Abstract
It has recently been shown that the transcription factor Erg, an Ets family member, drives constitutive expression of the intercellular adhesion molecule 2 (ICAM-2) in human umbilical vein endothelial cells (HUVECs) and that its expression is down-regulated by the pleiotropic cytokine tumor necrosis factor α (TNF-α). To identify other Erg target genes and to define its function in the endothelium, a combined approach of antisense oligonucleotides (GeneBloc) and differential gene expression was used. Treatment of HUVECs with Erg-specific GeneBloc for 24, 48, and 72 hours suppressed Erg mRNA and protein levels at all time points. Total RNA extracted from HUVECs treated withErg-specific or control GeneBloc was analyzed for differences in gene expression using high-density, sequence-verified cDNA arrays containing 482 relevant genes. Inhibition ofErg expression resulted in decreased expression ofICAM-2, as predicted. Four more genes decreased in Erg-deficient HUVECs were the extracellular matrix proteinsSPARC and thrombospondin, the adhesive glycoprotein von Willebrand factor, and the small GTPaseRhoA. Each of these molecules has been directly or indirectly linked to angiogenesis because of its role in vascular remodeling, adhesion, or shape change. Therefore, the role of Erg in vascular remodeling was tested in an in vitro model, and the results showed that HUVECs treated with Erg GeneBloc had a decreased ability to form tubulelike structures when grown on Matrigel. These results suggest that Erg may be a mediator of the TNF-α effects on angiogenesis in vivo.
Introduction
The endothelium plays a key role in a large number of diseases, among them cancer and metastasis, rheumatoid arthritis, atherosclerosis, and thrombosis, by regulating several key processes such as leukocyte recruitment, hemostasis, vascular permeability, and angiogenesis. During inflammation, several mediators can activate the endothelium to promote a proinflammatory, prothrombotic state and to regulate the angiogenic response. Angiogenesis (or new vessel formation) is a tightly regulated process that requires the integration of signals triggered by growth factors and adhesion events, namely modulation of cell–cell contact and interaction with the extracellular matrix.1 Angiogenesis is a critical aspect of the development, growth, and repair of new tissues; it occurs physiologically in circumstances such as wound healing and the menstrual cycle. It is also a key component of many diseases driven by tissue proliferation, such as cancer and rheumatoid arthritis.2,3 In such diseases, new vessel formation supports tissue growth and cellular infiltration, and it contributes to the destructive proliferation of the inflammatory tissue. The link between angiogenesis and inflammation has been suggested by various studies that focus on common features such as the cellular infiltrate, proliferation, and overlapping roles of inflammatory mediators and growth factors.4-6
One such mediator is the pleiotropic cytokine tumor necrosis factor α (TNF-α), which is involved in angiogenesis and in inflammation. In vivo, TNF-α was shown to be proangiogenic in rabbit corneal and chick chorioallantoic membrane models.4,7 The TNF-α effect on angiogenesis appears to be dose dependent: TNF-α is a potent inducer at low doses but an inhibitor at high doses.6 In vitro evidence is conflicting. TNF-α was shown to promote endothelial cell migration and vascular tube formation,7 but it was also shown to inhibit endothelial cell proliferation in vitro.8 9 The mechanisms by which TNF-α mediates these effects on cell proliferation and angiogenesis are unclear, though they appear to involve the regulation of gene expression.
The proinflammatory effects of TNF-α are much better understood. TNF-α activates the endothelium and induces up-regulation and down-regulation of gene expression. TNF-α–dependent pathways that lead to the up-regulation of the expression of proinflammatory genes such as intercellular adhesion molecule 1 (ICAM-1) and E-selectin have been extensively studied and shown to involve transcription factors such as NF-κB and AP-1.10,11 Less is known about the pathways that lead to down-regulation of constitutive endothelial gene expression. Recently, we have shown that TNF-α can down-regulate the expression of the adhesion molecule ICAM-2and of the transcription factor Erg, which drives basalICAM-2 expression, in human umbilical vein endothelial cells (HUVECs).12Erg is a member of the Ets family of transcription factors involved in embryonic development, inflammation, and cellular transformation.13-16Erg is a proto-oncogene, as shown by the ability of NIH 3T3 cells overexpressing Erg to form solid tumors in nude mice.17 The role of Erg in endothelial cells is as yet unknown. In vitro evidence suggests that Erg is also involved in the transcriptional regulation of von Willebrand factor (vWF), a multimeric glycoprotein required for platelet adhesion to the subendothelium.
To define the endothelial target genes regulated by the transcription factor Erg and thus provide new insight into the role of Erg in endothelial cells, we used an antisense approach to decrease levels of Erg mRNA and protein in HUVECs. The RNA obtained from these cells was analyzed to identify Erg target genes, using differential gene expression (DGE) technology. We show that Erg regulates several genes involved in vascular cell remodeling and shape change, angiogenesis, proliferation, and cell adhesion. We confirm that ICAM-2 andvWF are downstream targets of Erg and identifySPARC, thrombospondin (TSP-1), andRhoA as new Erg target genes. The expression of all of these genes is down-regulated by TNF-α. Finally, we show that the decrease of Erg expression by antisense treatment results in decreased endothelial cell proliferation and vascular tube formation in vitro. These results make Erg a candidate regulator of angiogenesis in vivo and suggest that Erg may represent a new link between the cytokine TNF-α and the regulation of angiogenesis.
Materials and methods
Design of Erg-specific GeneBlocs
GeneBlocs were obtained from Ribozyme Pharmaceuticals (Boulder, CO) and were synthesized by a modification of the method of Wincott.18 They were as follows: Erg GeneBloc 566, 5′-UCCCGCCTTTGGCCACACUGCAU-3′; Erg GeneBloc 573, 5′-UCUGCGCTCGTTCGTGGUCAUGU-3′; GeneBloc control 3.3 was randomized at every position—5′-(N)23-3′; Erg Mismatch GeneBloc 478, 5′-UCCGGCCTTAGGCCTCACUCCAU-3′, where the underlined positions indicate the sites of the 4 mismatches.
Delivery of GeneBlocs to HUVECs
HUVECs (BioWhittaker, Berkshire, United Kingdom) were seeded at 1 × 105 cells per well of a 35-mm, 6-well dish in EGM-2 medium (BioWhittaker) the day before transfection. GeneBloc (final concentration, 50 nM) and lipid NC388 (final concentration, 2 μg/mL; Atugen, Boulder, CO) were complexed in EGM-2 medium at 37°C for 30 minutes in polystyrene tubes. After mixing, the lipid–GeneBloc mix was added to each well and incubated for the times indicated. For initial optimization experiments, cells were seeded at 1 × 103in 96-well plates, and the lipid–GeneBloc mix was added as described. Lack of toxicity was determined by measuring c-Raf RNA after control GeneBloc treatment.
TaqMan and Lightcycler quantification of mRNA
Total RNA was prepared from HUVECs after GeneBloc delivery using the RNA purification kit for the 35-mm well or RNeasy extraction kit for the 96-well assays (both from Qiagen, United Kingdom). For TaqMan analysis, dual-labeled probes were synthesized with a reporter dye, covalently linked at the 5′ end and a quencher dye conjugated to the 3′ end. One-step reverse transcription–polymerase chain reaction (RT-PCR) amplifications were performed on the ABI Prism 7700 Sequence Detector (PE Biosystems, Foster City, CA) using 50-μL reactions consisting of 10 μL total RNA, 100 nM forward primer, 900 nM reverse primer, 100 nM probe, 1× TaqMan PCR reaction buffer (PE-Applied Biosystems, Foster City, CA), 5.5 mM MgCl2, 300 μM each dATP, dCTP, dGTP, and dTTP, 10 U RNase Inhibitor (Promega, Madison, WI), 1.25 U AmpliTaq Gold (PE-Applied Biosystems), and 10 U M-MLV Reverse Transcriptase (Promega). Thermal cycling conditions consisted of 30 minutes at 48°C, 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C, and 1 minute at 60°C. Standard curves were generated from serial dilutions of total HUVEC RNA; normalization was performed using β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels as controls in parallel TaqMan reactions. For each gene of interest, 5′ and 3′ primers and a fluorescence-labeled probe were designed as follows: Erg 5′ primer, GGAGTGGGCGGTGAAAGA; Erg 3′ primer, AAGGATGTCGGCGTTGTAGC; Erg probe, TGGCCTTCCAGACGTCAACATCTTGTTATT; ICAM-2 5′ primer, ATCTGTCCTGCTCTGCTTCATCT; ICAM-2 3′ primer, CGTAGGTGCCCATCCGC; ICAM-2 probe, CGGCCAGCACTTGCGCCA; Rho 5′ primer, TTAACGATGTCCAACCCGTCT; Rho 3′ primer, GGTGGGAGTGCAGAGGAGG; Rho probe, CAGGGTCCTTTTGACACTGCTCTAACAGCC; Actin 5′ primer, GCA TGG GTC AGA AGG ATT CCT AT; Actin 3′ primer, TGT AGA AGG TGT GGT GCC AGA TT; Actin probe, TCGAGCACGGCATCGTCACCAA; GAPDH primer–probe was purchased from PerkinElmer (catalog number, 4310884E). For analysis of gene expression using the Lightcycler (Roche Molecular Biochemicals, Mannheim, Germany), the following primers were used: vWF, 5′ CTTGGTCACATCTTCACATTCACT; vWF, 3′ GACACAGCTGCCTTCCAACAT; TSP, 5′ ATGCTGGTGGTAGACTAGGGTTGTTT; TSP, 3′ GAAGGAGGATGTCAGGGTGGTTT; SPARC, 5′ AATGTTT- GGATGGTTTGTTGTTCTGC; SPARC, 3′ ACGTTCTGGTTGGTGGATTCTGC; GAPDH, 5′ACCACAGTCCATGCCATCAC; GAPDH, 3′ACCACAGTCCATGCCATCAC. Real-time incorporation of SYBR Green I dye into a specific PCR product was measured in glass capillary tubes using the Lightcyler (Roche Molecular Biochemicals). A standard curve was generated for each primer pair using control HUVEC RNA, and values are represented as relative expression to GAPDH in each sample.
Western blot analysis
All nuclear extracts were prepared using the micropreparation technique.19 Protein extracts from supernatants were prepared using TCA precipitation. Briefly, an equal volume of 20% TCA was added to the cell supernatant, incubated on ice for 1 hour, and pelleted by centrifugation for 5 minutes. Pellets were washed in acetone, dried, and resuspended in water. HUVEC protein extracts were run on a 10% Bis-Tris NuPage (nuclear extracts) or 4% to 12% Tris-Glycine (supernatant extracts) polyacrylamide gel (Novex, San Diego, CA) following the manufacturer's protocols and were transferred onto nitrocellulose membranes using Novex X Cell-II module at 25 mA for 90 minutes. Nonspecific binding was blocked by incubation with 5% nonfat milk for 1 hour followed by primary antibody for 16 hours at 4°C. The following antibodies were used for Western blotting and were diluted as indicated: vWF (rabbit polyclonal A0082; Dako, Ely, United Kingdom), 1:100; SPARC (monoclonal ON1-1; Takara Biomedicals, Biowhittaker, Wokingham, United Kingdom), 1:1000; thrombospondin (TSP-B7; Sigma, St Louis, MO), 1:100; Erg (C20; Santa Cruz Biotechnology, Santa Cruz, CA), 1:1000; Oct-1 (C21; Santa Cruz), 1:100. After washes, the secondary antibody was applied (1:10 000 dilution; Sigma) for 1 hour at room temperature, and the signal was detected with the SuperSignal reagent (Pierce, United Kingdom).
Differential gene expression
Sequence-verified high-density cDNA arrays on nylon filters were prepared in house. Known human genes were selected because of their known or predicted role in inflammation and in the development atherosclerosis (for review, see Ross20). Genes were grouped in the following categories: inflammation, cell adhesion, apoptosis, cell cycle, cytoskeleton, extracellular matrix, signal transduction, transcription, growth factors, hemostasis, and lipid metabolism. One thousand forty EST clones were selected from the IMAGE library, representing 482 individual genes. Sequence-verified PCR products of 800-bp average length, corresponding to the 3′ end of each gene, were gridded using a KB robotic gridder (KayBee Engineering, Essex, United Kingdom) in duplicate onto nylon filters (Boehringer Mannheim, Mannheim, Germany) using an automated 384-pin gridding device (Gentix, Hants, United Kingdom). Filters were then denatured (0.5 M NaOH, 100 mM NaCl) for 2 minutes, neutralized (1 M Tris, pH 7.2, 100 mM NaCl) for 2 minutes, and UV cross-linked (Stratalinker; Stratagene, Palo Alto, CA). Total RNA was prepared from HUVECs treated withErg GeneBloc or a random control GeneBloc 3.3, as described. RNA was precipitated overnight and analyzed on a dimethyl sulfoxide–glyoxal–agarose gel to check the quantity and integrity of the RNA. Approximately 10 μg total RNA was used to generate duplicate radiolabeled cDNA probes using Superscript II (Stratagene) and α-33P dCTP (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). After purification on a G50 column (Amersham Pharmacia Biotech), probes were run on a 6% TBE urea gel (Novex) alongside known size markers, and the image data were captured using a Storm Scanner (Molecular Dynamics, Sunnyvale, CA) for quality control of probe size. All cDNAs in the array contained the M13 sequence; thus, an M13 oligonucleotide probe, end-labeled with γ-33P dATP, was hybridized to the membranes to control for differences in spot loading. For prehybridization, membranes were incubated in DIG Easy Hyb (Boehringer Mannheim) at 45°C for 90 minutes and were then hybridized overnight in DIG Easy Hyb containing probe at 45°C. Filters were washed 3 × 15 minutes in 0.1 × SSC, 0.1% sodium dodecyl sulfate at 68°C and exposed to phospho-screens for 2 days, and images were captured using a phosphor imager (Storm Scanner; Molecular Dynamics). Processing of the images, including spot finding, spot intensity quantitation, and local background subtraction, was performed using Glaxo Wellcome proprietary image analysis software (DGENT; GlaxoSmithKline, Uxbridge, Middlesex, United Kingdom). Raw values were normalized using M13 hybridization data and were compared in a 4-way analysis on duplicate probes and replica filters. The genes, which were consistently more than 1.4-fold different between the Erg GeneBloc and the control GeneBloc samples, were selected.
Matrigel morphogenetic assay
Aliquots of Matrigel (Becton Dickinson, Oxford, United Kingdom) were thawed on ice, and 200 μL was plated onto chilled 15-mm wells and incubated at 37°C for 30 minutes to form a gel as per the manufacturer's instructions. HUVECs in 6-well plates were treated with the Erg GeneBloc or the Mismatch GeneBloc for 24 hours as described, trypsinized, and counted, and 4 × 104 cells in EGM-2 were added to each well containing Matrigel. After 16 hours, the formation of tubulelike structures was analyzed using a Leica 550 image analyser. Dark-field, low-magnification images were processed to remove background and were skeletonized, and total curve length (in micromolars) and number of branches were calculated.
Results
Erg GeneBlocs decrease Erg mRNA and protein levels in HUVECs
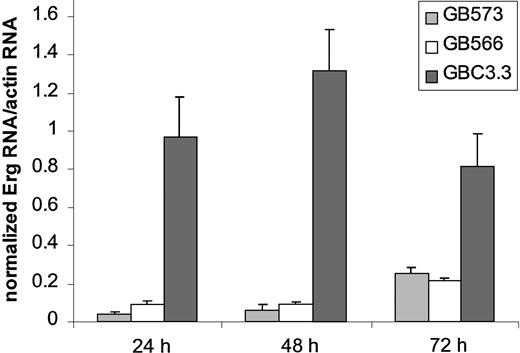
To suppress Erg expression in endothelial cells, antisense oligonucleotides (GeneBlocs) were designed. GeneBlocs are modified RNA oligonucleotides that are less susceptible to degradation and have reduced toxicity and increased target-binding affinity than traditional antisense oligonucleotides.18 Because otherEts family members are expressed in endothelial cells,Erg GeneBlocs were designed to target a region of human Erg not homologous to 5 closely related family members also found in endothelial cells: Ets-1, Ets-2, Fli-1, Nerf-1, andNerf-2.13 21-23 In initial studies, 8 Erg GeneBlocs were designed, and inhibition of Erg mRNA in HUVECs was assayed using TaqMan (data not shown). Two GeneBlocs (GB573 and GB566) gave more than 80% inhibition of Erg mRNA levels after 24 hours when compared to a random scrambled control GeneBloc (GBC3.3) (Figure 1). This inhibition lasted for at least 72 hours of continuous GeneBloc delivery.Erg mRNA was inhibited in a dose-dependent manner after treatment of HUVECs with GB566 from 0 nM to 25 nM (data not shown). Because the 2 GeneBlocs gave comparable results, only GeneBloc GB566 was used in all subsequent studies. To verify that Erg protein levels were also decreased after GeneBloc treatment, nuclear extracts from HUVECs treated with Erg GeneBloc (GB566), the scrambled control GeneBloc (GBC3.3), or the Erg Mismatch GeneBloc (GB478) were subjected to Western blot analysis. Expression levels of the ubiquitous transcription factor Oct-1 were also determined to normalize for loading differences (Figure2A-B). Densitometry was performed on the bands representing Erg and Oct-1, and theErg:Oct ratio was calculated. Results showed that Erg protein levels were decreased approximately 2.5-fold after 24 and 48 hours of Erg GeneBloc treatment and by 5-fold after 72 hours of treatment when compared with control GeneBloc treatment (Figure 2A).Erg Mismatch GeneBloc GB478 did not decrease Erg protein levels compared with random control GeneBloc GBC3.3 (Figure2B).
Erg GeneBlocs decrease Erg mRNA levels in HUVECs.
RNA was prepared from HUVECs treated with Erg-specific (GB573 and GB566) and random control (GBC3.3) GeneBlocs for 24, 48, and 72 hours. Shown are levels of Erg mRNA, normalized to actin mRNA, as measured by TaqMan analysis. A ratio of 1 represents Erg mRNA level in resting HUVECs.
Erg GeneBlocs decrease Erg mRNA levels in HUVECs.
RNA was prepared from HUVECs treated with Erg-specific (GB573 and GB566) and random control (GBC3.3) GeneBlocs for 24, 48, and 72 hours. Shown are levels of Erg mRNA, normalized to actin mRNA, as measured by TaqMan analysis. A ratio of 1 represents Erg mRNA level in resting HUVECs.
Erg protein levels are decreased in HUVECs treated with Erg GeneBlocs.
Nuclear extracts from cells treated with 50 nM Erg GeneBloc (GB566) or control GeneBloc (GBC3.3) for 24, 48, or 72 hours (A) and 50 nM control GeneBloc (GBC3.3), Mismatch GeneBloc (GB478), or Erg GeneBloc (GB566) for 24 hours (B) were generated as described (see “Materials and methods”). Western blot analysis with anti-Erg and anti-Oct monoclonal antibodies was performed, and the intensity of the bands was measured by densitometry. Levels of the transcription factor Oct were used to normalize for differences in loading. The ratio between Erg and Oct in the Erg GeneBloc and in the Control samples is shown below using data from densitometry.
Erg protein levels are decreased in HUVECs treated with Erg GeneBlocs.
Nuclear extracts from cells treated with 50 nM Erg GeneBloc (GB566) or control GeneBloc (GBC3.3) for 24, 48, or 72 hours (A) and 50 nM control GeneBloc (GBC3.3), Mismatch GeneBloc (GB478), or Erg GeneBloc (GB566) for 24 hours (B) were generated as described (see “Materials and methods”). Western blot analysis with anti-Erg and anti-Oct monoclonal antibodies was performed, and the intensity of the bands was measured by densitometry. Levels of the transcription factor Oct were used to normalize for differences in loading. The ratio between Erg and Oct in the Erg GeneBloc and in the Control samples is shown below using data from densitometry.
Differential gene expression of HUVECs treated with Erg GeneBloc
GeneBloc treatment significantly decreased Erg mRNA and protein levels at all time points tested. The model was, therefore, used in a DGE experiment between Erg GeneBloc–treated and control GeneBloc–treated HUVECs to identify differences in the gene expression pattern caused by the inhibition of Ergexpression. Total RNA was generated from cells treated withErg GeneBloc (GB566) or control GeneBloc (GBC3.3) for 24, 48, and 72 hours and was used to prepare radiolabeled cDNA probes for DGE analysis. Probes were hybridized to sequence-verified cDNA arrays containing clones corresponding to 482 individual genes. Genes belonged to several functional families, including inflammation, cell adhesion, and transcription (see “Materials and methods”). For each clone, duplicate spots were present in each array. Each hybridization was performed in duplicate so that for each clone at each time point at least 4 duplicate determinations were available. A 4-way comparison analysis was performed between control GeneBloc and ErgGeneBloc samples to identify genes that were decreased after the suppression of Erg expression. Only the genes that were consistently different by 1.4-fold or more in all 4 comparisons were considered. Several hits were identified that were consistently decreased in the samples treated with Erg GeneBloc (Table1). ICAM-2 and vWF were consistently decreased in the Erg GeneBloc samples. This confirms thatErg drives expression of these 2 genes, as suggested by previous reports showing that Erg cDNA can transactivate theICAM-2 and vWF promoters.12 21 Other genes decreased after Erg GeneBloc treatment wereSPARC, TSP, and RhoA. The expression of these 5 genes was decreased at all time points, though only 2 of the 4 RhoA replicates were decreased at 24 hours. These results suggest that the 5 genes identified by DGE are down-stream targets ofErg. Interestingly, a common functional theme is present in the DGE hits because 4 of the 5 genes identified are cell adhesion molecules, and RhoA is involved in signaling downstream of adhesion molecules.
Genes down-regulated by Erg GeneBloc treatment
| Genes . | Functional category . |
|---|---|
| ICAM-2 | Cell adhesion |
| RhoA | Cytoskeleton/signaling |
| SPARC | Cell adhesion/extracellular matrix |
| TSP-1 | Cell adhesion/extracellular matrix |
| vWF | Cell adhesion/hemostasis |
| Genes . | Functional category . |
|---|---|
| ICAM-2 | Cell adhesion |
| RhoA | Cytoskeleton/signaling |
| SPARC | Cell adhesion/extracellular matrix |
| TSP-1 | Cell adhesion/extracellular matrix |
| vWF | Cell adhesion/hemostasis |
RNA was prepared from HUVECs treated with 50 nM Erg GeneBloc (GB566) or a control GeneBloc (GBC3.3) was used to prepare cDNA probes for DGE analysis. Genes that were consistently down-regulated at 24, 48, and 72 hours are listed.
Real-time polymerase chain reaction analysis of differential gene expression hits
To validate the DGE results, 2 real-time PCR-based methods of RNA quantification—TaqMan (PerkinElmer) and Lightcycler (Roche)—were used. A new set of RNA was generated from cells treated with theErg GeneBloc 566 or with scrambled control GeneBloc. A new 4-base mismatch control (GB478), which did not inhibit ErgmRNA levels after 24, 48, or 72 hours of treatment (Figure3), was used in these experiments. To select the most appropriate control for normalization, a panel of 11 putative housekeeping genes was screened for variability across the different GeneBloc treatments. Expression of GAPDH was the least variable between treatments (data not shown) and was, therefore, used in subsequent experiments for normalization. Expression ofICAM-2 and RhoA was detected using the TaqMan system, whereas expression of SPARC, TSP, andvWF was detected using the light cycler real-time PCR. mRNA levels of all 5 genes analyzed were decreased to various degrees in HUVECs after 24 hours of treatment with the Erg GeneBloc (Figure 4). An additional control was represented by fibronectin mRNA, whose levels were unchanged after 24 hours of treatment and were up-regulated after 48 and 72 hours of Erg GeneBloc treatment (data not shown). Therefore, the results obtained by DGE on the 5 putative Erg target genes were confirmed at the 24-hour time-point on a separate set of samples by PCR-based quantification methods.
Treatment of HUVECs with the Erg mismatch control GeneBloc 478 does not reduce Erg mRNA levels.
RNA was prepared from HUVECs treated with Erg specific (GB566), Erg mismatch (GB478), and random control (GBC3.3) GeneBlocs for 24, 48, and 72 hours. Shown are levels of Erg mRNA, normalized to actin mRNA, as measured by TaqMan analysis. A ratio of 1 represents Erg mRNA level in untreated HUVECs. Erg expression is inhibited by the Erg-specific GeneBloc but not by the 2 control GB. At the 72-hour point, a small amount of toxicity is observed with the control GeneBloc treatment and may be caused by prolonged treatment with GeneBloc or lipid.
Treatment of HUVECs with the Erg mismatch control GeneBloc 478 does not reduce Erg mRNA levels.
RNA was prepared from HUVECs treated with Erg specific (GB566), Erg mismatch (GB478), and random control (GBC3.3) GeneBlocs for 24, 48, and 72 hours. Shown are levels of Erg mRNA, normalized to actin mRNA, as measured by TaqMan analysis. A ratio of 1 represents Erg mRNA level in untreated HUVECs. Erg expression is inhibited by the Erg-specific GeneBloc but not by the 2 control GB. At the 72-hour point, a small amount of toxicity is observed with the control GeneBloc treatment and may be caused by prolonged treatment with GeneBloc or lipid.
Erg GeneBloc decreases ICAM-2, RhoA, TSP, vWF, and SPARC mRNA levels in HUVECs.
RNA was prepared from HUVECs treated with Erg-specific (GB566) or control (GB478) GeneBloc. Real-time PCR analysis (TaqMan for ICAM-2 and RhoA or Lightcycler for TSP, vWF, and SPARC) was used to quantify mRNA levels. Results for each gene are shown as a ratio to GAPDH mRNA levels, which were identified as the least variable control genes in these cells (see “Results”). 100% represents Erg mRNA level in HUVECs treated with the mismatch control GeneBloc (GB478).
Erg GeneBloc decreases ICAM-2, RhoA, TSP, vWF, and SPARC mRNA levels in HUVECs.
RNA was prepared from HUVECs treated with Erg-specific (GB566) or control (GB478) GeneBloc. Real-time PCR analysis (TaqMan for ICAM-2 and RhoA or Lightcycler for TSP, vWF, and SPARC) was used to quantify mRNA levels. Results for each gene are shown as a ratio to GAPDH mRNA levels, which were identified as the least variable control genes in these cells (see “Results”). 100% represents Erg mRNA level in HUVECs treated with the mismatch control GeneBloc (GB478).
TNF-α regulation of Erg target genes in endothelial cells
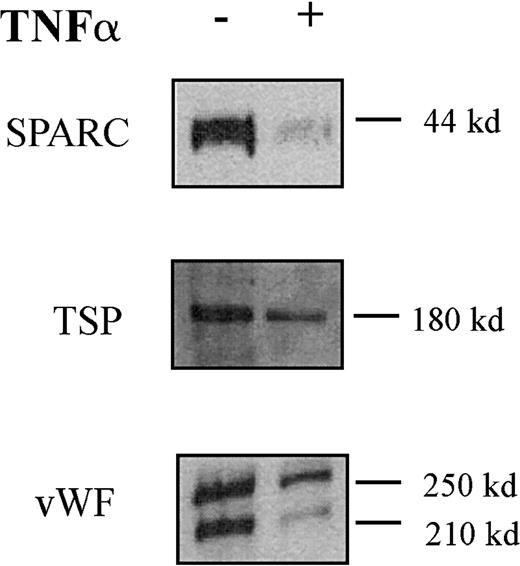
Expression of the Erg target genesICAM-212 and RhoA (A. Ridley, P. Thompson, personal communication) have previously been shown to be down-regulated by TNF-α in HUVECs. Hence, we looked at the effect of TNF-α on expression of the other Erg target genes identified by DGE, namely the secreted proteins vWF,SPARC, and TSP-1 (Figure5). HUVECs were treated with TNF-α (10 μg/mL) for 24 hours, and protein extracts were prepared from the cell supernatants. Western blot analysis was performed, and band intensity was measured by densitometry. Protein levels of SPARC, TSP, and vWF were all markedly decreased in response to 24 hours of TNF-α treatment (Figure 5). Therefore, TNF-α down-regulates the expression of Erg and of its target genes in endothelial cells.
TNF-α down-regulates SPARC, TSP, and vWF protein secretion by HUVECs.
Western blot analysis of SPARC, TSP, and vWF in the supernatant of HUVECs treated (+ lane) with TNF-α (10 ng/mL) for 24 hours or untreated (− lane). Sizes of the detected bands are reported on the right.
TNF-α down-regulates SPARC, TSP, and vWF protein secretion by HUVECs.
Western blot analysis of SPARC, TSP, and vWF in the supernatant of HUVECs treated (+ lane) with TNF-α (10 ng/mL) for 24 hours or untreated (− lane). Sizes of the detected bands are reported on the right.
Effect of Erg GeneBloc on endothelial cell tube formation
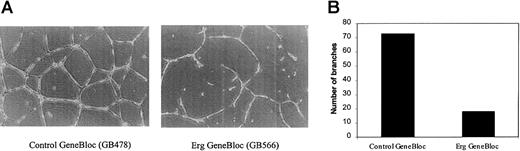
All the genes identified by DGE as downstream targets of the transcription factor Erg are associated with angiogenesis or vascular remodeling. SPARC and TSP-1 have been shown to modulate angiogenesis, cell shape change, motility, differentiation, and proliferation.24-27 RhoA is a key regulator of the actin cytoskeleton, mediating cell shape responses to the interaction between adhesion molecules and the surrounding extracellular matrix.28 vWF has recently been associated with angiogenesis through interaction with its endothelial receptor, GPIb.29 We tested the ability of the ErgGeneBloc to modulate endothelial tube formation in a 3-dimensional culture system. An identical number of HUVECs, treated withErg GeneBloc (GB566) or control GeneBloc (GB478) for 24 hours, was plated on Matrigel in 15-mm wells and allowed to form tubes over 16 hours. Figure 6A shows a representative photograph of HUVEC on Matrigel after treatment withErg GeneBloc (right panel) or the mismatch control GeneBloc (left panel). After treatment with Erg GeneBloc, a clear reduction in the amount of tube formation of HUVECs and in the number of branches formed is observed. The number of branches formed in each well was quantified by image analysis, and the results are shown in Figure 6B. A 3-fold reduction in the number of branches was observed after Erg GeneBloc treatment, indicating that Erggene expression is required for vascular remodeling, one of the key steps of angiogenesis.
Erg GeneBlocs reduce endothelial cell tube formation on Matrigel.
The phenotypic effect of the inhibition of Erg on vascular remodeling was tested in vitro using the Matrigel tube formation assay. HUVECs were treated with Erg GeneBloc (GB566) or the Control GeneBloc (GB478) for 24 hours as described. Then they were trypsinized and counted, and 4 × 104 cells/well were plated onto 15-mm wells containing Matrigel. After 16 hours, tubule formation was analyzed using a Leica 550 image analyser. (A) Low-magnification image (original magnification × 200) of tubes formed by HUVECs treated with control GeneBloc (left) or with Erg GeneBloc (right), showing a marked reduction in tube formation in the Erg GeneBloc-treated cells. (B) The number of branches in each sample was quantified using image analysis (results of one representative experiment are shown here).
Erg GeneBlocs reduce endothelial cell tube formation on Matrigel.
The phenotypic effect of the inhibition of Erg on vascular remodeling was tested in vitro using the Matrigel tube formation assay. HUVECs were treated with Erg GeneBloc (GB566) or the Control GeneBloc (GB478) for 24 hours as described. Then they were trypsinized and counted, and 4 × 104 cells/well were plated onto 15-mm wells containing Matrigel. After 16 hours, tubule formation was analyzed using a Leica 550 image analyser. (A) Low-magnification image (original magnification × 200) of tubes formed by HUVECs treated with control GeneBloc (left) or with Erg GeneBloc (right), showing a marked reduction in tube formation in the Erg GeneBloc-treated cells. (B) The number of branches in each sample was quantified using image analysis (results of one representative experiment are shown here).
Discussion
Inflammation, leukocyte recruitment into tissues, and angiogenesis are just a few of the processes regulated by the endothelium.3 In healthy conditions, a dynamic balance between inhibitors and activators of these key functions maintains endothelial homeostasis. The up-regulation of proinflammatory or proangiogenic factors is accompanied by the down-regulation of inhibitory factors. Consequently, understanding the mechanisms underlying up-regulation and down-regulation of gene expression is crucial to understanding endothelial activation and its role in health and disease.
Numerous stimuli affect endothelial function by modulating gene expression.30 Although many studies have focused on the up-regulation of gene expression induced by cytokines such as TNF-α, relatively little is known about the mechanisms underlying the down-regulation of gene expression. Several genes have been described in endothelial cells that are down-regulated in response to TNF-α: the antithrombotic molecules protein C, protein S, and thrombomodulin, the anti-inflammatory mediator eNOS, the adhesion molecule CD31, and many others.31-35 Various mechanisms have been implicated in the cytokine-induced down-regulation of gene expression.20,33,36,37 We have demonstrated that the transcription factor Erg, involved in basal endothelial gene expression, is down-regulated by TNF-α treatment, both in vitro and ex vivo. Erg was shown to transactivate the ICAM-2promoter12 and the vWF promoter,21and Erg protein isolated from endothelial cell nuclear extracts was shown to bind to the VE-cadherin and stromelysinpromoters.12,21,38 39
To identify new Erg target genes and gain some insight into its role in the endothelium, we designed a strategy to suppressErg expression in HUVECs using antisense oligonucleotides, and we analyzed the gene expression profile in the cells with reduced Erg expression. A specific Erg antisense (GeneBloc) decreased levels of Erg mRNA (by 80%-90%) and protein (by 60%-80%) in HUVECs over a period of 24 to 72 hours. RNA from HUVECs treated with either Erg GeneBloc or a control GeneBloc was used in a differential gene expression experiment to compare the gene expression profile in cells with reduced Erg expression with control HUVECs. DGE was performed on high-density cDNA arrays containing 482 known genes, representing genes involved in inflammation, cell adhesion, angiogenesis, hemostasis, cell cycle, apoptosis, lipid metabolism, signaling, and transcription. Several genes were identified that were reduced in the Erg GeneBloc–treated cells. After a stringent analysis of duplicates and replicates, and across all time points, ICAM-2, vWF, SPARC,TSP-1, and Rho A were consistently down-regulated after Erg GeneBloc treatment.
The results of this study suggest a new functional role for the transcription factor Erg in endothelial cells. The new genes identified as Erg targets have been clearly involved in angiogenesis, often displaying both pro- and antiangiogenic properties. SPARC was initially referred to as antiadhesin because it can disrupt cell–matrix interactions.40 It was shown to inhibit endothelial cell spreading on collagen40 and to alter endothelial morphology41 and barrier properties.42 Evidence for a role of SPARC in regulating angiogenesis comes from several studies. SPARC regulates endothelial cell proliferation, induces loss of focal adhesion (thus loosening adhesion to the extracellular matrix), and stimulates production of matrix-degrading enzymes such as MMPs and PAI-1.43 TSP-1 is also an extracellular matrix protein involved in the regulation of adhesion and angiogenesis.25,26 It inhibits angiogenesis both in vitro and in vivo,44 and it functions by binding to several molecules including αvβ3, CD36, TGFβ, SPARC, and heparin sulfate proteoglycans.25 vWF, a glycoprotein involved in platelet adhesion to the subendothelium,45 is expressed in human endothelial cells in vitro. Expression of vWF is increased by angiogenic growth factors fibroblast growth factor-2 (FGF-2) and vascular endothelial growth factor (VEGF)46 and is decreased by the proinflammatory cytokine interleukin-1β47 and TNF-α. Recently, vWF interaction with its receptor, GPIb, on endothelial cells was implicated in angiogenesis.29 RhoA is a member of the Ras superfamily involved in the reorganization of the actin cytoskeleton, as shown by micro-injection experiments.28,48 Rho GTPases are downstream of integrins in the signaling pathways that regulate cell proliferation and gene transcription.49 In endothelial cells, RhoA is thought to be involved in cell flattening and maintenance of barrier function.50 Of the Erg target genes identified here, ICAM-2 is the only gene that has not been linked to angiogenesis. The function of ICAM-2 in the endothelium is unclear, though it is implicated in transendothelial migration of eosinophils,51 neutrophils,52lymphocytes,53 dendritic cells,54 and monocytes.55 In confluent resting monolayers, ICAM-2 is expressed at the endothelial junctions, and TNF-α treatment decreases expression, particularly at the cell–cell contact. Another cell adhesion molecule of the immunoglobulin superfamily, CD31, involved in regulating endothelial proliferation and angiogenesis, was shown to be decreased by cytokines, albeit with a different mechanism than that described for ICAM-2.56 57 Studies are in progress to define the role of ICAM-2 in the endothelium and to determine whether ICAM-2 is, like the other Erg target genes, also involved in the regulation of angiogenesis.
Given the involvement of 4 Erg target genes in angiogenesis and cell proliferation, we investigated the role of Erg in vascular cell remodeling and adhesion using an in vitro model of endothelial cell differentiation. Endothelial cells plated on Matrigel undergo a morphologic rearrangement, and alignment of cells into tubulelike structures is complete by 16 hours.58 HUVECs treated with Erg GeneBloc for 24 hours were plated on Matrigel and allowed to form these tubular structures overnight. Cells in which Erg expression was decreased because of GeneBloc treatment formed significantly less tubulelike structures than cells treated with a specific control GeneBloc. The final alignment of endothelial cells into these structures on Matrigel may not itself require transcription59; however, in the experiment presented here, HUVECs were treated with antisense oligonucleotides for 24 hours before seeding on Matrigel. This time is sufficient to significantly reduce the transcription of Erg andErg target genes, as shown here. Therefore, it is the transcriptional effect that occurs before seeding onto Matrigel that is responsible for this phenotype. This assay clearly demonstrates a role for Erg as a direct modulator of this endothelial differentiation phenotype, a prerequisite for the in vivo process of angiogenesis. Further support for this model comes from recent studies in Xenopus embyros. A Xenopus homologue ofErg, Xl-erg, was micro-injected into embryos, and several developmental defects were observed, including eye malformations and ectopic endothelial differentiation,60suggesting that in Xenopus Erg may be involved in regulation of cell motility, adhesion, and angiogenesis. Other Etsfamily members, Ets-114,61 andFli-1,23 have been involved in the regulation of angiogenesis. It is unlikely that the effects observed in this study are attributable to the inhibition of Ets family members other than Erg because the GeneBloc used in this study was specific for Erg, and a 4-bp mismatch was sufficient to abrogate the effect.
All Erg target genes described here have been shown to be down-regulated in HUVEC by TNF-α treatment. This could be a direct consequence of the decrease in Erg expression caused by TNF-α, or it could be secondary to effects on other Erg target genes. Because Erg has been shown to transactivate theICAM-2 and vWF promoters,12,21 it is likely that, at least for these 2 genes, TNF-α down-regulation is a direct consequence of the decrease in Erg expression. Further studies will be required to define the role of Ergin the transcriptional regulation of SPARC, TSP, and RhoA. An interesting issue is the relation between the proinflammatory cytokine TNF-α, which down-regulates Erg, and the angiogenic process that is perturbed by decreasing Erg levels. TNF-α, like SPARC and TSP-1, seems to play a dual role in the regulation of angiogenesis. Whether TNF-α is proangiogenic or antiangiogenic may depend on the model system or on the microenvironment. In vitro, TNF-α has many antiangiogenic and antiproliferative properties. It inhibits the basic FGF (bFGF)–stimulated growth of endothelial cells4; however, it induces the expression of proangiogenic molecules such as bFGF, platelet-activated factor, and urokinase-type plasminogen activator, and it increases the transcription of VEGF receptor 2.62,63 In tumor endothelium, up-regulation of proinflammatory adhesion molecules in response to TNF-α is reduced in comparison to normal endothelium, and this is mediated by angiogenic factors such as FGF.5 Recently, in patients with rheumatoid arthritis, anti–TNF-α therapy was shown to decrease circulating levels of VEGF.64 Several studies provide evidence of a link between TNF-α and angiogenesis in vivo,4,7 but little is known about the mechanism of TNF-α regulation of angiogenesis. The results shown here suggest thatErg may be a key player in the TNF-α–dependent regulation of angiogenesis. HUVECs are commonly used as a model system for endothelial function. In future studies, however, it will be important to compare the effects of decreased Erg expression in endothelial cells from different vascular beds, where possible, because there can be variations in expression and regulation of genes depending on the endothelial cell type.65
In conclusion, we have demonstrated that the transcription factorErg regulates the expression of ICAM-2,SPARC, TSP-1, RhoA, and vWFand is a modulator of endothelial cell differentiation, which is a component of angiogenesis. Strategies aimed at the regulation ofErg may be of use in the therapy of diseases such as cancer, rheumatoid arthritis, osteoporosis, and wound healing.
We thank Francesco Falciani (GSK) and Eugenia Biguzzi (Policlinico Hospital, Milan, Italy) for preparation of the cDNA arrays; Mark Edbrooke (GSK) and Thale C. Jarvis (Atugen) for setting up a fruitful collaboration; Brian Hayes (GSK) and Justin Mason (Hammersmith Hospital, London) for assistance with the Matrigel assay; Anne Ridley and Paul Thompson (Ludwig Institute, London, United Kingdom) for a personal communication; Chantelle Ward (GSK) and Matthew Goulden (GSK) for help with real-time PCR set-up; and Callum J. Campbell (GSK) for helpful discussions.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anna M. Randi, Experimental Medicine, GlaxoSmithKline, Addenbrooke's Centre for Clinical Investigation, Addenbrooke's Hospital, Hills Road, Cambridge, United Kingdom, CB2 2 GG; e-mail: ar18946@gsk.com.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal