Abstract
Recent studies show that several Hox transcription factors are important for regulation of proliferation and differentiation in hematopoiesis. Among these is H0XA10, which is selectively expressed at high levels in the most primitive subpopulation of human CD34+ bone marrow cells. When overexpressed, H0XA10 increases the proliferation of early progenitor cells and can lead to the development of myeloid leukemia. To study the effects of H0XA10 on primitive hematopoietic progenitors in more detail, transgenic mice were generated with regulatable H0XA10 expression. The transgenic mouse model, referred to as tetO-HOXA10, contains theH0XA10 gene controlled by a tetracycline-responsive element and a minimal promoter. Thus, the expression of H0XA10 is inducible and reversible depending on the absence or presence of tetracycline or its analog, doxycycline. A retroviral vector containing the tetracycline transactivator gene (tTA) was used to induce expression of the H0XA10 gene in bone marrow cells from the transgenic mice. Reverse transcription–polymerase chain reaction analysis confirmed regulatable H0XA10 expression in several transgenic lines. H0XA10 induction led to the formation of hematopoietic colonies containing blastlike cells and megakaryocytes. Moreover, the induction of H0XA10 resulted in significant proliferative advantage of primitive hematopoietic progenitors (spleen colony-forming units [CFU-S12]), which was reversible on withdrawal of induction. Activation of H0XA10 expression intet0-H0XA10 mice will therefore govern proliferation of primitive myeloid progenitors in a regulated fashion. This novel animal model can be used to identify the target genes of HOXA10 and better clarify the specific role of HOXA10 in normal and malignant hematopoiesis.
Introduction
Hematopoiesis is a lifelong, dynamic process in which a small number of pluripotent hematopoietic stem cells (HSCs) residing in the bone marrow (BM), give rise to all the mature blood cells of both myeloid and lymphoid origin.1-3 This powerful proliferation and differentiation in hematopoiesis requires tight control to supply the host with all the different types of blood cells required for normal function. Much information has emerged on the role of regulatory factors and cytokines affecting hematopoiesis and several studies have shown that among candidate genes involved in the regulation are genes encoding various transcription factors (reviewed by Ogawa4 and Orkin5).
HoxA10 belongs to a large family of transcription factors that share a highly conserved DNA binding domain, the homeodomain. In mammals, 39 homeobox (Hox) genes are grouped together in 4 clusters, A to D,6-8 and over the last few years, accumulating evidence has shown that the Hox proteins are important in the mechanisms controlling differentiation and proliferation of hematopoietic cells.9 Several Hox genes from the A, B, and C clusters are expressed in a distinct pattern during hematopoiesis and aberrant Hox gene expression has also been documented in different types of leukemia.10-15 Direct effects of Hox gene function on hematopoiesis have been shown in various studies where the expression of Hox genes has been modulated by antisense oligonucleotides,16,17gene disruption by homologous recombination,18 or by overexpression.15,19-25 In mice, for example, the overexpression of HoxB4 leads to selective expansion of primitive progenitor cells without development of leukemia or other malignancies. In contrast, overexpression of HoxB3, HoxB8, HoxA9, and HoxA10 eventually leads to myeloid leukemia after a latency period of several months. This latency period can be shortened dramatically if the Hox cofactors, Pbx1 or Meis1, are co-overexpressed with their Hox partner.22,26-28 Other myeloid-specific effects of HOXA10 have also been documented in studies using constitutive overexpression of HOXA10 in mouse hematopoietic cells.25 In addition to these findings, expression of the H0XA10 gene is found in all types of human acute myeloid leukemia (except promyelocytic leukemia) but not in lymphoid leukemia, further implicating an important role for H0XA10 as a regulator of myeloid progenitors.
Because overexpression of H0XA10 in murine hematopoietic cells leads to increased proliferation of primitive myeloid progenitors and the generation of blast cell colonies in vitro but does not lead to the immediate development of leukemia in vivo,25 we decided to ask whether we could use the H0XA10 gene to generate a transgenic murine model with inducible and reversible proliferation of primitive hematopoietic progenitors. To achieve this, we used the tetracycline-transactivating system (tet-system), placing theH0XA10 gene under the control of the minimal promoter from human cytomegalovirus (hCMV) fused to the tet operator sequences (tetO).29 BM cells from the transgenic mice were transduced with a retroviral vector containing the tetracycline transactivator (tTA) to activate H0XA10 expression. The results show that regulatable short-term overexpression of H0XA10 alters the commitment and colony-forming ability of clonogenic progenitor cells and increases the proliferation of primitive spleen colony-forming unit (CFU-S) progenitors. It is noteworthy that the proliferative effects on primitive CFU-S progenitors are reversible on withdrawal of HOXA10 induction. Therefore, a novel animal model has been generated where regulatable expression of H0XA10 can be used to induce proliferation of primitive hematopoietic progenitors in vivo in a reversible fashion. This model can be used for biologic studies of normal and abnormal hematopoiesis and to identify hitherto unknown transcriptional targets of HOXA10.
Materials and methods
Generation and screening of transgenic animals
A 1.8-kb EcoRI fragment of the human H0XA10 complementary DNA (cDNA; provided by Dr Corey Largman, San Francisco, CA) was blunt ligated in the BamHI site in the pUHG10-3 plasmid (provided by Dr Herman Bujard, Heidelberg, Germany). The cassette was then cut out with AatII and NotI (Figure 1A) and microinjected into the pronucleus of fertilized oocytes from C57BL/6xCBA F1 mice and transplanted into pseudopregnant foster mothers of the same strain using standard techniques.30 Genomic DNA was isolated from tails of 82 mice and screened by polymerase chain reaction (PCR) using primers A10: TGGGCAATTCCAAAGGTG and BG: CAATCAAGGGTCCCCAAACTC amplifying a 950–base pair (bp) fragment from the expression cassette. The presence of the transgene was also verified by Southern blot analysis. Then, 10 μg DNA was digested with BamHI and a 742-bp XhoI/DraI fragment was used as a probe.31 Nineteen lines were positive. They were mated with C57BL/6xCBA F1 and 9 transmitted their transgene through germline. Background expression of H0XA10 was tested by isolating whole RNA (RNeasy, Qiagen, Stockholm, Sweden) from peripheral blood (and BM in some experiments) and performing reverse transcription (RT) PCR (RT-PCR) using the A10-BG primer pair. Template for the PCR was made by reverse transcription of 5 μg total RNA, primed with oligo dT primers. PCR analysis using hypoxanthine-guanine phosphoribosyltransferase (HPRT) specific primers (5′-3′: CACAGGACTAGAACACCTGC, 3′-5′: GCTGGTGAAAAGGAC CTC) was performed to evaluate possible genomic DNA contamination and to test the presence of cDNA. The same techniques were used when analyzing induction and leakiness of cultured transduced BM cells derived from thetet0-H0XA10 transgenic mice.
The
tetO-HOXA10 transgenic construct and characterization of transgenic mice. (A) The HOXA10 construct containing the human HOXA10 cDNA under the control of the PhCMV*-1, a minimal promoter from hCMV fused to the tetO sequences. The intron-polyA (pA) sequence is derived from the rabbitβ-globin gene. The picture is not drawn to scale. (B) Southern blot analysis showing 3 of the transgenictetO-HOXA10 mouse lines. Approximately 10 μg genomic DNA was digested with BamHI and hybridized with a HOXA10-specific probe. The band in the control lane corresponds to 10 copies (cop) of the construct mixed together with genomic DNA from wild-type control mice.
The
tetO-HOXA10 transgenic construct and characterization of transgenic mice. (A) The HOXA10 construct containing the human HOXA10 cDNA under the control of the PhCMV*-1, a minimal promoter from hCMV fused to the tetO sequences. The intron-polyA (pA) sequence is derived from the rabbitβ-globin gene. The picture is not drawn to scale. (B) Southern blot analysis showing 3 of the transgenictetO-HOXA10 mouse lines. Approximately 10 μg genomic DNA was digested with BamHI and hybridized with a HOXA10-specific probe. The band in the control lane corresponds to 10 copies (cop) of the construct mixed together with genomic DNA from wild-type control mice.
Retroviral vector production
The Neo–internal ribosomal entry site (IRES)–tTA (NIT)–GFP retroviral vector (kindly provided by Dr F. Gage, The Salk Institute, La Jolla, CA) contains the 5′ Moloney murine leukemia virus (Mo-MLV) long terminal repeat (LTR), which drives the expression ofneoR and tTA genes (neomycin resistance and tetracycline transactivator genes, respectively). To ensure efficient transcription of the tTA gene, there is an IRES between the neoR and tTA genes. Downstream from the tTA gene, the green fluorescent protein (GFP) gene is placed behind thePhCMV*-1/tetO promoter (Figure2A). The viral producer cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Cleveland, OH) supplemented with 1% penicillin/streptomycin (Gibco BRL) and 15% tetracycline-free fetal calf serum (FCStetfree; Clontech, Intermedica, Stockholm, Sweden). The NIT-GFP retroviral vector was transfected into amphotropic Phoenix cells (Dr Nolan, Stanford University Medical Center, Stanford, CA). After 48 hours, viral supernatants were collected and used to transduce ecotrophic GP+E-86 cells. Thereafter the cells were selected in the presence of 0.9 mg/mL G418 (Gibco BRL) for 14 days. The viral titer of the bulk GP+E-86/NIT-GFP producers cells was determined to be 0.6 to 0.8 × 106 by transfer of G418 resistance to NIH 3T3 cells.32 Absence of helper virus in the producer line was demonstrated by a marker rescue assay.33
The experimental design using the tTA vector.
(A) The NIT-GFP vector used in the experiment for transducingtetO-HOXA10 transgenic BM cells. LTR indicates long terminal repeats; neo, neomycin-resistance gene; IRES, internal ribosomal entry site; tTA, tetracycline transactivator; tetO, PhCMV*-1, minimal promoter fused to the tet operator (tetO) sequences; EGFP, enhanced green fluorescent protein; prestim, prestimulated; doxy, doxycycline; Liq, liquid; rad, cGy. (B) A schematic overview of the experimental design.
The experimental design using the tTA vector.
(A) The NIT-GFP vector used in the experiment for transducingtetO-HOXA10 transgenic BM cells. LTR indicates long terminal repeats; neo, neomycin-resistance gene; IRES, internal ribosomal entry site; tTA, tetracycline transactivator; tetO, PhCMV*-1, minimal promoter fused to the tet operator (tetO) sequences; EGFP, enhanced green fluorescent protein; prestim, prestimulated; doxy, doxycycline; Liq, liquid; rad, cGy. (B) A schematic overview of the experimental design.
Retroviral transduction of primary BM cells
The BM cells were harvested from 8- to 14-week-oldtetO-H0XA10 transgenic mice and C57BlxCBA control mice, 4 days after intravenous injection of 150 mg/kg body weight of 5-fluorouracil (5-FU; Nycomed, Stockholm, Sweden). Femurs and tibias were crushed carefully in a mortar in the presence of Iscoves modified Dulbecco medium (IMDM; Gibco BRL) containing 15% FCStetfree (Clontech), and then filtered through a 70-μm cell strainer. Single cell suspensions of approximately 5 × 105 cells/mL were cultured for 48 hours in IMDM containing 20% FCStetfree, 10−4β-mercaptoethanol (β-ME; Sigma Chemical, Stockholm, Sweden), 1%l-glutamine (Gibco BRL), 1% penicillin/streptomycin, 50 ng/mL human recombinant interleukin 6 (rhIL-6; Novartis, Basel, Switzerland), and rat recombinant stem cell factor (SCF; Amgen, Thousand Oaks, CA). The cells were then harvested, resuspended in the same medium supplemented with 4 μg/mL protamine sulfate, and plated at 5 × 105 cells/mL on mitomycin C–treated producer cells (24 hours before usage, the producer cells were treated with 5 μg/mL mitomycin C [Sigma] for 2 hours, washed 5 times in phosphate-buffered saline [PBS], and replated on gelatinized plates, 3 × 106 producer cells/plate). The BM cells were cocultivated for 48 hours. Nonadherent cells were recovered from the cocultures by repeated washes of the dishes and a final treatment with 1 mL cell dissociation buffer (Gibco BRL) for 1 minute at 37°C. The cells were washed once and then counted.
In vitro clonogenic progenitor assay
For myeloid clonogenic progenitor assays, cells were cultured in 35-mm Petri dishes (Stem Cell Technologies, Vancouver, BC, Canada) in a 1.1-mL mixture of 0.8% methylcellulose in alpha medium (Gibco BRL) supplemented with 30% FCS, 1% bovine serum albumin (BSA; Stem Cell Technologies), 10−4 β-ME, 5 U/mL human erythropoietin (hEpo; Janssen-Cilag, Sollentuna, Sweden), 2% spleen cell–conditioned medium (SCCM; Stem Cell Technologies) in the presence or absence of 1.4 mg/mL G418 and 2 μg/mL doxycycline (Sigma). Colonies were scored on days 10 to 12 of incubation as derived from macrophage colony-forming units (CFU-Ms), granulocyte-macrophage CFUs (CFU-GMs), erythroid burst-forming units (BFU-Es), or a mixed colony containing more than 2 cell types (CFU-Mix). The identification of colonies was confirmed by Wright-Giemsa staining of cytospin preparations of colonies. The transduction efficiency was estimated by FACS analysis of GFP expression, or in some experiments, colonies were picked randomly from the plates without G418, DNA was prepared, and the presence/absence of the retroviral vector was analyzed by PCR.
CFU-S12 assay
The NIT-GFP transduced BM cells derived from transgenic or control animals were injected into lethally irradiated (950 cGy, 110 cGy/min, 137Cs gamma rays) recipient C57BlxCBA mice either directly after retroviral cocultivation (day 0; d0) or after 7 days of culture (d7) in IMDM containing 30% FCStetfree, 1% BSA, 10−4 β-ME, 2% SCCM, 5 U/mL hEpo, and 1.4 mg/mL G418 in the presence or absence of 2 μg/mL doxycycline. The number of cells that each mouse received was adjusted to give 8 to 15 macroscopic colonies per spleen. The mice receiving doxycycline were given 0.2 mg/mL doxycycline plus 3% sucrose in the drinking water. Twelve days after injection, animals were killed and the number of macroscopic colonies on the spleen was evaluated. An aliquot of the d7 cultured cells was taken and used for preparations of total RNA (RNeasy, Qiagen) for Northern blot analysis to verify the presence or absence of the RNA transcripts of the H0XA10 transgene. The 742-bpXhoI/DraI fragment was used as a probe. RNA (20 μg) was used for blotting (PerfectHyb-Plus, Sigma). In some cases, well-isolated colonies were cut open with scissors and the cells were either transferred to microscopic slides and stained with Wright-Giemsa for morphologic analysis or lysed for isolation of DNA or RNA for subsequent PCR or RT-PCR analysis, respectively.
Flow cytometry
Flow cytometric analysis (FACS) was used to determine the level of GFP expression for measurements of transduction efficiency as well as measurement of inducibility and down-regulation of GFP expression in the absence or presence of doxycycline. Peripheral blood and, in some cases, BM cells, were treated with ammonium chloride (Stem Cell Technologies) prior to the FACS procedure. Results were analyzed with CellQuest software (Becton Dickinson, San Jose, CA).
Results
Generation and characterization of HOXA10 transgenic mice
The HOXA10 cDNA used in this study is complementary to a messenger RNA (mRNA), which represents the most abundant HOXA10 transcript in human BM cells.34 This cDNA was subcloned into the pUHG10-3 plasmid where it is placed under the control of the regulatable promoter, PhCMV*-1/tetO. The rabbit β-globin intron and poly A sequence is placed downstream of theHOXA10 gene to generate a stable mRNA from the transgene (Figure 1A). The expression of the HOXA10 cDNA is therefore dependent on the activation of PhCMV*-1/tetO by the tTA, which is composed of the tet repressor and the activation domain of viral protein VP16 of herpes simplex virus.29Transgenic mice bearing the tetO-HOXA10 construct were generated and 19 founders were shown to be positive when examined by PCR and Southern blot analysis. Nine founder mice transmitted thetetO-HOXA10 transgene through germline. In these mice the number of transgene copies ranged from a few to more than 50 as estimated by Southern blot analysis (Figure 1B and Table1).
Identification of inducible tet0-HOXA10 transgenic mice
| Mouse line . | Transgene copies . | Background expression . | Induction with tTA without doxycycline . | Leaky with tTA with doxycycline . |
|---|---|---|---|---|
| 2-5 | 60 | − | +++ | − |
| 6-2 | 10 | − | +++ | − |
| 12-5 | 5-10 | + | +++ | + |
| 13-1 | < 5 | + | ++ | + |
| 13-4 | 20 | − | + | − |
| 14-1 | 10 | + | ++ | + |
| 15-1 | 20 | − | +++ | + |
| 17-3 | 60 | − | +++ | − |
| 17-5 | < 5 | ++ | ++ | ++ |
| Mouse line . | Transgene copies . | Background expression . | Induction with tTA without doxycycline . | Leaky with tTA with doxycycline . |
|---|---|---|---|---|
| 2-5 | 60 | − | +++ | − |
| 6-2 | 10 | − | +++ | − |
| 12-5 | 5-10 | + | +++ | + |
| 13-1 | < 5 | + | ++ | + |
| 13-4 | 20 | − | + | − |
| 14-1 | 10 | + | ++ | + |
| 15-1 | 20 | − | +++ | + |
| 17-3 | 60 | − | +++ | − |
| 17-5 | < 5 | ++ | ++ | ++ |
Copy number in the tetO-HOXA10 transgenic mouse lines was estimated by Southern blot analysis. To induce transgene expression in hematopoietic cells, BM cells were transduced with the NIT-GFP vector and grown in liquid culture for 5 days with (uninduced) or without (induced) doxycycline. RNA from transduced cells, with and without induction, was analyzed by RT-PCR. The number of + signs indicates the level of HOXA10 transgene expression; + weak expression, ++ intermediate expression, and +++ strong expression.
HOXA10 is inducible in tetO-HOXA10 hematopoietic cells
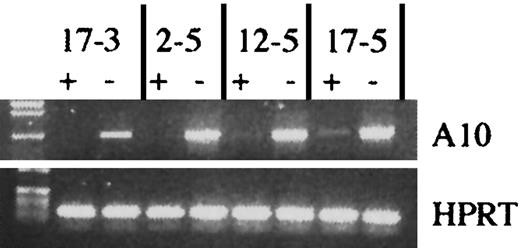
To identify which HOXA10 transgenic lines were optimal with regard to inducibility of HOXA10 expression in the hematopoietic system, we first asked whether there was any leakiness in the expression of the HOXA10 cDNA in peripheral blood cells from uninduced transgenic animals. Total RNA was isolated from peripheral blood cells and RT-PCR was performed. Five of the lines did not express HOXA10 RNA (no background), 3 showed low levels of the transcript, and one line had a somewhat higher expression level in the uninduced state (Table 1). Next, we asked whether the expression of HOXA10 could be induced in the presence of tTA and, furthermore, if it could be down-regulated in the presence of doxycycline. Figure 2 shows the retroviral vector (NIT-GFP) and the experimental design for induction of the transgene. Four days after treatment with 5-FU, BM cells were harvested from thetetO-HOXA10 mouse lines, prestimulated for 48 hours, and then cocultivated with NIT-GFP viral vector producer cells for an additional 48 hours. The NIT-GFP vector contains theneoR along with the tTA gene and thetetO-GFP, which acts as an inducible marker gene responsive to tTA. This results in a simultaneous expression of HOXA10and GFP genes in the absence of doxycycline and down-regulation of both these genes in the presence of doxycycline. After transduction the cells were grown in selective medium containing G418 (1.4 mg/mL) for 5 days in the presence or absence of doxycycline (2 μg/mL). Through RT-PCR amplification of the transgene, the effect of doxycycline on the expression of HOXA10 could be monitored. Five lines showed high induction of HOXA10 expression, whereas the others had moderate to low induction (Figure 3and data not shown). No direct correlation could be seen between levels of induction or leakiness and copy number of the transgene (Table 1). Three transgenic lines (2-5, 6-2, 17-3) were identified where the HOXA10 transgene was highly inducible, whereas no expression was detected in the uninduced state (no leakiness with or without tTA) in transgenic peripheral blood or NIT-GFP–transduced BM cells. These findings demonstrate that cells from these transgenic lines can be induced to express HOXA10 to high levels from a baseline of practically zero in the uninduced state.
Induction of transgenic HOXA10 expression in vitro.
Induction of HOXA10 transgene expression in 4 different transgenic mouse lines. tetO-HOXA10 BM cells were transduced with the NIT-GFP vector and then grown in selective medium in the presence (+) or absence (−) of doxycycline (2 μg/mL) for 5 days. RT-PCR was performed on RNA isolated from cultured cells using the A10 and BG primers amplifying a 950-bp band. For RNA/DNA control, HPRT primers were used. PCR was also performed on isolated RNA to verify the absence of genomic DNA contamination (data not shown).
Induction of transgenic HOXA10 expression in vitro.
Induction of HOXA10 transgene expression in 4 different transgenic mouse lines. tetO-HOXA10 BM cells were transduced with the NIT-GFP vector and then grown in selective medium in the presence (+) or absence (−) of doxycycline (2 μg/mL) for 5 days. RT-PCR was performed on RNA isolated from cultured cells using the A10 and BG primers amplifying a 950-bp band. For RNA/DNA control, HPRT primers were used. PCR was also performed on isolated RNA to verify the absence of genomic DNA contamination (data not shown).
Induction of HOXA10 expression leads to proliferation of blastlike colonies in vitro
Because HOXA10 expression is inducible and can be regulated in BM cells derived from the tet0-HOXA10 transgenic mice, we wanted to analyze whether the induction would affect proliferation and differentiation of hematopoietic progenitors in vitro. The experimental design is described in Figure 2B. BM cells, harvested fromtetO-HOXA10 transgenic mouse lines 2-5 and 17-3 and control C57BL/6xCBA mice, were transduced with the NIT-GFP vector and plated in methylcellulose cultures for myeloid colony formation in the presence or absence of doxycycline (2 μg/mL). The proportion of G418-resistant colonies following transduction was 19.2% ± 4.1% and 19.8% ± 4.8% for transgenic and control cells, respectively (values are means ± SD from 4 independent experiments). The plating efficiency was similar for both types of cells and was not affected by addition of doxycycline (about 50 colonies/1000 cells). However, when G418-resistant methylcellulose colonies were analyzed on the plates after Wright-Giemsa staining, a clear difference was observed between the groups. Approximately 30% of the transduced progenitors derived from the transgenic mice formed large colonies containing megakaryocytes and blast cells in the absence of doxycycline (Figure 4A,B). This colony type was not seen in the presence of doxycycline nor in the transduced colonies derived from control mice. As previously described, the appearance of this special colony type was accompanied by a lower frequency of the CFU-GM and CFU-Mix colonies.25 Another striking feature was the absence of unilineage macrophage colonies among transduced transgenic progenitors in the absence of doxycycline as compared to approximately 20% frequency of CFU-M colonies in the presence of doxycycline and in the control settings (Figure 4A). The effects of homeobox transcription factors have been shown to be concentration dependent during development,35-37 and the same phenomenon could apply to the hematopoietic system as well. In light of this, it is noteworthy that the expression levels of HOXA10 that can be transiently induced are more than sufficient to generate a striking proliferative effect in the most primitive progenitors (megakaryocyte-blast progenitors) and to influence commitment in less primitive progenitors.
Induced expression of HOXA10 affects colony formation in methylcellulose culture.
(A) Depicted is the frequency of CFU-M, CFU-GM, CFU-Mix, and the unique megakaryocytic-blast colonies. BM cells from tetO-HOXA10 and control mice were transduced with the NIT-GFP vector and plated out on methylcellulose. On day 11 to 12, colonies (n = 434 for A10−doxy, n = 346 for A10+doxy, n = 158 for control cells) were scored on the plates. The morphology of a number of the colonies was further verified by picking single G418-resistant colonies for examination by Wright-Giemsa staining. Results are expressed as means ± SD from 4 independent experiments. (B) Day 12 methylcellulose colonies, derived from NIT-GFP–transduced tetO-HOXA10 progenitor cells grown in the absence of doxycycline. (Bi) Colony expressing the GFP marker gene (original magnification × 40). (Bii) Wright-Giemsa staining of cytospin preparation of methylcellulose colony containing megakaryocytes (Meg) and blastlike (b) cells (CFU-Meg/blast) formed by cells overexpressing HOXA10 (original magnification × 200).
Induced expression of HOXA10 affects colony formation in methylcellulose culture.
(A) Depicted is the frequency of CFU-M, CFU-GM, CFU-Mix, and the unique megakaryocytic-blast colonies. BM cells from tetO-HOXA10 and control mice were transduced with the NIT-GFP vector and plated out on methylcellulose. On day 11 to 12, colonies (n = 434 for A10−doxy, n = 346 for A10+doxy, n = 158 for control cells) were scored on the plates. The morphology of a number of the colonies was further verified by picking single G418-resistant colonies for examination by Wright-Giemsa staining. Results are expressed as means ± SD from 4 independent experiments. (B) Day 12 methylcellulose colonies, derived from NIT-GFP–transduced tetO-HOXA10 progenitor cells grown in the absence of doxycycline. (Bi) Colony expressing the GFP marker gene (original magnification × 40). (Bii) Wright-Giemsa staining of cytospin preparation of methylcellulose colony containing megakaryocytes (Meg) and blastlike (b) cells (CFU-Meg/blast) formed by cells overexpressing HOXA10 (original magnification × 200).
HOXA10 expression can be regulated in vivo
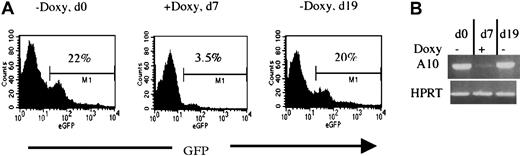
To verify whether the HOXA10 induction can be applied in vivo, 2 lines (2-5 and 17-3) were also tested for regulatable expression of HOXA10 and GFP in mice receiving transplants of NIT-GFP–transducedtetO-HOXA10 BM cells. Directly after the transduction, 106 cells were injected into lethally irradiated recipients. By adding or removing doxycycline from the drinking water of the mice, both HOXA10 and GFP expression in peripheral blood cells could be regulated as measured by RT-PCR and FACS analysis. Representative results from a recipient of transduced BM from the 2-5 line are shown in Figure 5. Induction of both GFP and HOXA10 expression was seen in the absence of doxycycline (d0). Doxycycline was then administrated for 7 days and the expression of both HOXA10 and GFP was clearly down-regulated (d7). Withdrawal of doxycycline led to up-regulation of both the genes when measured 12 days later (d19). These results show that the expression of HOXA10 can clearly be regulated in vivo by administration and withdrawal of doxycycline to the transgenic mice.
Regulation of HOXA10 and GFP expression in vivo.
Directly after NIT-GFP transduction, 106tetO-HOXA10 cells were injected into lethally irradiated recipients. The expression of GFP and HOXA10 was then monitored by FACS and RT-PCR, respectively. Representative results from one recipient mouse receiving transplants of NIT-GFP–transduced BM cells from line 2-5 are presented. (A) Percentage of GFP-expressing cells in the peripheral blood of the recipient 10 weeks after transplantation, in the absence (d0 and d19) or presence (d7) of doxycycline. (B) HOXA10 expression levels in the peripheral blood at the indicated time points. The levels were measured by performing RT-PCR on total RNA isolated from leukocytes. Absence of DNA contamination was also verified (data not shown).
Regulation of HOXA10 and GFP expression in vivo.
Directly after NIT-GFP transduction, 106tetO-HOXA10 cells were injected into lethally irradiated recipients. The expression of GFP and HOXA10 was then monitored by FACS and RT-PCR, respectively. Representative results from one recipient mouse receiving transplants of NIT-GFP–transduced BM cells from line 2-5 are presented. (A) Percentage of GFP-expressing cells in the peripheral blood of the recipient 10 weeks after transplantation, in the absence (d0 and d19) or presence (d7) of doxycycline. (B) HOXA10 expression levels in the peripheral blood at the indicated time points. The levels were measured by performing RT-PCR on total RNA isolated from leukocytes. Absence of DNA contamination was also verified (data not shown).
Induction of HOXA10 expression leads to increased proliferation of CFU-S12 progenitors
Because blastlike colonies could be generated in methylcellulose on HOXA10 induction, we wanted to determine whether primitive myeloid progenitors (CFU-S12) could be stimulated to proliferate in a regulatable manner. Constitutive overexpression of HOXA10 in murine BM cells has been shown to increase proliferation of primitive CFU-S12 progenitors when cultured for 7 days after the retroviral transduction.25 Therefore, we transduced BM cells from the tetO-HOXA10 mice as well as wild-type control mice with the NIT-GFP retroviral vector. The transduced cells were injected into lethally irradiated recipients either directly after transduction or after 7 days in selective liquid medium (1.4 mg/mL G418) with or without doxycycline as shown in Figure 2B. The frequency of day 12 CFU-S colonies was similar for all groups when measured directly after the retroviral transduction (Table2). However, induced expression of HOXA10 during the 7-day culture increased the day 12 CFU-S content of the culture approximately 3-fold as compared to transduced control cells or transgenic cells grown in the presence of doxycycline (Table 2 and Figure 6). The HOXA10 expression in the 7-day culture was verified by Northern blot analysis (Figure 6C). These findings demonstrate that transient induction of HOXA10 increases the proliferation of primitive myeloid progenitors with CFU-S12 biopotency.
Number of CFU-Sd12 colonies before and after HOXA10 induction
| Mouse line . | Number of spleen colonies/104 input cells with doxycycline . | Number of spleen colonies/104 input cells without doxycycline . |
|---|---|---|
| HOXA10 2-5 | 7.8 ± 0.8 (n = 6) | 7.2 ± 1.5 (n = 6) |
| HOXA10 17-3 | 7.4 ± 1.5 (n = 5) | 7.0 ± 0.7 (n = 5) |
| Control mice | Not done | 7.2 ± 1.6 (n = 5) |
| Mouse line . | Number of spleen colonies/104 input cells with doxycycline . | Number of spleen colonies/104 input cells without doxycycline . |
|---|---|---|
| HOXA10 2-5 | 7.8 ± 0.8 (n = 6) | 7.2 ± 1.5 (n = 6) |
| HOXA10 17-3 | 7.4 ± 1.5 (n = 5) | 7.0 ± 0.7 (n = 5) |
| Control mice | Not done | 7.2 ± 1.6 (n = 5) |
Number of day 12 CFU-S macroscopic spleen colonies derived from 104 input BM cells that were transduced with NIT-GFP and injected immediately after the viral coculture. Doxycycline was added to the drinking water (0.2 mg/mL in 3% sucrose) of the corresponding recipient mice. Results are expressed as means ± SD from 2 independent experiments. Numbers in parentheses indicate the number of spleens analyzed.
HOXA10 induction increases the number of CFU-Sd12 colonies.
(A) Average number of day 12 CFU-S colonies derived from BM cells (tetO-HOXA10 and wild-type mice) that were grown for 7 days after NIT-GFP transduction in selective medium (G418, 1.4 mg/mL) in the presence or absence of doxycycline before injection into irradiated recipients. Data from 2 transgenic lines are shown as the day 12 CFU-Sd12 content per 104 starting day 0 cells. Results (number of colonies [col] ± SD) from line 2-5 as well as the wild-type (wt) mice are based on 2 independent experiments and findings from the 17-3 transgenic line represent one experiment. Doxy indicates doxycycline. The number of spleens (n) analyzed in each group is indicated. (B) Transgene expression in day 12 spleen colonies derived from NIT-GFP–transduced tetO-HOXA10 progenitors. (Bi) Expression of the GFP marker gene on induction (original magnification × 40). (Bii) HOXA10 expression as measured by RT-PCR is detected in the absence of doxycycline, whereas expression is absent in the presence of doxycycline. The HPRT control lane contains a mixture of cDNA and genomic DNA, where the DNA gives rise to the larger band, which is absent in the other lanes, demonstrating lack of contamination by genomic DNA. (C) Northern blot analysis verifies the induction of HOXA10 RNA in NIT-GFP–transducedtetO-HOXA10 BM cells cultured for 7 days in the absence of doxycycline (−Doxy). In contrast, no detectable HOXA10 transgene expression is seen in cells grown in the presence of doxycycline (+Doxy). Total RNA (20 μg) was loaded from each sample and theXhoI/DraI HOXA10 probe was used for hybridization. The control lane contains RNA from NIT-GFP–transduced normal BM cells. Time of exposure was 24 hours for HOXA10 and 10 hours for glyceraldehyde 3-phosphate dehydrogenase (G3PDH).
HOXA10 induction increases the number of CFU-Sd12 colonies.
(A) Average number of day 12 CFU-S colonies derived from BM cells (tetO-HOXA10 and wild-type mice) that were grown for 7 days after NIT-GFP transduction in selective medium (G418, 1.4 mg/mL) in the presence or absence of doxycycline before injection into irradiated recipients. Data from 2 transgenic lines are shown as the day 12 CFU-Sd12 content per 104 starting day 0 cells. Results (number of colonies [col] ± SD) from line 2-5 as well as the wild-type (wt) mice are based on 2 independent experiments and findings from the 17-3 transgenic line represent one experiment. Doxy indicates doxycycline. The number of spleens (n) analyzed in each group is indicated. (B) Transgene expression in day 12 spleen colonies derived from NIT-GFP–transduced tetO-HOXA10 progenitors. (Bi) Expression of the GFP marker gene on induction (original magnification × 40). (Bii) HOXA10 expression as measured by RT-PCR is detected in the absence of doxycycline, whereas expression is absent in the presence of doxycycline. The HPRT control lane contains a mixture of cDNA and genomic DNA, where the DNA gives rise to the larger band, which is absent in the other lanes, demonstrating lack of contamination by genomic DNA. (C) Northern blot analysis verifies the induction of HOXA10 RNA in NIT-GFP–transducedtetO-HOXA10 BM cells cultured for 7 days in the absence of doxycycline (−Doxy). In contrast, no detectable HOXA10 transgene expression is seen in cells grown in the presence of doxycycline (+Doxy). Total RNA (20 μg) was loaded from each sample and theXhoI/DraI HOXA10 probe was used for hybridization. The control lane contains RNA from NIT-GFP–transduced normal BM cells. Time of exposure was 24 hours for HOXA10 and 10 hours for glyceraldehyde 3-phosphate dehydrogenase (G3PDH).
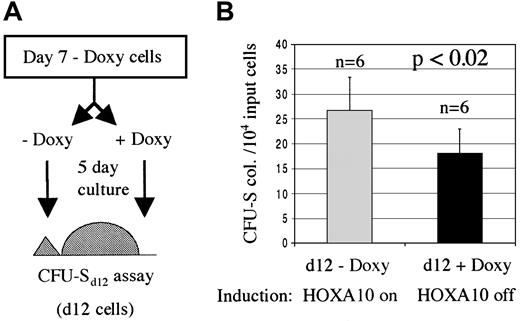
HOXA10-induced proliferation of primitive myeloid progenitors is reversible
To verify that the enhanced proliferation of primitive progenitors was a direct result of HOXA10 expression, we asked whether these effects could be reversed by down-regulating the expression of HOXA10 following the initial induction. For this, an aliquot was taken from the NIT-GFP–transduced tetO-HOXA10 BM cells that had been cultured for 7 days in selective medium in the absence of doxycycline and were used in the previously described CFU-S12 assay. These cells were split into 2 cultures for an additional 5 days where the HOXA10 expression was either down-regulated (+ doxycycline) or maintained in an induced state (− doxycycline). The cells were then injected into lethally irradiated recipient mice and 12 days later the spleens of these mice were harvested and the colonies enumerated. Mice receiving cells with up-regulated HOXA10 expression had approximately 40% higher number of colonies in their spleens as compared with mice that were injected with cells grown in the presence of doxycycline for the last 5 days (Figure 7). These results indicate a direct and reversible effect of induced HOXA10 expression on the proliferation of primitive myeloid progenitors.
The inducible proliferative effect on CFU-Sd12 progenitors is reversible.
HOXA10 production was induced for 7 days and then either down-regulated (+Doxy) or the induction was kept on (−Doxy). (A) An aliquot of transduced transgenic cells that had been cultured for 7 days in the absence of doxycycline was taken and split into 2, 5-day cultures with or without doxycycline. The cells were then injected into lethally irradiated recipients for a CFU-Sd12 assay. (B) Number of day 12 spleen colonies per 10 000 input cells. Results are shown as number of colonies ± SD from 1 of 2 experiments. Doxy indicates doxycycline; n, number of mice.
The inducible proliferative effect on CFU-Sd12 progenitors is reversible.
HOXA10 production was induced for 7 days and then either down-regulated (+Doxy) or the induction was kept on (−Doxy). (A) An aliquot of transduced transgenic cells that had been cultured for 7 days in the absence of doxycycline was taken and split into 2, 5-day cultures with or without doxycycline. The cells were then injected into lethally irradiated recipients for a CFU-Sd12 assay. (B) Number of day 12 spleen colonies per 10 000 input cells. Results are shown as number of colonies ± SD from 1 of 2 experiments. Doxy indicates doxycycline; n, number of mice.
Discussion
The development of inducible gene expression systems that function well in vivo to demonstrate biologic effects has been problematic. However, the advent of the tetracycline system has changed the possibilities considerably because background levels are relatively low and the inducibility high, often reaching several orders of magnitude above background in tissue culture cells.29 Several reports have also demonstrated that the tetracycline system functions in vivo, including studies that demonstrate biologic effects,29,38-43 for example, induction of reversible acute B-cell leukemia by BCR-ABL144 and an inducible block in transforming growth factor-β (TGF-β) signaling using a dominant-negative TGF-β receptor mutant.45 Here, we present a novel transgenic mouse model in which the expression of HOXA10 can be regulated on demand. On expression of tTA through a retroviral vector, the HOXA10 transgene is effectively induced in the absence of doxycycline, whereas the transgene is silent in the presence of doxycycline. Induction of HOXA10 expression generates characteristic blast cell colonies that also contain megakaryocytes and formation of pure macrophage colonies is completely blocked. Furthermore, induced expression of HOXA10 leads to increased proliferation of primitive myeloid progenitors as measured by increased numbers of CFU-S colonies. Interestingly, the latter effect was found to be reversible on down-regulation of HOXA10 in the presence of doxycycline. The down-regulation led to approximately 40% decrease in the number of CFU-S colonies when compared to constitutively induced HOXA10 expression. The reason as to why this decrease, although significant, is relatively moderate, could be that during the initial 7-day culture in the absence of doxycycline, high levels of CFU-S progenitors have accumulated. Therefore, even though the addition of doxycycline for the last 5 days of culture down-regulates the proliferation of these progenitor cells, there might still remain relatively high numbers of them, giving rise to spleen colonies in the irradiated recipients. Of course, we cannot exclude that there may be some other secondary factors contributing to these findings. However, these data strongly support the general hypothesis that HOXA10 by itself directly regulates proliferation of primitive myeloid progenitors.25 Our results demonstrate that the levels of HOXA10 that can be induced in the tet0-HOXA10 mice are high enough to alter lineage decisions and increase the proliferative capabilities of the target cells. It is noteworthy that the development of single lineage macrophage colonies is completely inhibited in the presence of inducible HOXA10, whereas macrophages can develop from HOXA10-induced CFU-GM bipotential progenitors. More insight into the mechanism of HOXA10 action and its interplay with other factors that control myeloid development is required to understand this apparent discrepancy.
Whether the levels of HOXA10 in this inducible model are sufficient to give rise to acute myeloid leukemia, as reported by Thorsteinsdottir and colleagues,25 is currently under investigation. In preliminary experiments, mice receiving transplants of NIT-GFP–transduced tetO-HOXA10 BM cells have been monitored for up to a year without signs of leukemic development (data not shown). However, the long latency period observed before the onset of the leukemia25 strongly argues for the need of a secondary mutation(s), and indeed, here the genetic background of the mice might play a role as well. Simultaneous overexpression of cofactors of HOXA10 might facilitate the leukemic progression and this might even prove to be necessary for the leukemia to appear if the levels of HOXA10 expression do not reach the threshold needed. Should this be the case, then these mice could provide an important background to screen for genes that can collaborate and cause myeloproliferation or leukemia. Another way to enhance the potential leukemic development is to sort for transduced tetO-HOXA10 BM cells before transplantation, thereby increasing the overall impact of induced HOXA10 expression. We are currently addressing these questions.
The effects of HOXA10 and closely related transcription factors on proliferation and differentiation of primitive hematopoietic progenitors have been documented, but the molecular mechanisms causing these effects are still poorly understood. Although most downstream target genes of Hox transcription factors remain elusive, a few examples have been emerging. The target genes even include other homeobox transcription factors, generating complex cross-regulatory and autoregulatory pathways.46-49 Recent findings also suggest the involvement of Hox transcription factors in various other pathways; that is, HoxA5 affects expression of p5350 and the progesterone receptor,51 and HoxA9 can interact with Smad proteins to repress TGF-β–induced transcription of theosteopontin gene.52 With regard to target genes of HOXA10, the cyclin-dependent kinase inhibitor, p21waf1/cip1 has been suggested as a transcriptional target of HOXA10 in differentiating myelomonocytic cells.53 In a recent study by Eklund and coworkers,54 a regulatory role for HOXA10 in myelopoiesis is revealed. The study shows that HOXA10 can repress transcription of the NCF2 and CYBB genes, which code for p67phox and gp91phox, respectively. These respiratory burst oxidase components are expressed only in myeloid cells that have differentiated beyond the promyelocyte stage. HOXA10- mediated repression of NCF2 and CYBB transcription might therefore contribute to the differentiation block seen in myeloid leukemia caused by overexpression of HOXA10. The repression involves recruitment of histone deacetylases, which results in tightly bound DNA/histone complex and decreased access for transcription. This repression mechanism has also been reported by Saleh and colleagues for HOX-PBX complexes.55 Retrovirally engineered overexpression studies on the role of cofactors of Hox proteins, Meis1 and PBX, have revealed their importance for the onset of leukemia.22 26-28 However, the formation and action of the transcriptional protein complex are not well understood. Therefore, even though data are emerging on the role and molecular actions of Hox transcription factors, the picture still remains largely unclear and complex. The new animal model described here can be used in future experiments to reveal new target genes of HOXA10 and unravel new and hitherto unknown regulatory pathways that mediate the effects of HOXA10 on proliferation and commitment of hematopoietic progenitors.
The availability of new transgenic animals, in which tTA expression is restricted to certain tissues or even lineages, will enable studies where HOXA10 can be induced in a tissue-specific manner. Therefore, thetet0-HOXA10 mouse should be of value to many investigators who aim to study the effects of HOXA10 on the development, differentiation, and proliferation of cells in vivo.
We would like to thank Kristina Sundgren and Eva Gynnstam for expert animal care, Lilian Wittman for help with animal experiments, Rusty Gage for providing the NIT-GFP vector, and Sten Eirik Jacobsen and members of the Department of Stem Cell Biology, Lund University, for helpful advice and discussions.
Supported by grants from Cancerfonden, Sweden; Barncancerfonden, Sweden; Astra Draco (now AstraZeneca); and the Donation Funds Lund University Hospital (to S.K.); and by a clinical research award (ALF) from Lund University Hospital to S.K. and N.L. J.M.B. was supported by a graduate student award from the Medical Faculty, Lund University.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Stefan Karlsson, Molecular Medicine and Gene Therapy, Lund University Hospital, BMC A12, 221 84 Lund, Sweden; e-mail: stefan.karlsson@molmed.lu.se.




![Fig. 6. HOXA10 induction increases the number of CFU-Sd12 colonies. / (A) Average number of day 12 CFU-S colonies derived from BM cells (tetO-HOXA10 and wild-type mice) that were grown for 7 days after NIT-GFP transduction in selective medium (G418, 1.4 mg/mL) in the presence or absence of doxycycline before injection into irradiated recipients. Data from 2 transgenic lines are shown as the day 12 CFU-Sd12 content per 104 starting day 0 cells. Results (number of colonies [col] ± SD) from line 2-5 as well as the wild-type (wt) mice are based on 2 independent experiments and findings from the 17-3 transgenic line represent one experiment. Doxy indicates doxycycline. The number of spleens (n) analyzed in each group is indicated. (B) Transgene expression in day 12 spleen colonies derived from NIT-GFP–transduced tetO-HOXA10 progenitors. (Bi) Expression of the GFP marker gene on induction (original magnification × 40). (Bii) HOXA10 expression as measured by RT-PCR is detected in the absence of doxycycline, whereas expression is absent in the presence of doxycycline. The HPRT control lane contains a mixture of cDNA and genomic DNA, where the DNA gives rise to the larger band, which is absent in the other lanes, demonstrating lack of contamination by genomic DNA. (C) Northern blot analysis verifies the induction of HOXA10 RNA in NIT-GFP–transducedtetO-HOXA10 BM cells cultured for 7 days in the absence of doxycycline (−Doxy). In contrast, no detectable HOXA10 transgene expression is seen in cells grown in the presence of doxycycline (+Doxy). Total RNA (20 μg) was loaded from each sample and theXhoI/DraI HOXA10 probe was used for hybridization. The control lane contains RNA from NIT-GFP–transduced normal BM cells. Time of exposure was 24 hours for HOXA10 and 10 hours for glyceraldehyde 3-phosphate dehydrogenase (G3PDH).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/98/12/10.1182_blood.v98.12.3301/6/m_h82311785006.jpeg?Expires=1767727147&Signature=qdkvRulxLbTGrTlt-4CenZQn89VWUalNNknDXUCN0RMdTe4eHWsRdp1NzA9jBAt~VkmbeIu4QM0KrPKtyo9iZMmbCvHllip1o6L2Fea~wfddWz8abF0doMBi~whMi7~FuPpvlqJRv-aE9cPqmf6h9lpDMrarQ9ot-tVHqFosZSkff4faZ3JSGiSdJYuIThLlEWU~QpF6WxDb74-kMIGlvcOkXn7r78sA8A0gxqwrl3XY7UNkBO~lPb0XbyCy6Ku64XOFH2775dK4f1qWXxGDjlLqOKbwbc2d-J~AGbdvbBJQxEJsgjOdliNXb60M8nvXHClgd6JriI47wmfoYkZheg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal