone marrow fromFancg+/− mice orFancg−/− mice. Cells were plated in methylcellulose in the presence of the indicated concentration of MMC. Colonies were enumerated at day 7.
Cellular sensitivity of Fancg−/−cells to ionizing radiation
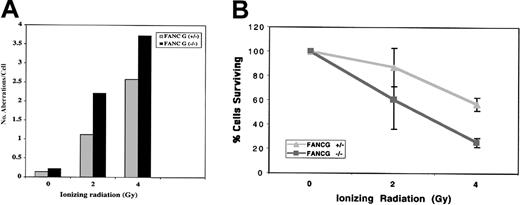
Recent studies suggest that FA patients and FA cells are also sensitive to IR,5,30,33 although the relative IR sensitivity of FA cells from different FA subtypes is unknown. The human FANCG/XRCC9 complementary DNA was originally cloned by its ability to functionally complement the MMC- and IR-sensitive Chinese hamster cell line, UV-40, suggesting that the FANCG gene product may function directly in the normal cellular response to MMC or IR damage.34 We therefore tested the sensitivity of theFancg−/− primary splenocytes to IR (Figure4). Compared withFancg+/− splenocytes,Fancg−/− splenocytes had a slight but reproducible increase in IR sensitivity based on their IR dose-dependent chromosome breakage (Figure 4A).
Differential IR sensitivity of
Fancg+/− andFancg−/− primary splenic lymphocytes. (A) Analysis of chromosome breakage following exposure to variable amounts of IR. Results shown are representative of 3 separate experiments. (B) Analysis of cell viability by the trypan blue assay following IR exposure. Based on this assay, theFancg+/− and Fancg−/−cells had approximately equal viability (95%-98% viable) in the absence of IR exposure. Data shown are representative of 3 separate experiments.
Differential IR sensitivity of
Fancg+/− andFancg−/− primary splenic lymphocytes. (A) Analysis of chromosome breakage following exposure to variable amounts of IR. Results shown are representative of 3 separate experiments. (B) Analysis of cell viability by the trypan blue assay following IR exposure. Based on this assay, theFancg+/− and Fancg−/−cells had approximately equal viability (95%-98% viable) in the absence of IR exposure. Data shown are representative of 3 separate experiments.
To further demonstrate the differential IR sensitivity ofFancg+/− and Fancg−/−splenocytes, we exposed cells to variable amounts of IR and measured cell viability by the trypan blue exclusion assay (Figure 4B). By this assay, Fancg−/− cells had decreased survival following IR, confirming their enhanced IR sensitivity.
Disruption of the FA pathway inFancg−/−cells
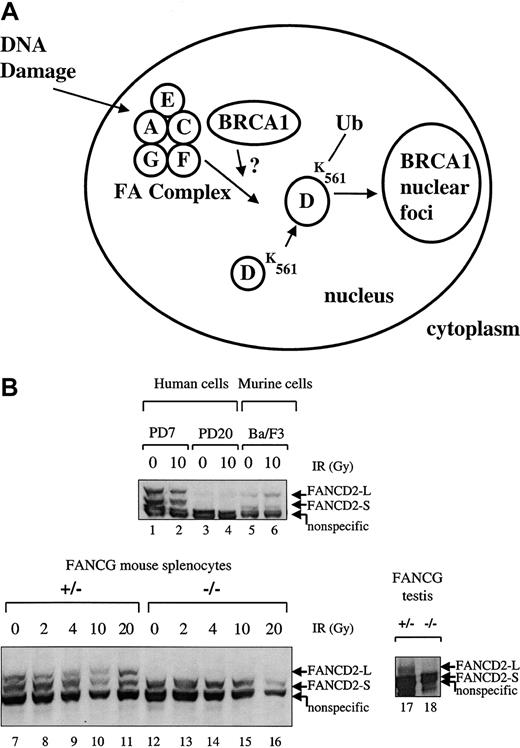
Recent studies have demonstrated that the FA proteins cooperate in a common biochemical pathway leading to the generation of FANCD2/BRCA1 foci.15 Five of the cloned human FA proteins (FANCA, FANCC, FANCE, FANCF, FANCG) interact in a multisubunit protein complex,17,20,21 and this complex is required for the DNA damage–inducible monoubiquitination of the downstream FANCD2 protein.15 We therefore tested theFancg−/− splenocytes for the integrity of the FA pathway (Figure 5).
Primary splenic lymphocytes from FA-G mice are defective in IR-inducible Fancd2 monoubiquitination.
(A) Schematic representation of the human FA pathway. The FA protein complex, containing the FANCA, FANCC, FANCF, FANCG, and perhaps FANCE proteins, is required for the monoubiquitination of the FANCD2-S protein (155 kd) to the FANCD2-L protein (162 kd). Monoubiquitination is increased in response to DNA damage by IR. Ubiquitinated (Ub) FANCD2 is targeted to foci containing the BRCA1 protein and other proteins involved in DNA repair. (B) Primary splenic lymphocytes (splenocytes) were isolated from mice with the indicatedFancg genotype. Cells were untreated or exposed to IR (2, 4, 10, or 20 Gy), as indicated. Cell lysates were prepared, and total cellular proteins were electrophoresed, transferred to nitrocellulose, and immunoblotted with an anti-Fancd2 antiserum. This polyclonal antiserum cross-reacts with the human and murine Fancd2protein and also recognizes a nonspecific 150 kd protein in both human and murine lysates, as indicated. Alternatively, whole tissue extracts were prepared from testes (lanes 17,18) from aFancg+/− mouse or aFancg−/− mouse. Cells examined were PD7 (wild-type human lymphoblasts), PD20 (human FA-D2 lymphoblasts), Ba/F3 (murine interleukin-3–dependent lymphocytes), or murine splenocytes (lanes 7-16). Both FANCD2-S and FANCD2-L isoforms are absent from the PD-20 (FA-D2) lymphoblasts, as previously described.
Primary splenic lymphocytes from FA-G mice are defective in IR-inducible Fancd2 monoubiquitination.
(A) Schematic representation of the human FA pathway. The FA protein complex, containing the FANCA, FANCC, FANCF, FANCG, and perhaps FANCE proteins, is required for the monoubiquitination of the FANCD2-S protein (155 kd) to the FANCD2-L protein (162 kd). Monoubiquitination is increased in response to DNA damage by IR. Ubiquitinated (Ub) FANCD2 is targeted to foci containing the BRCA1 protein and other proteins involved in DNA repair. (B) Primary splenic lymphocytes (splenocytes) were isolated from mice with the indicatedFancg genotype. Cells were untreated or exposed to IR (2, 4, 10, or 20 Gy), as indicated. Cell lysates were prepared, and total cellular proteins were electrophoresed, transferred to nitrocellulose, and immunoblotted with an anti-Fancd2 antiserum. This polyclonal antiserum cross-reacts with the human and murine Fancd2protein and also recognizes a nonspecific 150 kd protein in both human and murine lysates, as indicated. Alternatively, whole tissue extracts were prepared from testes (lanes 17,18) from aFancg+/− mouse or aFancg−/− mouse. Cells examined were PD7 (wild-type human lymphoblasts), PD20 (human FA-D2 lymphoblasts), Ba/F3 (murine interleukin-3–dependent lymphocytes), or murine splenocytes (lanes 7-16). Both FANCD2-S and FANCD2-L isoforms are absent from the PD-20 (FA-D2) lymphoblasts, as previously described.
As previously described,15 the FA protein complex (Fanconi A/C/E/F/G protein complex) is required for the monoubiquitination of the FANCD2 protein (Figure 5A). MurineFancg+/− splenocytes express both forms of theFancd2 protein (Fancd2-S and Fancd2-L) (Figure 5B, lanes 7-11). In contrast, Fancg−/−splenocytes failed to express the Fancd2-L isoform either in the absence or the presence of IR (lanes 12-16). The Fancg−/− splenocytes did express the Fancd2-S isoform (lanes 12-16). Taken together, these results demonstrate that the Fancg−/− splenocytes have a disruption of the FA pathway.
Fancg−/−mice have germ cell defects and decreased fertility
Several attempts to inbreed the homozygousFancg−/− mice have not yielded any offspring over a period of 6 months. Cross-breeding ofFancg+/− and Fancg−/−mice has resulted in a decreased frequency of pregnancies and reduced litter sizes, as compared with Fancg+/−inbreeds, suggesting that both male and femaleFancg−/− homozygote mutant mice have impaired fertility (data not shown).
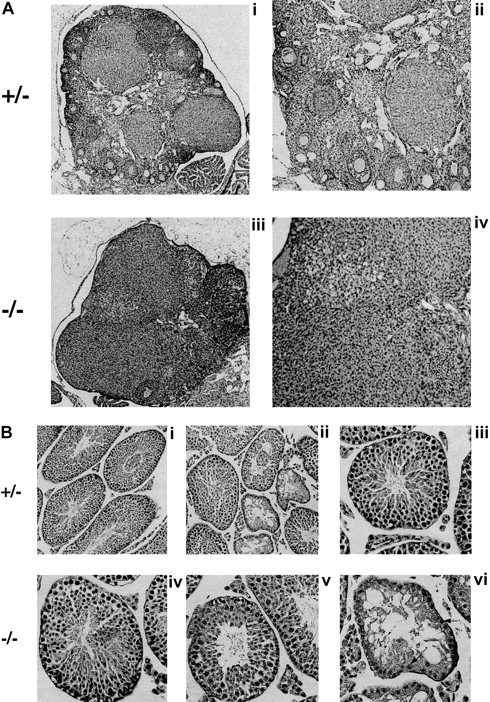
To investigate the cause of the infertility, we performed measurements and histologic evaluation of the reproductive organs of 6- to 8-week-old mice (Figure 6). The ovaries of Fancg−/− mutant mice were abnormal compared with those of littermate controls. Mutant ovaries were almost completely devoid of follicles (Figure 6A). The testes ofFancg−/− mutant mice were also abnormal. The testicular weight of Fancg−/− homozygote males 6 weeks and older was markedly reduced (218 ± 47 mg per testis) compared with littermate Fancg+/− heterozygote controls (575 ± 81 mg per testis, P < .01). Similar to the testicular histology observed for theFancc−/− animal, theFancg−/− testis had a mosaic pattern of the seminiferous tubules, with the appearance of both normal and abnormal tubules. The abnormal tubules were devoid of spermatocytes and contained only Sertoli cells (Figure 6B).
Fancg −/− mice have germ cell defects and decreased fertility.
(A) Histology of mutant and control ovaries (hematoxylin and eosin staining). Ovary of an 8-week-old mutant or control mouse. The mutant ovary was almost completely devoid of follicles (iii, × 60; iv, × 125). The control ovary had an abundance of developing follicles at various stages (i, × 60; ii, × 125). (B) Histology of mutant and control testis (hematoxylin and eosin staining). A mosaic pattern of seminiferous tubules devoid of germ cells and normal tubules can be seen in the mutant testis (iv, × 250; v and vi, × 320). A control testis is shown at the same magnification (i, ii, iii). Arrows indicate Sertoli cells.
Fancg −/− mice have germ cell defects and decreased fertility.
(A) Histology of mutant and control ovaries (hematoxylin and eosin staining). Ovary of an 8-week-old mutant or control mouse. The mutant ovary was almost completely devoid of follicles (iii, × 60; iv, × 125). The control ovary had an abundance of developing follicles at various stages (i, × 60; ii, × 125). (B) Histology of mutant and control testis (hematoxylin and eosin staining). A mosaic pattern of seminiferous tubules devoid of germ cells and normal tubules can be seen in the mutant testis (iv, × 250; v and vi, × 320). A control testis is shown at the same magnification (i, ii, iii). Arrows indicate Sertoli cells.
In addition, we tested protein extracts from the testes ofFancg−/− mice and littermate controls for expression of the Fancd2 protein (Figure 5B, lanes 17, 18). Interestingly, the Fancg−/− mutant testis expressed only the short (nonubiquitinated) isoform of theFancd2 protein (lane 18), while theFancg+/− sibling control testis expressed bothFancd2-S and Fancd2-L isoforms (lane 17). Taken together, these data show that an intact FA pathway and the activation of Fancd2 to the monoubiquitinated isoform of Fancd2correlates with the ability of the germ cells to differentiate into mature spermatocytes.
Discussion
In the current study, we used homologous recombination to disrupt the murine Fancg gene and to generate a murine model of FA subtype G. The cellular and gross phenotype of theFancg−/− mice was similar to the phenotype observed for the previously described Fancc−/−mice.22 23 Primary cells from theFancg−/− mice demonstrated spontaneous chromosome breakage that was increased in response to the DNA cross-linking agents, MMC and DEB. While theFancg−/− mice had few gross phenotypic abnormalities, the mice were infertile and had clear histologic abnormalities of the gonads.
While the bone marrow cellularity and peripheral blood erythroid mass (hematocrit) for the Fancg−/− mice were normal, the bone marrow progenitor cells were sensitive to MMC. Decreased survival of Fancg−/− bone marrow progenitor cells was observed in methylcellulose culture. Similar MMC sensitivity has previously been reported for bone marrow fromFancc−/− mice.
Taken together, our data suggest that theFancg−/− mice exhibit minor hematologic abnormalities characteristic of FA patients during the preanemic phase. However, in contrast to human FA patients,Fancg−/− mice do not develop progressive bone marrow failure, at least not during the first year of life. The biological significance of the difference between human and murine FA is unclear. It is possible that additional redundant antiapoptotic signaling pathways exist in the murine system, which partially compensate for the loss of the FA pathway in murine bone marrow cells.
It will be interesting to determine whether in vivo administration of low doses of MMC to the Fancg−/− mice versus sibling control mice will result in bone marrow failure. Also of interest will be whether retroviral gene transfer of the wild-type human FANCG complementary DNA corrects the MMC sensitivity of the bone marrow progenitors, leading to enhanced survival of transduced cells in vivo.
The primary splenic lymphocytes from theFancg−/− mice also had increased sensitivity to IR, based on the chromosome breakage assay and survival studies. Increasing evidence suggests that FA cells are mildly sensitive to IR, although they are more sensitive to DNA cross-linking agents. For example, nonmyeloablative doses of IR have recently been shown to enhance the selection of wild-type hematopoietic cells over FA hematopoietic cells in murine competitive repopulation models.30 The IR sensitivity of primaryFancg−/− cells in our study is consistent with other recent studies describing the FA molecular pathway.15 IR activates the dose-dependent and time course–dependent monoubiquitination of the FANCD2 protein in normal (non-FA) cells. Monoubiquitinated FANCD2 is targeted to BRCA1 nuclear foci, which appear to play a role in the repair of DNA double-strand breaks. Interestingly, the FA protein complex, including the FANCA, FANCC, FANCE, FANCF, and FANCG proteins, is required for the IR-dependent monoubiquiitination of FANCD2. Human FA cells, defective in the genes encoding any one of these FA proteins, fail to form FANCD2/BRCA1 foci in response to IR, perhaps accounting for the IR sensitivity of the Fancg−/− cells observed in the current study.
Other mouse models with enhanced IR sensitivity have defects in the immune system. Specifically, mice with targeted disruptions of the Ku, DNA-PK, or ATM gene35-37 have cellular and humoral immunodeficiency. We therefore analyzed T cells and B cells fromFancg−/− mice versusFancg+/− sibling controls (data not shown). For single-cell suspensions prepared from thymus, spleen, or peripheral lymph nodes, there was no difference in the expression of T-cell surface antigens (CD3, CD4, CD8) or B-cell surface antigens (CD19, immunoglobulin M, B220) between Fancg−/− andFancg+/− mice. Consistent with these studies, human FA patients are not immunodeficient and appear to have normal immunoglobulin diversity.
The Fancg−/− mice are infertile, and the gonads from these animals have striking pathology. Ovaries from theFancg−/− females are small and have decreased cellularity. Testes from the Fancg−/− males are small and have decreased cellularity and spermatogenesis. While the molecular basis of this gonadal histology is unknown, it may result from a defect in the activation of the Fancd2 protein. Recent studies demonstrate that the activated human FANCD2 protein normally binds to meiotic chromosomes in the axial (unsynapsed) regions of synaptonemal complexes.15 FANCD2 binding correlates with the increase in DNA double-strand breaks, resulting from crossing-over events during meiosis. Based on this model,15 a disruption of the FA pathway, caused by a targeted disruption of the Fancg gene, may prevent normalFancd2 activation and may block the normal binding ofFancd2 to meiotic chromosomes in theFancg−/− animals. Absence of Fancd2staining of meiotic chromosomes may lead to accelerated apoptosis of differentiating spermatocytes.
Finally, the similarity between the Fancc−/−mouse23,38 and the Fancg−/− mouse in this study provides further genetic evidence of a common FA molecular pathway.15,39 Previous studies have demonstrated that FANCC and FANCG interact in a nuclear protein complex, along with other FA proteins (FANCA and FANCF). FANCE is probably a subunit of this nuclear complex as well, based on in vitro binding studies. Other proteins may also be part of this nuclear complex, such as the recently cloned FANCC-interacting protein, FAZF.40 The FA protein complex (A/C/E/F/G) is required for DNA damage–inducible monoubiquitination and activation of the downstream FANCD2 protein. Activated FANCD2 interacts with chromatin and may be required for DNA repair. That disruption of the Fancc or Fancggenes yields the same phenotype suggests that the major (or sole) function of the Fancc and Fancg proteins is to cooperate in the FA protein complex. For instance, if theFancc or Fancg proteins had additional functions beyond their role in the FA protein complex, one might expect additional unique phenotypic characteristics for theFancc−/− and Fancg−/−knockout animals. Disruption of the murine Fanca gene also results in a similar phenotype,41 further supporting a unified function of these proteins (A/C/G) in the FA protein complex. Whether any of the FA genes have additional functions outside of the FA pathway will require the generation of mice with double or triple gene disruptions. Also, it will be interesting to determine whether targeted disruption of Fancd2, the putative substrate of the FA protein complex, yields a similar phenotype to the Fancc, Fanca, and Fancg knockout models.
We thank En Li for providing 129/SvJae embryonic stem cells; Ramnik Xavier and Robin Mayfield for their help in histology analysis; and Stephen Conley and Michelle Forestall for preparing the histology slides.
Supported by National Institutes of Health grants RO1-HL52725-04, RO1-DK43889-09, and PO1-HL54785-04 (A.D.D.). Y.Y. is supported by a postdoctoral fellowship from the Cancer Research Institute. B.S. is supported by RO1-AI27849 and UO1-HL66678.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alan D. D'Andrea, Dana-Farber Cancer Institute, Dept of Pediatric Oncology, Harvard Medical School, 44 Binney St, Boston, MA 02115; e-mail: alan_dandrea@dfci.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal