Abstract
Immunoglobulin G (IgG) receptors (FcγRs) on myeloid cells are responsible for the internalization of immune complexes. Activation of the oxidase burst is an important component of the integrated cellular response mediated by Fc receptors. Previous work has demonstrated that, in interferon-γ–primed U937 cells, the high-affinity receptor for IgG, FcγRI, is coupled to a novel intracellular signaling pathway that involves the sequential activation of phospholipase D (PLD), sphingosine kinase, and calcium transients. Here, it is shown that both known PLD isozymes, PLD1 and PLD2, were present in these cells. With the use of antisense oligonucleotides to specifically reduce the expression of either isozyme, PLD1, but not PLD2, was found to be coupled to FcγRI activation and be required to mediate receptor activation of sphingosine kinase and calcium transients. In addition, coupling of FcγRI to activation of the nicotinamide adenine dinucleotide phosphate (reduced form) (NADPH) oxidase burst was inhibited by pretreating cells with 0.3% butan-1-ol, indicating an absolute requirement for PLD. Furthermore, use of antisense oligonucleotides to reduce expression of PLD1 or PLD2 demonstrated that PLD1 is required to couple FcγRI to the activation of NADPH oxidase and trafficking of internalized immune complexes for degradation. These studies demonstrate the critical role of PLD1 in the intracellular signaling cascades initiated by FcγRI and its functional role in coordinating the response to antigen-antibody complexes.
Introduction
Receptors for the constant region (Fc) of immunoglobulins play a pivotal role linking the humoral and cellular arms of the immune system. On leukocytes, aggregation of receptors (FcγRs) for immunoglobulin G (IgG) leads to a number of cellular responses, including the internalization of immune complexes, release of proteases, activation of the respiratory burst, and release of cytokines. Receptor aggregation can ultimately lead to targeted cell killing through antibody-directed cellular cytotoxicity.1,2 These Fc receptors, therefore, play critical roles in host defense mechanisms against invading pathogens, in autoimmune diseases,3 and in cancer surveillance.4 We have recently reported that, in cytokine-primed U937 cells, aggregation of the high-affinity receptor for IgG (FcγRI)5 activates, through nonreceptor tyrosine kinases, a novel signaling pathway that involves the sequential activation of phospholipase D (PLD) and sphingosine kinase.6 This pathway is necessary for efficient intracellular trafficking of FcγRI-internalized immune complexes to lysosomes for degradation and release of calcium from intracellular stores.6 7
Phosphatidylcholine-specific PLD (PC-PLD) catalyzes the hydrolysis of the terminal diester bond of phosphatidylcholine to liberate phosphatidic acid and choline.8 PC-PLD was first identified in plants but has subsequently been shown to be highly conserved across all species and present in large amounts in bacteria, yeast, and mammalian cells.9,10 In mammalian cells, activation of PC-PLD has been proposed to control signal transduction pathways regulating a wide range of physiological processes, including membrane trafficking and cytoskeletal reorganization,11-17 mitogenesis,18,19neuronal and cardiac stimulation,20,21phagocytosis,22 the respiratory burst in neutrophils,23,24 inflammation, and diabetes.25
The immediate products of PLD hydrolysis of phosphatidylcholine are phosphatidic acid and choline.8 A role for phosphatidic acid as a key intracellular signaling molecule has been proposed as it has been shown to directly activate protein kinases,18,19,26,27 protein tyrosine phosphatase,28-30 phospholipase C,31phosphoinositol-4-kinase,32 sphingosine kinase,33 and small molecular weight guanosine triphosphatase–activating proteins.34 Phosphatidic acid has also been shown to promote the release of calcium from intracellular compartments35,36 and, in neutrophils, to activate the oxidative burst through nicotinamide adenine dinucleotide phosphate (reduced form) (NADPH) oxidase.24,27Phosphatidic acid itself can also act as a precursor for other intracellular signaling molecules. Thus, phosphatidic acid can be converted into diacyl glycerol (DAG) by phosphatidic acid–phosphohydrolase10,23 or to the mitogen lyso–phosphatidic acid (LPA) by phospholipase A2.10,23DAG is an established activator of conventional and novel protein kinase C (PKC) isoforms37,38 and LPA, which, following release from cells, acts on G-protein–coupled receptors to further stimulate cells or adjacent cells.39 Phosphatidic acid can also be subject to acid hydrolases followed by lipo-oxygenase, leading to the formation of oxygen radicals and lipid peroxides that cause tissue damage.40
In mammalian cells, 2 isoforms of PLD (PLD1 and PLD2) have been cloned, sequenced, and characterized.17,41,42 Furthermore, PLD1 is expressed as 2 splice variants, namely, PLD1a and PLD1b.43 Both PLD1 and PLD2 use phosphatidylcholine as substrate. In previous studies, we have shown that coupling of FcγRI to PLD activation results in activation of sphingosine kinase6 7 and calcium transients. However, in these previous studies, the nature of the PLD isozyme activated by FcγRI was not defined, and the relationship of the activation of PLD to the various signaling enzyme cascades following FcγRI aggregation was unknown.
Here, we demonstrate that coupling of FcγRI to NADPH oxidase has an absolute requirement on the activation of PLD. Although both isozymes of PLD (PLD1 and PLD2) are present in U937 cells, only PLD1, but not PLD2, functionally couples FcγRI to intracellular effectors, such as the activation of sphingosine kinase and cytosolic calcium transients. PLD1, but not PLD2, is also required for FcγRI-mediated activation of the oxidative burst and trafficking of immune complexes.
Materials and methods
Cell culture
U937 cells were cultured in RPMI 1640 (Gibco, Rockville, MD) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, 10 U/mL penicillin, and 10 μg/mL streptomycin at 37°C in 6.8% carbon dioxide in a water-saturated atmosphere. The cells were treated with interferon (IFN)–γ (200 ng/mL) (Bender Wien, Vienna, Austria) for 16 hours. Antisense oligonucleotides were purchased from Oswell DNA Service (Southampton, United Kingdom); 24-mers were synthesized and capped at either end by the phosphothiorate linkages (first 2 and last 2 linkages); these 24-mers corresponded to the reverse complement of the first 8 amino acids for either PLD1 or PLD2. The sequences of the oligonucleotides were as follows: 5′CCGTGGCTCGTTTTTCAGTGACAT3′ for PLD1 and 5′GAGGCTCTCAGGGGTCGCCGTCAT3′ for PLD2. Cells were incubated in 10 μM oligonucleotide for a total of 36 hours (20 hours prior to, and then for the duration of culture with IFN-γ).
Reverse-transcriptase–polymerase chain reaction
Cells were either primed with IFN-γ or differentiated to a macrophage phenotype with dibutyryl cyclic adenosine monophosphate (dbcAMP)6 and messenger RNA (mRNA)–isolated (Quiagen midi kit for mRNA extraction). Specific forward and reverse primers were designed for either PLD1 or PLD2: PLD1 forward, GTGGGCTCACCATGAGAAGC; PLD1 reverse, GCAATGTCATGCCAGGGCATC; PLD2 forward, CTGCACTTTACTTACAGGACCCTG; and PLD2 reverse, CTGCTCATAGATATTGGCGTTGC.
The PLD1 primers were designed against an overlapping region in the sequence of both PLD1 isoforms to yield a fragment of approximate 640 base pairs (bp) for PLD1a and another fragment of approximate 520 bp for PLD1b.43 Specific primers designed for PLD2 would yield a 450-bp fragment. The reaction was carried out as described previously.43
Receptor aggregation
Cells were harvested by centrifugation and then incubated at 4°C for 45 minutes with 1 μM human monomeric IgG (Serotec, Oxford, United Kingdom) to occupy surface FcγRI in the presence or absence of inhibitors or alcohols. Excess unbound ligand was removed by dilution and centrifugation of the cells. Cells were resuspended in ice-cold RPMI 1640/10 mM Hepes/0.1% bovine serum albumin (BSA) (RHB medium), and surface immune complexes were formed by incubating with cross-linking antibody (sheep antihuman IgG; 1:50) in the continued presence of inhibitors or alcohols. Cells were then warmed to 37°C for the times specified in each assay as described previously.44 45
Immunoprecipitation of PLDs
PLD1 and PLD2 were immunoprecipitated from cell lysates prior to Western blot analysis of the desired proteins. Rabbit polyclonal antibody (2 μg), either anti-PLD1 or anti-PLD2 (QCB, Hopkinton, MA), were incubated with 50 μL 50% protein A-agarose and 450 μL buffer for 2 hours on a rocking platform at 4°C in order to form precipitating complexes. Then, the antibody–protein A–agarose mix was washed to remove unbound antibody. Following this, 500 μL cell lysate containing 200 μg protein was mixed with the precipitating (antibody–protein A–agarose) complex and placed in a tumbler at 4°C for 4 hours. Following incubation, the precipitating complex was centrifuged and washed prior to addition of Laemlli buffer for loading onto sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Gel electrophoresis and Western blots
Proteins were resolved on 8% polyacrylamide gels (SDS-PAGE) under denaturing conditions and then transferred to 0.45-μm nitrocellulose membranes. After blocking overnight at 4°C with 5% nonfat milk in Tris-buffered saline and 0.1% Tween 20 and washing, the membranes were incubated with the relevant antibodies for 4 hours at room temperature. The membranes were washed extensively in the washing buffer and bands visualized by means of the appropriate horseradish peroxidase–conjugated secondary antibody and ECL Western Blotting Detection System (Amersham, Buckinghamshire, United Kingdom).
Measurement of phospholipase D activity
PLD activity was measured as previously described in Melendez et al,7 by means of the transphosphatidylation assay. Briefly, U937 cells were labeled (106 cells/mL) with [3H] palmitic acid (5 μCi/mL [185 kBq/mL]) (Amersham) in the cell culture medium for 16 hours. Following washing, the cells were incubated at 37°C for 15 minutes in RHB medium containing butan-1-ol (0.3% final). Following FcγRI aggregation, cells were incubated for a further 30 minutes and then extracted by Bligh-Dyer phase separation. The accumulated phosphatidylbutanol was assayed as described previously.7
Measurement of rate of trafficking of immune complexes
Trafficking of immune complexes was measured with a protocol similar to that used in previous studies.44,45 FcγRI was aggregated as described above, but surface immune complexes were formed by using radiolabeled cross-linking antibody ([125I]-rabbit antihuman IgG; 1:50) (R&D Systems, Abington, United Kingdom). Supernatant trichloroacetic acid (TCA)–soluble counts were measured to provide the rate of intracellular trafficking.45 46 The results were expressed as a percentage of the total cell surface counts at time zero.
Oxidase assays
Whole cell superoxide production following FcγRI aggregation or N-formyl-1-methionyl-1-leucyl-1-phenylalamine (f-MLP) stimulation was measured in IFN-γ–primed U937 cells, pretreated or not with butan-1-ol, butan-2-ol, or antisense oligonucleotides for PLD1 or PLD2.
Cells were assayed in RPMI–1% FCS without phenol red placed in a 96-well plate. For each well, 200 000 cells suspended in 80 μL were mixed with 20 μL luminol-based substrate (Diogenes, National Diagnostics, Atlanta, GA) at the same time as the cross-linking antibody, or f-MLP (1 μM). Luminescence was measured with a luminometer (Wallac 1420 Multilabel counter, Cambridge, United Kingdom).
Cytosolic calcium assays
Cytosolic calcium was measured as described previously except the cuvette buffer was calcium supplemented (final concentration, 1.5 mM Ca++).7 Briefly, cells were loaded with 1 μg/mL Fura-2–AM (Molecular Probes, Leiden, The Netherlands) and 1 μM human monomeric IgG (Serotec) in phosphate-buffered saline (PBS), 1.5 mM Ca++, and 1% BSA. After removal of excess reagents by dilution and centrifugation, the cells were resuspended in 1.5 mM calcium-supplemented PBS and warmed to 37°C in the cuvette. Cell surface–bound IgG was aggregated by the addition of goat anti–human IgG (1:50 dilution) (Sigma, Poole, United Kingdom). Fluorescence was measured at 340 and 380 nm, and the background-corrected 340:380 ratio was calibrated as previously described.6
Sphingosine kinase assays
Activation of sphingosine kinase was measured as described previously.7 33 Briefly, cells were resuspended in ice-cold 0.1 M phosphate buffer (pH 7.4) containing 20% glycerol, 1 mM mercaptoethanol, 1 mM EDTA, phosphatase inhibitors (20 mM ZnCL2, 1 mM sodium orthovanadate, and 15 mM sodium fluoride), protease inhibitors (10 μg/mL leupeptin, 10 μg/mL aprotinin, and 1 mM phenylmethyl sulfonyl fluoride), and 0.5 mM 4-deoxypyridoxine, disrupted by freeze-thawing and centrifuged at 105 000g for 90 minutes at 4°C. Supernatants were assayed for sphingosine kinase activity by incubating with sphingosine (Sigma) and [γ32P]–adenosine triphosphate (2 μCi, 5 mM [74 kBq]) for 30 minutes at 37°C, and products were separated by thin-layer chromatography on silica gel G60 (Whatman, Maidstone, United Kingdom) by means of chloroform/methanol/acetic acid/water (90:90:15:6) and visualized by autoradiography. The radioactive spots corresponding to sphingosine phosphate were scraped and counted in a scintillation counter. The activity of sphingosine kinase following in vitro activation by phosphatidic acid was measured in cell lysates by addition of L-α-phosphatidic acid (1,2-diacyl-sn-glycero-3-phosphate) (Sigma Aldrich, Paris, France) at 10 mol 1% Triton X-100.
Results
Both PLD1 and PLD2 are expressed in U937 cells, but only PLD1 is coupled to FcγRI activation
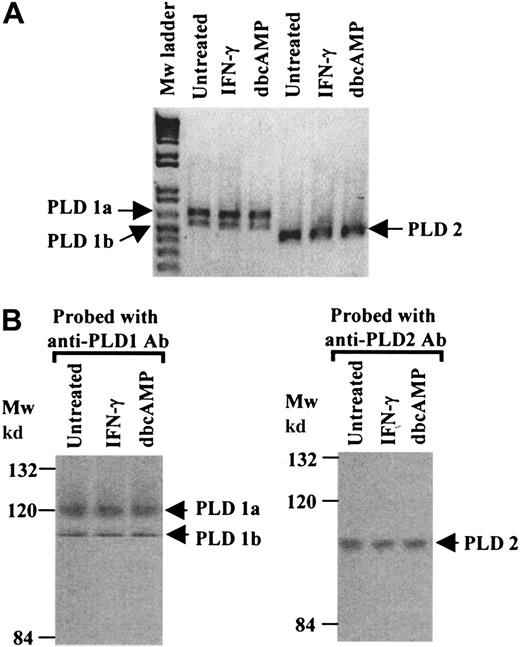
PLD expression profiles in the human monocytic cell line U937.
The isozymes of PLD expressed in U937 cells were determined by means of reverse-transcriptase–polymerase chain reaction (RT-PCR), Northern analysis, and Western analysis. Relative levels of expression were compared in untreated cells, cytokine (IFN-γ)–primed cells, and cells differentiated to a macrophage phenotype by means of dbcAMP.47
RT-PCR analysis of mRNAs extracted from untreated, IFN-γ–primed or dbcAMP-differentiated cells revealed that both known PLD isozymes, PLD1 and PLD2, were present. In addition, both splice variants of PLD1 (PLD1a and PLD1b) were present.43 The profile of the RT-PCR products was not altered by treating cells with IFN-γ or following differentiation (Figure 1A). At the protein level, Western blot analysis of immunoprecipitated PLDs revealed immunoreactive bands corresponding to the predicted molecular weights for PLD1a, PLD1b, and PLD2. The PLD expression profile did not alter following priming of cells with IFN-γ or cell differentiation by dbcAMP (Figure 1B).
PLD expression profiles in U937 cells.
(A) RT-PCR was performed with mRNA extracted from untreated, IFN-γ–primed and dbcAMP-differentiated U937 cells. Specific primers for PLD1 (which yield 2 fragments corresponding to PLD1a, 640bp, and PLD1b, 520bp) and primers specific for PLD2 amplifying a 450-bp fragment, were used. The results shown are typical from 3 separate experiments. (B) Western blot analysis of immunoprecipitates of PLD1 or PLD2, from cell lysates from untreated, IFN-γ–primed and dbcAMP-differentiated U937 cells, were resolved by SDS/PAGE 8% polyacrylamide gels. Proteins were transferred to nitrocellulose and probed with anti-PLD1 or anti-PLD2 antibodies. The results shown are typical from 3 separate experiments. Mw indicates molecular weight.
PLD expression profiles in U937 cells.
(A) RT-PCR was performed with mRNA extracted from untreated, IFN-γ–primed and dbcAMP-differentiated U937 cells. Specific primers for PLD1 (which yield 2 fragments corresponding to PLD1a, 640bp, and PLD1b, 520bp) and primers specific for PLD2 amplifying a 450-bp fragment, were used. The results shown are typical from 3 separate experiments. (B) Western blot analysis of immunoprecipitates of PLD1 or PLD2, from cell lysates from untreated, IFN-γ–primed and dbcAMP-differentiated U937 cells, were resolved by SDS/PAGE 8% polyacrylamide gels. Proteins were transferred to nitrocellulose and probed with anti-PLD1 or anti-PLD2 antibodies. The results shown are typical from 3 separate experiments. Mw indicates molecular weight.
FcγRI aggregation stimulates PLD1.
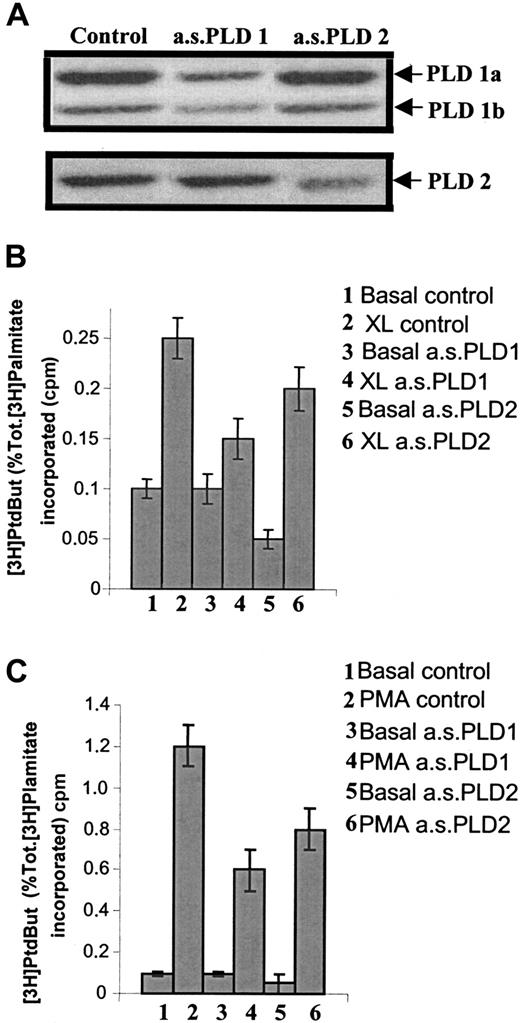
As both isozymes for PLD are expressed in U937 cells, experiments were performed to examine their respective roles, in particular, their activities following FcγRI aggregation. For this purpose, specific antisense oligonucleotides were designed against each of the PLD isozymes to specifically knock down the expression of each enzyme (ie, antisense to PLD1 and antisense to PLD2). We have previously shown that U937 cells are sensitive to antisense manipulation.6 48IFN-γ–primed cells were treated with 1 of the 2 antisense oligonucleotides, and PLD activity was assayed in unstimulated cells to measure basal levels of activity or after stimulation with either FcγRI activation by immune complexes or with phorbolmyristate acetate (PMA) treatment (PMA was used as control). The specificity of the antisense oligonucleotides on relative PLD isozyme expression was checked by Western analysis (Figure2A). Thus, in cells treated with antisense to PLD1, there was a reduction in PLD1 immunoreactivity whereas PLD2 immunoreactivity was unaffected. Conversely, in cells treated with antisense to PLD2, there was a reduction in PLD2 immunoreactivity whereas PLD1 immunoreactivity remained unchanged. Each antisense oligonucleotide, therefore, acted as an internal control for the other.
Use of antisense oligonucleotides to reduce specific expression of either PLD1 or PLD2 demonstrates that only PLD1 is coupled to FcγRI aggregation.
(A) Western blot analysis of immunoprecipitates of either PLD1 or PLD2 to assess expression of either isozyme in IFN-γ–primed U937 cells following treatment for 36 hours with antisense oligonucleotides (10 μM) specific for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2), and control cells (control). The results shown are typical from 3 separate experiments. (B) PLD activity following FcγRI aggregation in IFN-γ–primed U937 cells pretreated with 10 μM antisense oligonucleotides for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2). 1. Basal level (basal control); 2. FcγRI aggregation (XL control); 3. basal level in cells pretreated with antisense PLD1 (basal a.s.PLD1); 4. FcγRI aggregation in cells pretreated with antisense PLD1 (XL a.s.PLD1); 5. basal level in cells pretreated with antisense PLD2 (basal a.s.PLD2); 6. FcγRI aggregation in cells pretreated with antisense PLD2 (XL a.s.PLD2). Results are the mean ± SD for triplicate measurements and are representative of the results from 3 separate experiments. PtdBut indicates phosphatidylbutanol. (C) PLD activity following PMA stimulation (1 μM) in IFN-γ–primed U937 cells pretreated with antisense oligonucleotides (10 μM) for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2). 1. Basal level (basal control); 2. PMA stimulation (PMA control); 3. basal level in cells pretreated with antisense PLD1 (basal a.s.PLD1); 4. PMA stimulation in cells pretreated with antisense PLD1 (PMA a.s.PLD1); 5. basal level in cells pretreated with antisense PLD2 (basal a.s.PLD2); 6. PMA stimulation in cells pretreated with antisense PLD2 (PMA a.s.PLD2). Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments. Tot. indicates total.
Use of antisense oligonucleotides to reduce specific expression of either PLD1 or PLD2 demonstrates that only PLD1 is coupled to FcγRI aggregation.
(A) Western blot analysis of immunoprecipitates of either PLD1 or PLD2 to assess expression of either isozyme in IFN-γ–primed U937 cells following treatment for 36 hours with antisense oligonucleotides (10 μM) specific for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2), and control cells (control). The results shown are typical from 3 separate experiments. (B) PLD activity following FcγRI aggregation in IFN-γ–primed U937 cells pretreated with 10 μM antisense oligonucleotides for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2). 1. Basal level (basal control); 2. FcγRI aggregation (XL control); 3. basal level in cells pretreated with antisense PLD1 (basal a.s.PLD1); 4. FcγRI aggregation in cells pretreated with antisense PLD1 (XL a.s.PLD1); 5. basal level in cells pretreated with antisense PLD2 (basal a.s.PLD2); 6. FcγRI aggregation in cells pretreated with antisense PLD2 (XL a.s.PLD2). Results are the mean ± SD for triplicate measurements and are representative of the results from 3 separate experiments. PtdBut indicates phosphatidylbutanol. (C) PLD activity following PMA stimulation (1 μM) in IFN-γ–primed U937 cells pretreated with antisense oligonucleotides (10 μM) for either PLD1 (a.s.PLD1) or PLD2 (a.s.PLD2). 1. Basal level (basal control); 2. PMA stimulation (PMA control); 3. basal level in cells pretreated with antisense PLD1 (basal a.s.PLD1); 4. PMA stimulation in cells pretreated with antisense PLD1 (PMA a.s.PLD1); 5. basal level in cells pretreated with antisense PLD2 (basal a.s.PLD2); 6. PMA stimulation in cells pretreated with antisense PLD2 (PMA a.s.PLD2). Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments. Tot. indicates total.
Treatment of cells with the antisense oligonucleotide to PLD1 resulted in no change in basal activity. However, following aggregation of FcγRI, the increase in PLD activity was significantly reduced compared with the control cells (P < .01) (Figure 2B). The reduction in the increase after FcγRI activation was 77% ± 8% in cells treated with antisense PLD1 compared with control cells and was proportional to the observed reduction in protein expression by Western analysis. In contrast, treatment of cells with the antisense oligonucleotide to PLD2 significantly reduced basal PLD activity (P < .01). FcγRI-mediated activation of PLD was marginally reduced in cells treated with the antisense to PLD2, but this reduction was entirely accounted for by the reduction in basal levels; the increment over the basal level was identical in control (untreated) cells and those pretreated with PLD2 antisense oligonucleotide (Figure 2B). In contrast to PLD activation by FcγRI, PLD activity stimulated by PMA was significantly reduced in cells pretreated with either of the 2 antisense oligonucleotides, indicating that PMA is able to stimulate both forms of PLD (Figure 2C). Thus, PMA-stimulated PLD activity was reduced by 50% ± 5% in cells pretreated with PLD1 antisense and by 33% ± 5% in cells pretreated with PLD2 antisense. A combination of treatment with both antisense oligonucleotides (PLD1 and PLD2) proved toxic to the cells.
These data demonstrate that PLD2 contributes to the basal, unstimulated PLD activity in these cells, whereas PLD1, but not PLD2, is coupled to FcγRI activation.
PLD1 but not PLD2 is required to couple FcγRI to intracellular signaling cascades
As the coupling of FcγRI to sphingosine kinase6 and cytosolic calcium transients6 7requires PLD activation, the nature of the isozyme involved in this coupling was investigated by means of antisense oligonucleotides to specifically downregulate either PLD1 or PLD2.
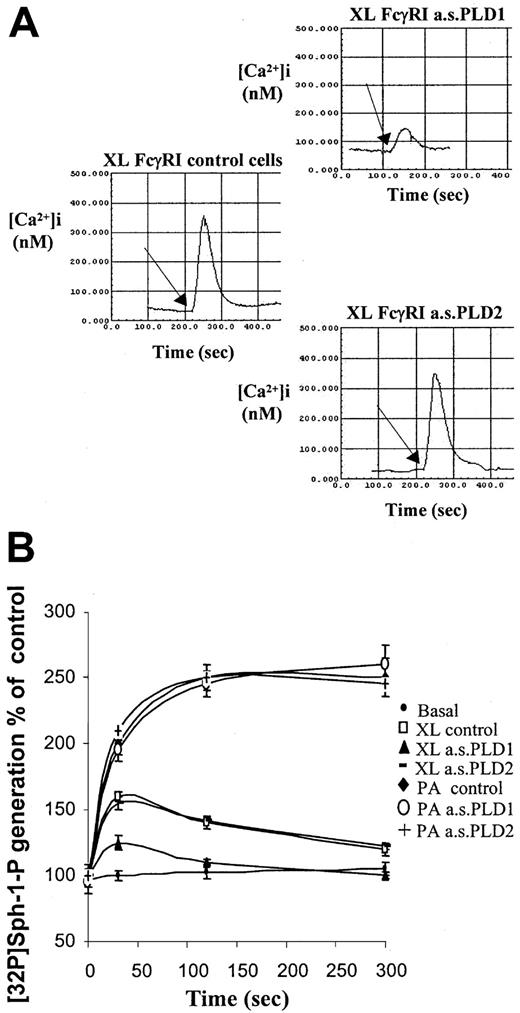
Previously, we have shown that aggregation of FcγRI in cytokine-primed cells results in PLD-dependent release of calcium from intracellular stores and subsequent cytosolic calcium transients.7 Here, the specific dependence for this response on PLD1 and not PLD2 is shown. Reduction in expression of PLD1 by pretreatment of cells with antisense PLD1 oligonucleotide resulted in an attenuation of peak cytosolic calcium spike observed after aggregation of FcγRI (Figure 3A). Reducing expression of PLD2 had no effect on the calcium transients compared with controls.
Coupling of FcγRI to downstream intracellular signaling pathways requires PLD1 and not PLD2.
(A) Intracellular cytosolic calcium changes following aggregation of FcγRI. Responses were compared in control cells and cells pretreated with antisense oligonucleotides (10 μM) to either PLD1 or PLD2. Traces shown are as follows: left, XL FcγRI control cells = FcγRI aggregation in IFN-γ–primed control cells; upper right panel, FcγRI aggregation in IFN-γ–primed cells pretreated with antisense to PLD1 (XL FcγRI a.s.PLD1); lower right panel, FcγRI aggregation in IFN-γ–primed cells pretreated with antisense to PLD2 (XL FcγRI a.s.PLD2). The arrow marks the addition of the goat antihuman IgG antibody to create cell surface immune complexes. Traces are typical from fura-2–loaded cells from 3 separate experiments. (B) FcγRI coupling to sphingosine kinase. Following aggregation of FcγRI in IFN-γ–primed U937 cells, cells were harvested at given time points to measure sphingosine kinase activity. Sphingosine kinase activity was assayed from basal control cells (basal control); following FcγRI aggregation in control cells (XL control) and in cells pretreated with antisense oligonucleotides (10 μM) for either PLD1 (XL a.s.PLD1) or PLD2 (XL a.s.PLD2). Lysates from these cells were treated with phosphatidic acid (L-α-phosphatidic acid (1,2-diacyl-sn-glycero-3-phosphate) in vitro to ensure sphingosine kinase activity (P.A.a.s.PLD1 and P.A.a.s.PLD2).33 Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments.
Coupling of FcγRI to downstream intracellular signaling pathways requires PLD1 and not PLD2.
(A) Intracellular cytosolic calcium changes following aggregation of FcγRI. Responses were compared in control cells and cells pretreated with antisense oligonucleotides (10 μM) to either PLD1 or PLD2. Traces shown are as follows: left, XL FcγRI control cells = FcγRI aggregation in IFN-γ–primed control cells; upper right panel, FcγRI aggregation in IFN-γ–primed cells pretreated with antisense to PLD1 (XL FcγRI a.s.PLD1); lower right panel, FcγRI aggregation in IFN-γ–primed cells pretreated with antisense to PLD2 (XL FcγRI a.s.PLD2). The arrow marks the addition of the goat antihuman IgG antibody to create cell surface immune complexes. Traces are typical from fura-2–loaded cells from 3 separate experiments. (B) FcγRI coupling to sphingosine kinase. Following aggregation of FcγRI in IFN-γ–primed U937 cells, cells were harvested at given time points to measure sphingosine kinase activity. Sphingosine kinase activity was assayed from basal control cells (basal control); following FcγRI aggregation in control cells (XL control) and in cells pretreated with antisense oligonucleotides (10 μM) for either PLD1 (XL a.s.PLD1) or PLD2 (XL a.s.PLD2). Lysates from these cells were treated with phosphatidic acid (L-α-phosphatidic acid (1,2-diacyl-sn-glycero-3-phosphate) in vitro to ensure sphingosine kinase activity (P.A.a.s.PLD1 and P.A.a.s.PLD2).33 Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments.
Previous studies have shown that sphingosine kinase is activated by FcγRI aggregation in cytokine-primed U937 cells and that PLD activation is necessary for coupling the receptor to this kinase.7 Pretreating cells with the antisense oligonucleotide to PLD1 to knock down isozyme expression significantly reduced the peak activation of sphingosine kinase following aggregation of FcγRI in IFN-γ–primed cells by 70% ± 6% (P < .01). The reduction in the peak activation was proportional to the loss of PLD enzyme expressed in these cells as assessed by Western analysis. Reduction in expression of PLD2 had no effect on the ability of FcγRI to couple to sphingosine kinase activation.
To ensure that the loss of sphingosine kinase activity after FcγRI activation in cells treated with the antisense oligonucleotide to PLD1 was a feature of the loss of coupling of the receptor and not some direct effect of the PLD1 antisense oligonucleotide on sphingosine kinase, enzyme activity was measured in lysates following activation of sphingosine kinase with exogenous phosphatidic acid (L-α-phosphatidic acid (1,2-diacyl-sn-glycero-3-phosphate). Addition of phosphatidic acid to the cell lysates from control cells or cells treated with either antisense PLD1 or antisense PLD2 resulted in an identical increase in sphingosine kinase activity (Figure 3B). These data indicate that the reduction in sphingosine kinase activity following FcγRI activation resulting from PLD1 antisense reflects blockage of this pathway and uncoupling of FcγRI to sphingosine kinase activation (Figure 3B).
Thus, in keeping with the observed coupling of FcγRI to PLD1 but not PLD2, receptor-mediated activation of sphingosine kinase and cytosolic calcium transients were found to be attenuated in cells following the specific downregulation of PLD1.
PLD1 but not PLD2 functionally couples FcγRI to intracellular effectors
FcγRI is functionally coupled to NADPH oxidase through PLD activation.
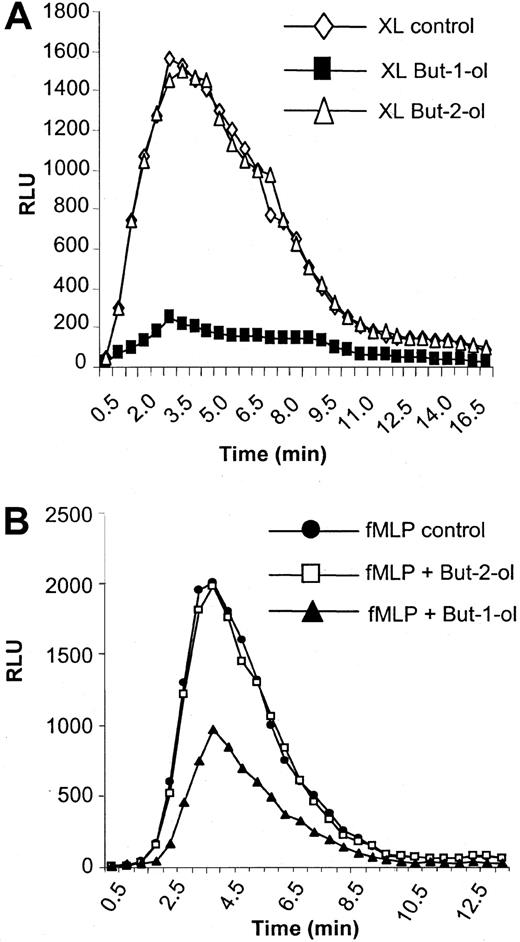
Fc receptors are coupled to the oxidative burst, and activation of NADPH oxidase is an important functional consequence of aggregation of these receptors by opsonized particles or immune complexes to assist in destruction of pathogens.49,50 Here, we show an absolute requirement for PLD in the coupling of FcγRI aggregation to the activation of NADPH oxidase in IFN-γ–primed U937 cells. Thus, formation of surface immune complexes and warming to 37°C result in a transient activation of NADPH oxidase as measured by the oxidative burst (Figure 4A). Pretreatment of cells with 0.3% butan-1-ol completely abolished this response whereas pretreatment with 0.3% butan-2-ol had no effect and cells demonstrated a normal response (Figure 4A). Butan-1-ol but not butan-2-ol can act as an acceptor for the phosphatidyl moiety, thereby generating phosphatidylbutanol instead of phosphatidic acid.7 As butan-2-ol cannot act as an acceptor, it serves as a control for nonspecific effects of the alcohol. This absolute dependence on PLD activation was not observed for other receptors known to be coupled to NADPH oxidase in these cells. Thus, f-MLP also initiates an oxidase burst in these cells. However, in contrast to FcγRI, pretreatment of cells with 0.3% butan-1-ol decreased the oxidase burst activated by f-MLP by only about 50% (Figure 4B). Again, 0.3% butan-2-ol was without effect on this response.
NADPH oxidase activity stimulated by FcγRI aggregation.
NADPH oxidase activity stimulated by FcγRI aggregation has an absolute dependence on PLD. (A) FcγRI-mediated activation of the oxidase burst in control cells (XL control) or cells pretreated for 20 minutes with either 0.3% butan-1-ol (XL but-1-ol) or 0.3% butan-2-ol (XL but-2-ol). The results shown are typical from 3 separate experiments. RLU = relative luminescence units. (B) Activation of oxidase by 1 μM f-MLP stimulation (fMLP control) in control cells and in cells pretreated for 20 minutes with either 0.3% butan-1-ol (fMLP but-1-ol) or 0.3% butan-2-ol (fMLP but-2-ol). The results shown are typical of 3 separate experiments. RLU = relative luminescence units.
NADPH oxidase activity stimulated by FcγRI aggregation.
NADPH oxidase activity stimulated by FcγRI aggregation has an absolute dependence on PLD. (A) FcγRI-mediated activation of the oxidase burst in control cells (XL control) or cells pretreated for 20 minutes with either 0.3% butan-1-ol (XL but-1-ol) or 0.3% butan-2-ol (XL but-2-ol). The results shown are typical from 3 separate experiments. RLU = relative luminescence units. (B) Activation of oxidase by 1 μM f-MLP stimulation (fMLP control) in control cells and in cells pretreated for 20 minutes with either 0.3% butan-1-ol (fMLP but-1-ol) or 0.3% butan-2-ol (fMLP but-2-ol). The results shown are typical of 3 separate experiments. RLU = relative luminescence units.
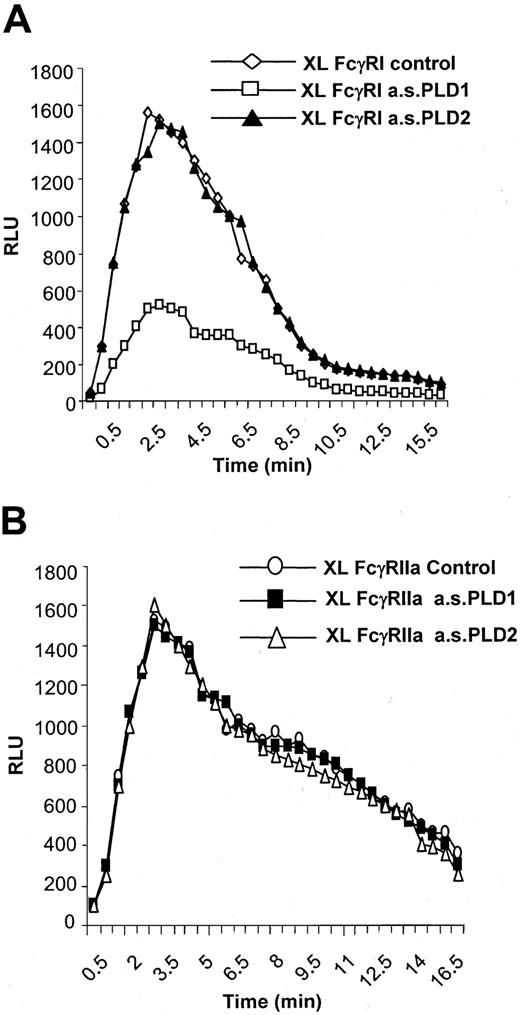
PLD1 and not PLD2 couples FcγRI to the activation of NADPH oxidase.
A role for PLD1 but not PLD2 in mediating the activation of NADPH oxidase by FcγRI was demonstrated. Treatment of IFN-γ–primed U937 cells with the antisense oligonucleotide to PLD1 to specifically knock down expression of this isozyme resulted in an attenuation of the activation of NADPH oxidase by FcγRI aggregation (peak activity, approximately 30% of control) (Figure5A). Similar treatment of cells with the antisense oligonucleotide to PLD2 did not alter FcγRI activation of NADPH oxidase compared with control cells despite decreasing PLD2 expression (Figure 5A). Treatment of cells with either antisense oligonucleotide (PLD1 or PLD2) did not alter the response of NADPH oxidase to activation by the low-affinity IgG receptor FcγRIIa, which has previously been shown to be coupled to phospholipase C and is independent of PLD activation (Figure 5B).6
FcγRI-mediated activation of NADPH oxidase.
FcγRI-mediated activation of NADPH oxidase is dependent on PLD1 and not PLD2. (A) Superoxide production in response to FcγRI in control cells (XL control) compared with cells pretreated with antisense oligonucleotide (10 μM) to either PLD1 (XL a.s.PLD1) or PLD2 (a.s.PLD2). The trace results shown are typical from 3 separate experiments. (B) Superoxide production in response to FcγRIIa in control cells (XL FcγRIIa control) compared with cells pretreated with antisense (10 μM) to PLD1 (FcγRIIa a.s.PLD1) or PLD2 (FcγRIIa a.s.PL2). FcγRIIa was specifically aggregated by means of an anti-FcγRII–specific monoclonal antibody.7 The trace results shown are typical from 3 separate experiments.
FcγRI-mediated activation of NADPH oxidase.
FcγRI-mediated activation of NADPH oxidase is dependent on PLD1 and not PLD2. (A) Superoxide production in response to FcγRI in control cells (XL control) compared with cells pretreated with antisense oligonucleotide (10 μM) to either PLD1 (XL a.s.PLD1) or PLD2 (a.s.PLD2). The trace results shown are typical from 3 separate experiments. (B) Superoxide production in response to FcγRIIa in control cells (XL FcγRIIa control) compared with cells pretreated with antisense (10 μM) to PLD1 (FcγRIIa a.s.PLD1) or PLD2 (FcγRIIa a.s.PL2). FcγRIIa was specifically aggregated by means of an anti-FcγRII–specific monoclonal antibody.7 The trace results shown are typical from 3 separate experiments.
PLD1 is necessary for trafficking of immune complexes for degradation.
Formation of surface immune complexes on myeloid cells results in their rapid internalization44 and trafficking to lysosomes for degradation.45 We have previously shown that endocytosis (the initial internalization of immune complexes to early endosomes) mediated by FcγRI is independent of PLD activation but that subsequent intracellular trafficking of immune complexes is significantly delayed in cells treated with 0.3% butan-1-ol.7 Here, using antisense oligonucleotides to downregulate either PLD1 or PLD2, we demonstrate that FcγRI is functionally coupled to PLD1 and not PLD2 to mediate the intracellular trafficking of immune complexes.
Trafficking of immune complexes to lysosomes for degradation can be readily monitored by means of radiolabeled immune complexes and the appearance of TCA-soluble counts in the cells over time.46Following FcγRI aggregation in cytokine-primed U937 cells, almost 50% of the initial radiolabel internalized as immune complexes appears as TCA-soluble counts in the supernatant of these cells after 120 minutes' incubation at 37°C.7 47
Consistent with previous findings that PLD activation is not necessary for the initial endocytosis of immune complexes mediated by FcγRI,44 downregulation of either PLD1 or PLD2 by pretreating cells with either antisense oligonucleotide did not alter the rate of initial endocytosis of radiolabeled immune complexes (data not shown). However, pretreatment of cells with the antisense PLD1 oligonucleotide significantly changed the rate of appearance of TCA-soluble counts in the supernatant of the cells whereas pretreatment with antisense PLD2 did not. In control and antisense PLD2-treated cells, 50% ± 2% and 49 ± 2% of the initial internalized counts appeared in the supernatant in the TCA-soluble fraction after 120 minutes' incubation whereas for cells treated with antisense PLD1, only 24% ± 1% of counts appeared in this fraction (Figure6). These data demonstrate that PLD1 but not PLD2 activation is required to mediate the trafficking of FcγRI-internalized immune complexes in U937 cells.
Effect of PLD1 on coupling of FcγRI to trafficking of immune complexes.
PLD1 functionally couples FcγRI to trafficking of immune complexes. Trafficking of radiolabeled immune complexes is monitored by the appearance of TCA-soluble counts in cell supernatants. Following aggregation of FcγRI, radiolabeled surface–bound counts are rapidly internalized. During 120 minutes incubation, appearance of radiolabel in the supernatant as TCA-soluble counts (XL control) is compared between control cells and cells pretreated with antisense oligonucleotides (10 μM) to either PLD1 (XL a.s.PLD1) or PLD2 (XL a.s.PLD2). Results shown for each time point are the counts in the incubation supernatant soluble in TCA expressed as a percentage of the total counts bound at time zero. Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments.
Effect of PLD1 on coupling of FcγRI to trafficking of immune complexes.
PLD1 functionally couples FcγRI to trafficking of immune complexes. Trafficking of radiolabeled immune complexes is monitored by the appearance of TCA-soluble counts in cell supernatants. Following aggregation of FcγRI, radiolabeled surface–bound counts are rapidly internalized. During 120 minutes incubation, appearance of radiolabel in the supernatant as TCA-soluble counts (XL control) is compared between control cells and cells pretreated with antisense oligonucleotides (10 μM) to either PLD1 (XL a.s.PLD1) or PLD2 (XL a.s.PLD2). Results shown for each time point are the counts in the incubation supernatant soluble in TCA expressed as a percentage of the total counts bound at time zero. Results are the mean ± SD for triplicate measurements and are representative of the results from at least 3 separate experiments.
Discussion
In this study, we have shown that FcγRI is functionally coupled to PLD1 but not PLD2 in IFN-γ–primed U937 cells even though both enzymes are expressed in these cells. Further, we show that PLD1 but not PLD2 is required for FcγRI-mediated activation of the NADPH oxidase burst and intracellular trafficking of immune complexes for degradation.
Two forms of PLD have been characterized in mammalian cells, and PLD1 exists as a number of splice variants although the significance of these is not known. Here we show that both isozymes are expressed in U937 cells and that priming with IFN-γ or differentiation with dbcAMP did not significantly alter the expression levels of the 2 enzymes. Specific roles for the 2 enzymes, PLD1 and PLD2, are unclear. Cell lines appear to differ in their expression of the different isozymes.51 Thus, a number of cell lines have been reported to express both isozymes although some cells appear to express one isozyme or the other exclusively. Thus, Jurkat cells only express PLD2 whereas 2 human monoblastic cell types, THP1 and HL60, express predominantly PLD1 in resting state; this contrasts with our report here for U937, cells which are also monoblastic in nature. The expression of PLD1 and PLD2 in U937 cells appeared to be relatively stable. Neither mRNA nor protein levels were altered by priming with IFN-γ or differentiation with dbcAMP. This contrasts with observations made for HL60 cells, where differentiation to a neutrophil phenotype with dbcAMP over 3 days resulted in the upregulation of PLD1 and a 20-fold induction of PLD2.52 In these cells, despite the large increase in PLD2 expression, the coupling of f-MLP to the oxidase burst correlated with PLD1 not PLD2 in these differentiated cells.53
In vitro studies have shown that the activity of these 2 enzymes is regulated differently. Both PLD1 and PLD2 have an absolute requirement for phosphatidylinositol(4,5)P2 (PIP2) for activation.41,42 Activity of PLD1 has been shown to be stimulated in vitro by 3 additional factors; adenosine diphosphate–ribosylation factor (ARF), Rho, and PKC54through protein-protein interactions. In contrast, PLD2 is insensitive to these 3 factors and, in the absence of other factors apart from PIP2, PLD2 is very active.17 Consistent with these in vitro observations, our data here indicate that, within the intact U937 cell, PLD2 contributes to the basal activity of PLD whereas PLD1 is coupled to cell activation through FcγRI aggregation by immune complexes. Recent data studying PLD1 activation in intact cells suggest that mechanisms similar to those observed in vitro may occur within cells55,56 although there is a growing body of evidence that ARF6, not ARF1, is responsible for coupling receptors to PLD activation.20,57-59 Our recent studies have demonstrated a role for ARF6 and PKCα in coupling FcγRI to the activation of PLD1 through protein-protein interactions.59Although FcγRI is coupled to signaling cascades through the activation of tyrosine kinases, tyrosine phosphorylation of PLD1 is not thought to play a role in regulating the enzyme.9 10
Consistent with the finding that FcγRI specifically activates PLD1 and not PLD2, antisense knock-down experiments demonstrated a specific role for PLD1 and not PLD2 in coupling this receptor to sphingosine kinase activation and calcium transients. The immediate product of PLD is phosphatidic acid, and this has been shown in vitro to directly activate sphingosine kinase.33 Furthermore, here we demonstrate that PLD1 but not PLD2 is required for the functional coupling of FcγRI to cellular effectors, such as NADPH oxidase activation and intracellular vesicular trafficking of immune complexes from endosomes to lysosomes for degradation. High local production of phosphatidic acid has been proposed to alter membrane properties and facilitate membrane fusion and budding events,13,60 which are important in vesicular trafficking within the cell. Recent work has demonstrated that phagocytosis, which similarly depends on membrane fusion events and vesicular trafficking, is dependent on PLD activation.24
The regulation of assembly of the subunits of NADPH oxidase to form active enzyme and the subsequent oxidase burst is complex. Here, we demonstrate a surprising absolute requirement on PLD activity for the coupling of FcγRI to NADPH oxidase activation. Thus, treatment of cells with butan-1-ol completely abolished the oxidase burst in response to FcγRI aggregation by immune complexes. By contrast, the oxidase burst in response to f-MLP was reduced by only about 50%. Receptor-coupled activation of oxidase assembly is regulated through the phosphorylation of the p47phox component, and PKCs are widely recognized as playing a major role in this phosphorylation event.61 Previous work has shown that, in these IFN-γ–primed U937 cells, the PKC isozymes δ, ε, and ζ are activated by FcγRI62 and, surprisingly, all PKC activity lies downstream of PLD activation.59 Thus, PLD may couple FcγRI to NADPH oxidase through the activation of these PKC isozymes. However, there is also growing evidence that phosphatidic acid can itself activate NADPH oxidase through both kinase-dependent and kinase-independent mechanisms. Thus, data have implied that a phosphatidic acid–dependent kinase is able to phosphorylate p47phox and p22phox to regulate NADPH oxidase assembly.63,64 In addition, recent data using a cell-free system revealed that phosphatidic acid and diacyl glycerol were able to activate NADPH oxidase in a kinase-independent manner.65
Our findings that PLD1-mediated generation of phosphatidic acid drives these monocyte biological responses demonstrates the pivotal role of PLD1 in the intracellular signaling cascades initiated by FcγRI and in the functional coupling of this receptor to provide a coordinated response that can ultimately lead to targeted cell killing through antibody-directed cellular cytotoxicity, an important host defense mechanism to combat invading pathogens and important in cancer surveillance.
We thank Drs Andrew Morris and Michael Frohman, Department of Pharmacology, State University of New York, for helpful advice in discussions of material in this manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Janet M. Allen, Inpharmatica, 60 Charlotte Street, London WIT 2NU, England, United Kingdom; e-mail:janet.allen@inpharmatica.co.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal