Abstract
The clinical importance of HLA class II gene disparity in unrelated stem cell transplantation is not entirely known. The impact was evaluated of matching donors and recipients for HLA-DR, HLA-DQ, and HLA-DP genes on clinical outcome after stem cell transplantation for chronic myeloid leukemia (CML) performed between 1988 and 1997. HLA-DRB1, -DQA1, -DQB1, -DPA1, and -DPB1 alleles were identified in 831 transplant pairs using a combination of sequence-specific oligonucleotide probes, sequence-specific priming, and sequencing methods. Among the 831 pairs, 696 (84%) were HLA-A and -B serologically matched; of these, 565 (81%) were also matched for HLA-DRB1. HLA-DRB1 matching correlated with significantly improved survival (relative risk [RR], 1.29 [95% confidence interval (CI), 1.02-1.64; P = .04]) independently of HLA-DQA1 or HLA-DQB1 (RR, 1.01 [95% CI, 0.81-1.26; P = .94]) and HLA-DPA1 or HLA-DPB1 (RR, 1.11 [95% CI, 0.84-1.48;P = .46]). Single-locus HLA-DQ or HLA-DP disparity was not associated with significantly poorer survival. For patients who underwent transplantation in the first chronic phase (CP) from HLA-A, B matched donors, the presence of DRB1 allele mismatching was independently associated with increased incidence of grades III-IV acute graft-versus-host disease (GVHD). No significant associations of class II allele mismatching with risk for delayed engraftment or chronic GVHD disease were detected. This study clearly demonstrates the importance of precise matching of HLA-DRB1 alleles for successful transplantation. Furthermore, a good-risk population of patients whose transplantations were performed in the first CP of disease from HLA-A, B, DRB1 matched unrelated donors can be shown to have superior survival.
Introduction
Hematopoietic stem cell transplantation is a well-established therapeutic modality for malignancies of the bone marrow.1-12 Only 30% of patients in need of a transplant, however, will have a genotypically matched sibling to serve as the donor. Transplantation from unrelated volunteer donors has been facilitated by the establishment of donor registries worldwide.13-18 The challenge of unrelated donor transplantation is the need to overcome the histocompatibility barrier to achieve engraftment, minimize graft-versus-host disease (GVHD), and facilitate immunologic tolerance.19 20
Historically, the selection of stem cell donors was based on serologic methods using highly selected antisera to identify the phenotypes of HLA antigens.21,22 Recently, the application of DNA typing methods has disclosed extensive diversity among human populations for the HLA class II genes HLA-DR, HLA-DQ, and HLA-DP.23,24 A single HLA-DR antigen can be encoded by one of several unique HLA-DRB1 alleles. Clinical experience has demonstrated significant increases in severe GVHD and graft failure, and lower survival in recipients of HLA phenotype-matched unrelated donor stem cell transplants compared to genotype-matched sibling transplants.25 Disparity for HLA class II alleles between phenotypically matched unrelated donors and recipients may account for the increased posttransplantation complications. We assessed the impact of donor matching for HLA class II genes on clinical outcome in patients who underwent stem cell transplantation for the treatment of chronic myeloid leukemia (CML) facilitated by the U.S. National Marrow Donor Program (NMDP). Patients and donors were retrospectively characterized for alleles encoded by HLA-DR, -DQ, and -DP, and they provided a unique clinical population with which to evaluate the biologic significance of multilocus class II allele disparity.
Patients, materials, and methods
Study patients and donors
To ensure no identifiable bias was introduced into the data set, the 831 cases retrospectively HLA typed for this study were compared with other CML transplants (excluding blast phase) facilitated by the NMDP in the same transplant centers during the same time period from February 1988 through April 1997 (n = 736). The other CML cases were not included in this study because a blood sample was unavailable for donor or recipient (n = 522), alleles could not be fully resolved (n = 126), or typing was scheduled for a later date (n = 88). Both groups had comparable rates of survival, relapse, relapse-free survival, severe acute GVHD, and chronic GVHD in univariate and multivariate analyses (data not shown). These results indicate that no identifiable bias was introduced by the exclusion of the cases. Of the 831 included patients, 70% underwent transplantation in the first chronic phase (CP), 23% underwent it in the accelerated phase (AP), and 8% underwent it in the second CP after blast phase. Characteristics of the 831 patients and donors are summarized in Table1.
Demographics of the study population (n = 831)
| Characteristics . | No. of patients (%) . |
|---|---|
| HLA-A, -B serologic matches | 696 (84) |
| T-cell depleted | 177 (21) |
| Year of transplantation | |
| 1988-1989 | 73 (9) |
| 1990-1993 | 383 (46) |
| 1994-1997 | 375 (45) |
| First chronic phase | 578 (70) |
| Accelerated phase | 189 (23) |
| Second chronic phase | 64 (8) |
| Gender (recipient–donor) | |
| Female/female | 171 (21) |
| Female/male | 176 (21) |
| Male/female | 187 (23) |
| Male/male | 297 (36) |
| Conditioning regimen | |
| Cy + TBI | 458 (55) |
| Cy + TBI + other | 246 (30) |
| Cy + Bu | 102 (12) |
| Other combination | 25 (3) |
| Recipient CMV status at transplantation | |
| Positive | 401 (48) |
| Negative | 425 (51) |
| Missing/undetermined | 5 (1) |
| Characteristics . | No. of patients (%) . |
|---|---|
| HLA-A, -B serologic matches | 696 (84) |
| T-cell depleted | 177 (21) |
| Year of transplantation | |
| 1988-1989 | 73 (9) |
| 1990-1993 | 383 (46) |
| 1994-1997 | 375 (45) |
| First chronic phase | 578 (70) |
| Accelerated phase | 189 (23) |
| Second chronic phase | 64 (8) |
| Gender (recipient–donor) | |
| Female/female | 171 (21) |
| Female/male | 176 (21) |
| Male/female | 187 (23) |
| Male/male | 297 (36) |
| Conditioning regimen | |
| Cy + TBI | 458 (55) |
| Cy + TBI + other | 246 (30) |
| Cy + Bu | 102 (12) |
| Other combination | 25 (3) |
| Recipient CMV status at transplantation | |
| Positive | 401 (48) |
| Negative | 425 (51) |
| Missing/undetermined | 5 (1) |
Categories are based on combinations of Cy, TBI, VP16, BU, Ara-C, ALG, and Thiotepa. In some patients, other drugs were also used but were ignored for purposes of this classification.
The median recipient age was 35 years (range, 2-63 years). The median donor age was 38 years (range, 18-56 years). The median disease duration was 1.4 years (range, 0.2-18.2 years); the diagnosis date was missing for 2 cases.
Cy indicates cyclophosphamide; TBI, total body irradiation; Bu, busulfan; ALG, antilymphocyte globulin.
Typing and matching of HLA class II alleles
At the time of the transplantation, approximately 60 mL whole blood from each patient and each donor was submitted to the NMDP Research Sample Repository at the Blood Centers of the Pacific, Irwin Center (San Francisco, CA). These blood samples were separated into peripheral blood mononuclear cells (to generate B lymphoblastoid cell lines), granulocytes, and serum and were cryopreserved for future studies. DRB1, DQA1, DQB1, DPA1, and DPB1 alleles of each sample were identified using locus-specific or one or more group-specific polymerase chain reaction amplifications followed by hybridization with sequence-specific oligonucleotide probes. Additional assays using sequence-specific priming, restriction fragment length polymorphism analysis, and direct DNA sequencing of amplified DNA were used as needed to aid in allele identification (T Williams et al, manuscript in preparation; B Schmeckpeper et al, manuscript in preparation). Each sample was tested at the allele level by 2 independent laboratories in a masked fashion. Allele assignment results were compared with the confirmatory typing reported by the transplant center at time of transplantation. Discrepant typings were reviewed, and, if required, further testing was used to identify the correct allele. Consensus typing was used for this study to determine the match status of each donor-recipient pair. Once the typings were validated, retrospective allele typing data were stored in a separate Sybase database at the NMDP for use in correlating transplantation outcome with degree of HLA match.26 This report is confined to the correlation of allele-level HLA class II donor-recipient identity with outcome of unrelated donor transplantation for CML. High-resolution typing HLA-A, -B, and -C donor-recipient alleles were not available at the time of analysis for every patient who underwent CML transplantation.
For the engraftment endpoint, mismatch was defined as disparity in the donor that could be recognized by the recipient; for the GVHD endpoint, mismatch was defined as disparity in the host that could be recognized by the donor. For survival, relapse, and relapse-free survival, mismatches recognized by either donor-versus-host or host-versus-donor were included. Any allele mismatch detected for HLA-DQA1 or HLA-DQB1 was scored as a mismatch at HLA-DQ; similarly, any mismatch for HLA-DPA1 or HLA-DPB1 was scored as a mismatch for HLA-DP.
Transplantation procedures
Seventy-five transplant centers contributed clinical data to this analysis. Patients underwent transplantation between 1988 and 1997, and the centers used their local protocols for conditioning regimen and GVHD prophylaxis. The specific approach to acute GVHD prophylaxis was left to the discretion of investigators and individual NMDP centers. Thirty-nine different approaches were used. The heterogeneity of the prophylaxis regimens, the small number of patients receiving each regimen, and the relatively minor differences between the regimens prompted us to classify all GVHD prophylaxis regimens into 2 categories—T-depleted and T-replete. We then analyzed the effect of these 2 general acute GVHD prophylaxis approaches on outcome. Of the 831 patients in this study, 654 (79%) received T-replete grafts, and 177 (21%) received T-depleted grafts. Data on each transplantation were provided by the individual center to the NMDP on standardized forms and included pretransplantation information, peritransplantation events, and short- and long-term follow-up. Follow-up data were provided at 100 days, 6 months, and annually after transplantation. Transplant data were validated for accuracy and consistency at the NMDP using cross-validation of fields and validation of laboratory values against range values. Inconsistencies were queried and corrected with the reporting center. Table 1 summarizes the demographics and treatment regimens administered to the study patients.
Clinical endpoints
The primary endpoint of this study was survival. Neutrophil engraftment, grades III-IV acute GVHD, chronic GVHD, and hematologic relapse were secondary endpoints. Time to neutrophil engraftment was defined as the first day of 3 consecutive daily laboratory counts with a level of 5 × 108/L neutrophils or higher. Maximum overall grade (I-IV) of acute GVHD was reported by the individual centers, as was the maximum severity of skin, liver, and gastrointestinal (GI) involvement (stage 0-4). To decrease potential center-to-center variation in GVHD scoring for the current study, GVHD grade was calculated from the stages for skin, liver, and GI tract. A stage of 4 for skin or of 2 or higher for liver or GI tract was defined as clinically significant grades III-IV acute GVHD.12 The distinction between extensive and limited chronic GVHD was determined at the individual transplant centers. Relapse was defined as hematologic evidence of CML recurrence.
Statistical analysis
Overall and relapse-free survival rates were calculated using the Kaplan and Meier method27 and were compared using the log-rank statistic.28 Rates of engraftment, GVHD, and relapse were calculated by cumulative incidence29 treating death as a competing risk. Cumulative incidence rates were compared using the maximum likelihood estimate for the variance.30
The proportional hazards regression model31 was used for multivariate analysis of survival, GVHD, and relapse. Multivariate analysis of engraftment was performed using logistic regression and was restricted to evaluable patients surviving at least 21 days. To increase the precision of the relative risks (for factors other than class II allele matching), the 736 patients with CML not retrospectively typed for HLA class II genes were included in each regression analysis. An indicator term was added to the model to distinguish these patients as a separate group. The models also included indicators for CML stage, transplant center, HLA-DRB1 matching, HLA-DQ matching, and HLA-DP matching. Other risk factors were included in the model if they demonstrated a statistically significant (Wald χ2; P ≤ .05) association with outcome. Factors considered for the model included recipient and donor age, sex, cytomegalovirus (CMV) serology, race, interval from diagnosis to transplantation, serologic matching for HLA-A and -B, year of transplantation, use of T-cell depletion, and transplant center. Indicators for mismatching for both alleles at a single locus were also considered for HLA-DRB1, -DQ, and -DP.
To assess further the possible effects of HLA-DQ and HLA-DP disparities on clinical outcome, additional models were fit controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP combined as a continuous variable. Mismatches for 2 alleles at the same locus were counted twice. Models were also fit to test interactions of the class II loci with each other, serologic matching for HLA-A and -B, and the use of T-cell depletion. P values from the tests of interactions were adjusted for multiple comparisons by the method of Bonferroni. Primary causes of death were compared between DRB1 matches and mismatches using the likelihood ratio χ2 test with P values adjusted by Bonferroni. All analyses were performed using SAS statistical software (version 6.12; SAS Institute, Cary, NC).
Results
HLA matching
Eight different combinations of matching among HLA-DRB1, HLA-DQ, and HLA-DP were identified in the study population (Table2). Most (n = 491; 59%) were allele matched at HLA-DRB1 and HLA-DQ (DQA1, DQB1) but were mismatched at HLA-DP (DPA1, DPB1, or both). The second most frequently observed pattern included a match at HLA-DRB1 but a mismatch at HLA-DQ and HLA-DP (n = 102; 12%). These findings support previous reports of relatively weak linkage disequilibrium between HLA-DP and HLA-DR/DQ and relatively strong linkage disequilibrium between HLA-DR and HLA-DQ.32
Extent of matching for HLA-DRB1, HLA-DQ, and HLA-DP in the study population
| HLA-DRB1 . | HLA-DQ* . | HLA-DP† . | Number (%) . |
|---|---|---|---|
| Match | Match | Match | 79 (10) |
| Match | Match | Mismatch | 491 (59) |
| Match | Mismatch | Match | 8 (1) |
| Mismatch | Match | Match | 6 (1) |
| Match | Mismatch | Mismatch | 102 (12) |
| Mismatch | Match | Mismatch | 60 (7) |
| Mismatch | Mismatch | Match | 6 (1) |
| Mismatch | Mismatch | Mismatch | 79 (10) |
| Total | 831 (100) |
| HLA-DRB1 . | HLA-DQ* . | HLA-DP† . | Number (%) . |
|---|---|---|---|
| Match | Match | Match | 79 (10) |
| Match | Match | Mismatch | 491 (59) |
| Match | Mismatch | Match | 8 (1) |
| Mismatch | Match | Match | 6 (1) |
| Match | Mismatch | Mismatch | 102 (12) |
| Mismatch | Match | Mismatch | 60 (7) |
| Mismatch | Mismatch | Match | 6 (1) |
| Mismatch | Mismatch | Mismatch | 79 (10) |
| Total | 831 (100) |
HLA-DQ mismatch defined as disparity at DQA1 or DQB1.
HLA-DP mismatch defined as disparity at DPA1 or DPB1.
At the DRB1 locus, 81% of donor-recipient pairs were allele matched, 14% were serologically matched but allele mismatched, and only 4% were serologically mismatched for DR1 to DR16 antigens. The low number of serologically mismatched donor-recipient pairs (n = 35) precluded meaningful analysis of the effects of this variable on outcome.
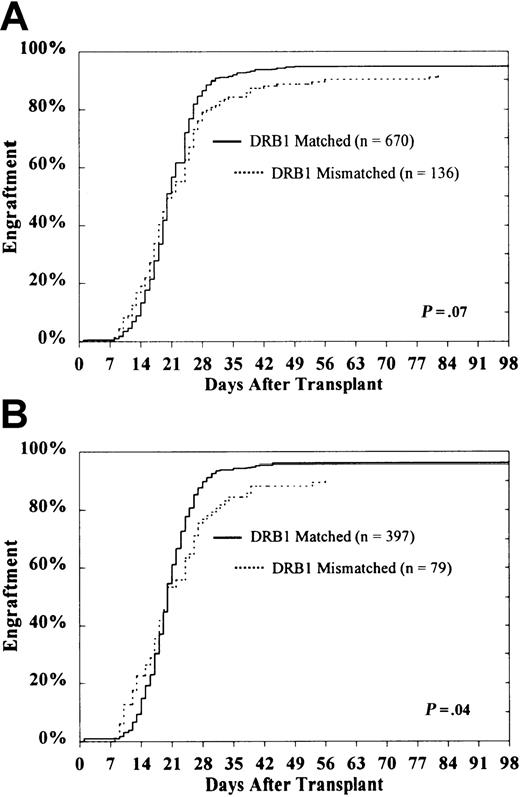
Neutrophil engraftment
No effect of class II disparity on delayed engraftment was detected (Figure 1). Among evaluable patients surviving at least 21 days (n = 806), the cumulative incidence of engraftment in HLA-DRB1–matched and HLA-DRB1–mismatched patients was 95% ± 1% and 92% ± 3%, respectively (P = .07) (Figure 1A). In transplants already mismatched for HLA-DRB1, the presence of an additional HLA-DQ or HLA-DP disparity was not associated with significantly reduced engraftment (HLA-DQ, 92% ± 4% vs 92% ± 5%; HLA-DP, 91% ± 4% vs 96% ± 6%).
Probability of engraftment according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Probability of engraftment according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Among evaluable HLA-A, B matched patients who underwent transplantation in the first CP of their disease (n = 476), the probability of engraftment for patients who were HLA-DRB1–matched was 96% ± 2% compared with 91% ± 5% for DRB1-mismatched patients (P = .04) (Figure 1B). Restricting the subset further to those mismatched for HLA-DRB1 (n = 79), the presence of an additional HLA-DQ or HLA-DP disparity was not predictive of engraftment (HLA-DQ, 88% ± 6% vs 94% ± 7%; HLA-DP, 89% ± 6% vs 100%).
Multivariable models confirmed the lack of independent association between class II disparity and delayed engraftment (data not shown). Logistic regression models for the analysis of the full data set (which controlled for the stage of the disease at the time of transplantation), recipient and donor sex (male recipient–female donor), recipient race (Hispanic recipient), use of T-cell depletion of the stem cell product, and transplant center showed no significant effects of class II allele mismatching on engraftment (data not shown).
Multivariable models for the analysis of the subset of first CP patients receiving transplants from HLA-A, B matched donors yielded similar results. Mismatching for HLA-DRB1, -DQ, and DP conferred an odds ratio (OR) of 0.61 (95% confidence interval [CI] 0.20-1.87;P = .38), an OR of 0.50 (95% CI, 0.18-1.39;P = .18), and an OR of 0.70 (95% CI, 0.20-2.50;P = .59), respectively.
No significant effects of 2-allele compared to 1-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected in either the entire study population or in the subset of patients in first CP undergoing transplantation from HLA-A, B serologically matched donors (data not shown). A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect (data not shown).
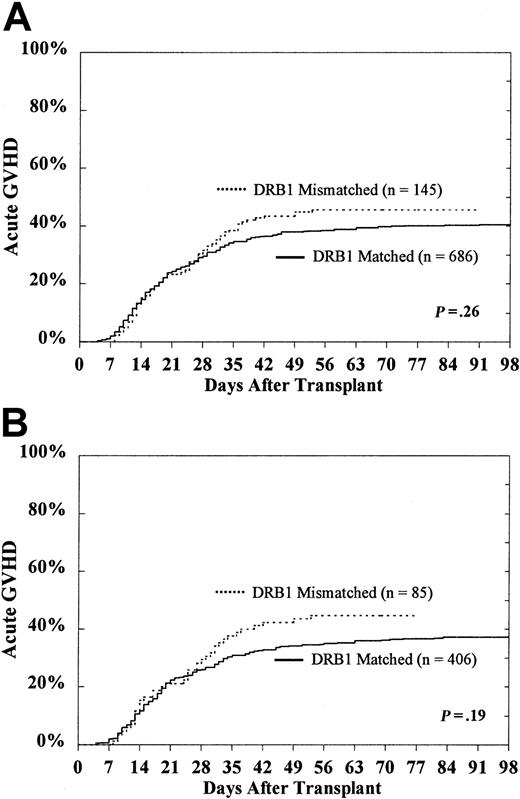
Acute graft-versus-host disease
For the entire study population of 831 patients, the cumulative incidence of grades III-IV acute GVHD in HLA-DRB1–matched and HLA-DRB1–mismatched patients was 41% ± 4% and 46% ± 8%, respectively (P = .26) (Figure2A). In transplants already mismatched for HLA- DRB1, the presence of an additional HLA-DQ or HLA-DP disparity was not associated with significantly increased risk for grades III-IV acute GVHD (HLA-DQ, 44% ± 11% vs 48% ± 12%; HLA-DP, 44% ± 9% vs 52% ± 18%)
Probability of grades III-IV acute GVHD according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Probability of grades III-IV acute GVHD according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Among the 491 HLA-A, B matched patients who underwent transplantation during the first CP of disease, the probability of grades III-IV acute GVHD for those who were HLA-DRB1–matched was 37% ± 5% compared with 45% ± 10% for those who were HLA-DRB1 mismatched (P = .19) (Figure 2B). Restricting the subset further to those mismatched for HLA-DRB1 (n = 85), the presence of an additional HLA-DQ or HLA-DP disparity was not predictive of grades III-IV acute GVHD (HLA-DQ, 41% ± 14% vs 49% ± 15%; HLA-DP, 42% ± 12% vs 52% ± 20%).
No independent effect of class II mismatching on increased risk for grades III-IV acute GVHD in the full data set was detected. Multivariate regression models for analysis of the full data set controlled for disease stage at the time of transplantation, serologic mismatching for HLA-A and B, use of T-cell depletion of the stem cell product, donor age, year of transplantation, and transplant center. Mismatching for HLA-DRB1, DQ, and DP conferred RR of 1.25 (95% CI, 0.92-1.70; P = .15), RR of 0.96 (95% CI, 0.72-1.27; P = .75), and RR of 1.06 (95% CI, 0.82-1.36;P = .66), respectively.
When the subset of patients in first CP undergoing transplantation from HLA-A, B matched donors was examined in a multivariable model, however, an effect of HLA-DRB1 disparity on grades III-IV acute GVHD was noted (RR, 1.62; 95% CI, 1.07-2.45; P = .02]). Mismatching for HLA-DQ or -DP did not confer risk to acute GVHD (RR, 0.67 [95% CI, 0.45-1.00; P = .05] and RR, 1.00 [95% CI, 0.72-1.38;P = .98], respectively).
No significant effects of 2-allele compared to 1-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected in either the entire study population or in the subset of patients in first CP undergoing transplantation from HLA-A, B serologically matched donors. A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect. These results suggest that in a homogeneous group of HLA-A, B matched patients undergoing transplantation during first CP, HLA-DRB1 mismatch may be associated with increased risk for acute GVHD.
Chronic graft-versus-host disease
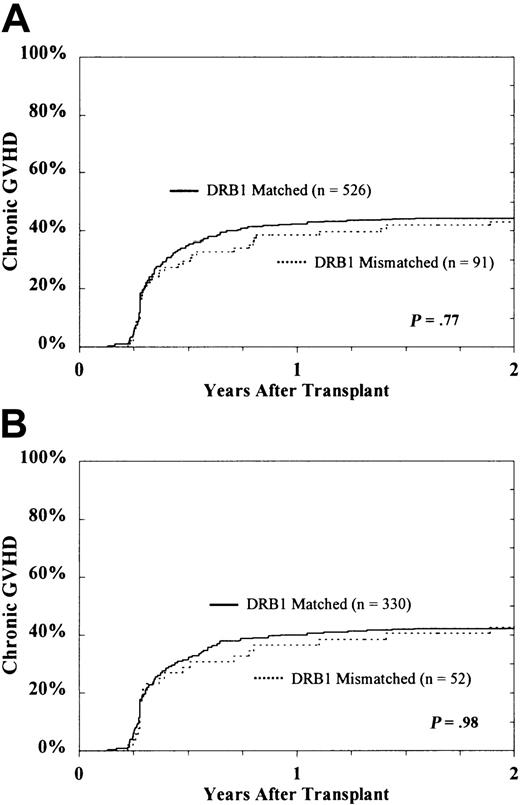
There was no association of class II disparity with increased risk for chronic or extensive chronic GVHD (Figure3). Among evaluable patients surviving at least 80 days (n = 617), the cumulative incidences of chronic GVHD in HLA-DRB1–matched and HLA-DRB1–mismatched patients were 64% ± 4% and 55% ± 9%, respectively (P = .06). For extensive chronic GVHD, the corresponding rates were 45% ± 4% and 43% ± 9% (P = .77). In transplants already mismatched for HLA-DRB1, the presence of an additional HLA-DQ or HLA-DP disparity was not associated with significantly increased risk for extensive chronic GVHD (HLA-DQ, 35% ± 13% vs 51% ± 13%; HLA-DP, 40% ± 11% vs 56% ± 20%).
Probability of extensive chronic GVHD according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Probability of extensive chronic GVHD according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Among the 382 evaluable HLA-A, B matched patients whose transplantations were performed in the first CP of disease, the cumulative incidence of chronic GVHD for those who were HLA-DRB1–matched was 63% ± 5% compared to 56% ± 12% for those who were HLA-DRB1–mismatched (P = .29). The cumulative incidence of extensive chronic GVHD was 43% ± 5% for the HLA-DRB1 matches and 43% ± 13% for the mismatches. Restricting the subset further to those mismatched for HLA-DRB1 (n = 52), the presence of an additional HLA-DQ or HLA-DP disparity was not predictive of extensive chronic GVHD (HLA-DQ, 40% ± 18% vs 44% ± 17%; HLA-DP, 41% ± 15% vs 47% ± 23%).
Multivariate regression models for extensive chronic GVHD controlled for disease stage at the time of transplantation, use of T-cell depletion of the stem cell product, donor sex, year of transplantation, and transplant center. No significant effects of class II allele mismatching on extensive chronic GVHD risk were detected. Mismatching for HLA-DRB1, -DQ, and -DP conferred RR of 1.14 (95% CI, 0.77-1.69;P = .51), RR of 0.87 (95% CI, 0.61-1.25;P = .46), and RR of 1.09 (95% CI, 0.82-1.45;P = .55), respectively. A multivariable model for the analysis of the subset of patients in first CP receiving transplanted tissue from HLA-A– and -B–matched donors yielded no significant effects of class II allele mismatching on extensive chronic GVHD (data not shown).
No significant effects of 2-allele compared to 1-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected in either the entire study population or in the subset of patients in first CP from HLA-A, B serologically matched donors. A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect.
Hematologic relapse
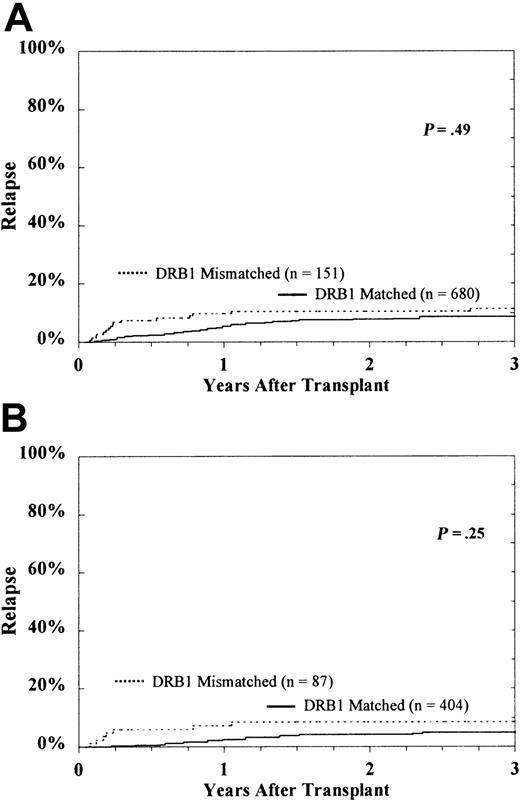
The cumulative incidence of hematologic relapse in HLA-DRB1–matched and HLA-DRB1–mismatched patients at 5 years was 9% ± 2% and 11% ± 5%, respectively (P = .49) (Figure 4). Interestingly, in transplantations with donor-recipient pairs already mismatched for HLA-DRB1, the presence of an additional HLA-DQ disparity had an effect on increased relapse, but there was no detectable effect of HLA-DP mismatching (DQ, 16% ± 8% vs 5% ± 5%, P = .02; DP, 12% ± 5% vs 8% ± 15%).
Probability of hematologic relapse according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Probability of hematologic relapse according to DRB1 match status among the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Among the 491 HLA-A and -B–matched patients who underwent transplantation in the first CP, the probability of relapse was 5% ± 2% for HLA-DRB1 matches and 8% ± 6% for DRB1 mismatches (P = .25). Restricting the subset further to those mismatched for HLA-DRB1 (n = 87), the presence of an additional HLA-DQ disparity was associated with a higher risk for relapse (15% ± 10% vs 0%). The small number of HLA-DRB1–matched, HLA-DQ–mismatched donor-recipient pairs precluded analysis of this subset (Table 2). A disparity for HLA-DP in this group was not associated with relapse (8% ± 6% vs 11% ± 20%).
Multivariate regression models for the analysis of the full data set controlled for disease stage at the time of transplantation, serologic mismatching for HLA-A and B, use of T-cell depletion of the stem cell product, donor age, year of transplantation, and transplant center. Disparity at HLA-DQ was associated with increased risk for relapse (RR, 2.39 [95% CI, 1.35-4.24; P = .003]). Mismatching for HLA-DRB1 and DP conferred RR of 0.95 (95% CI, 0.49-1.85;P = .89) and RR of 0.65 (95% CI, 0.33-1.29;P = .22), respectively.
A multivariable model for the analysis of the subset of patients in first CP receiving transplanted tissue from HLA-A, B matched donors yielded a significant effect of DQ (RR, 3.16 [95% CI, 1.19-8.45;P = .02]). Mismatching for HLA-DRB1 and -DP conferred RR of 1.42 (95% CI, 0.47-4.33; P = .54) and RR of 0.46 (95% CI, 0.15-1.44; P = .18), respectively.
No significant effects of 2-allele compared to 1-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected in either the 831 patients or in the subset of patients in first CP undergoing transplantation from HLA-A, B serologically matched donors. A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect.
Relapse-free survival
A strong association between HLA-DRB1 matching and improved relapse-free survival was observed. Five-year relapse-free survival rates for HLA-DRB1–matched and –mismatched patients were 35% ± 4% and 24% ± 7%, respectively (log-rank,P < .0001). Among 491 HLA-A, B matched patients who underwent transplantation in the first CP, the corresponding 5-year relapse-free survival rates were 43% ± 5% and 25% ± 10% (log-rank, P < .0001). Among the entire population of 831 patients, mismatching for HLA-DQ did not significantly affect relapse-free survival rates among HLA-DRB1 matched patients (29% ± 11% vs 35% ± 4%) or HLA-DRB1 mismatched (21% ± 9% vs 28% ± 11%). Similar results were observed for HLA-DP mismatching (34% ± 4% vs 38% ± 11% and 23% ± 7% vs 33% ± 27%). In the subset of 491 patients in first CP who received transplanted tissue from HLA-A, B matched donors, HLA-DQ disparity was not significantly associated with lower survival in either HLA-DRB1 matches (38% ± 16% vs 44% ± 6%) or HLA-DRB1 mismatches (24% ± 12% vs 27% ± 14%). Similar results were observed for HLA-DP disparity in this subset of patients (43% ± 6% vs 43% ± 15% and 24% ± 10% vs 33% ± 31%).
An independent effect of HLA-DRB1 mismatching on lower relapse-free survival was observed in the entire study population. Multivariate regression models adjusted for CML stage at the time of transplantation, serologic mismatching for HLA-A and B, HLA-DRB1, patient age, donor age, CMV seropositivity of the patient, time interval from diagnosis to transplantation, and transplant center. Mismatching for HLA-DRB1 conferred RR of 1.31 (95% CI, 1.04-1.66;P = .02), whereas the impact of HLA-DQ or DP disparity was not significant (RR, 1.09 [95% CI, 0.87-1.35;P = 0.46]; RR, 1.08 [95% CI, 0.82-1.42;P = .60], respectively). No significant effects of 1-allele compared to 2-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected. A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect.
A multivariable model for the analysis of the subset of patients in first CP receiving transplants from HLA-A, B matched donors demonstrated an independent effect of HLA-DRB1 disparity on relapse-free survival (RR, 1.63 [95% CI, 1.17-2.27;P = .004]). Mismatching for HLA-DQ and -DP conferred RR of 0.95 (95% CI, 0.69-1.30; P = .73) and RR of 1.14 (95% CI, 0.78-1.68; P = .50), respectively.
Survival
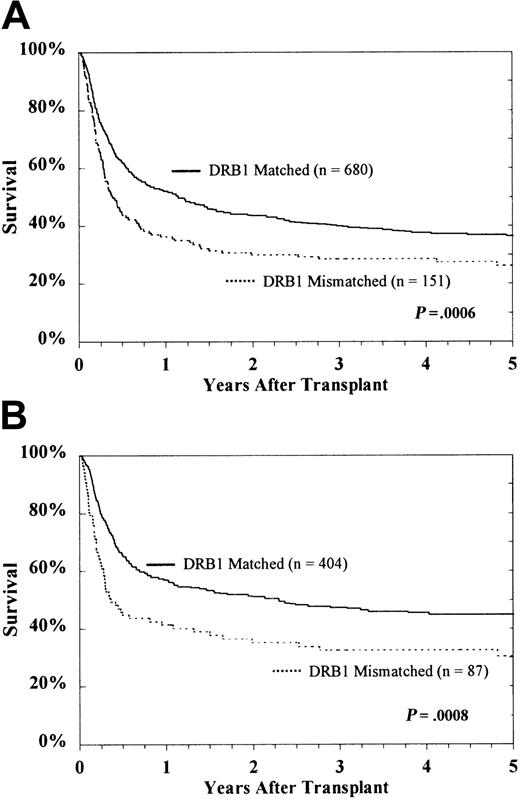
Analysis of the entire study population revealed a deleterious effect of HLA-DRB1 mismatching on survival (Figure5). HLA-DQ and HLA-DP disparity did not significantly increase mortality. Five-year survival rates for HLA-DRB1–matched and –mismatched patients were 36% ± 4% and 26% ± 7%, respectively (P = .0006). Among HLA-A, B matched patients who underwent transplantation in the first CP, 5-year survival rates were 45% ± 5% for HLA-DRB1 matches and 30% ± 10% for HLA-DRB1 mismatches (P = .0008). Among the entire population of 831 patients, mismatching at HLA-DQ did not significantly affect survival among HLA-DRB1 matched patients (30% ± 11% vs 38% ± 4%) or HLA-DRB1 mismatched (26% ± 10% vs 28% ± 11%) patients. Similar results were observed for HLA-DP (36% ± 4% vs 41% ± 11% and 26% ± 8% vs 33% ± 27%). In the subset of patients in first CP who received transplanted tissue from HLA-A, B matched donors, the presence of HLA-DQ mismatching was not associated with lower survival in either HLA-DRB1 matches (38% ± 16% vs 46% ± 6%) or HLA-DRB1 mismatches (34% ± 14% vs 27% ± 14%). Similar results were also observed for HLA-DP in this subset of patients (45% ± 6% vs 43% ± 15% and 30% ± 11% vs 33% ± 31%).
Kaplan-Meier survival of the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Kaplan-Meier survival of the entire study population.
CML patients (A) and subset of HLA-A, B matched patients in first CP (B).
Improved survival was observed with HLA-DRB1 allele matching independent of contributions from HLA-DQ and HLA-DP (RR, 1.29 [95% CI, 1.02-1.64; P = .04]) (Table3). Multivariate regression models for the full data set were adjusted for CML stage at the time of transplantation, serologic mismatching for HLA-A and -B, patient age, donor age, CMV seropositivity of the patient, time interval from diagnosis to transplantation, and transplant center. Increased mortality was not associated with disparity for HLA-DQ (RR, 1.01 [95% CI, 0.81-1.26; P = .94]) or HLA-DP (RR, 1.11 [95% CI, 0.84-1.48; P = .46]). No significant effects of 1-allele compared with 2-allele mismatching at HLA-DRB1, HLA-DQ, or HLA-DP were detected. A regression model controlling for HLA-DRB1 and treating the total number of mismatches at HLA-DQ and HLA-DP as a continuous variable showed no significant effect.
Multivariate model for survival according to matching for HLA-DRB1, HLA-DQ, and HLA-DP
| Disparities HLA locus . | RR . | 95% CI . | P . |
|---|---|---|---|
| DRB1 | 1.29 | 1.02 -1.64 | 0.04 |
| DQ3-150 | 1.01 | 0.81 -1.26 | 0.94 |
| DP3-151 | 1.11 | 0.84 -1.48 | 0.46 |
| Disparities HLA locus . | RR . | 95% CI . | P . |
|---|---|---|---|
| DRB1 | 1.29 | 1.02 -1.64 | 0.04 |
| DQ3-150 | 1.01 | 0.81 -1.26 | 0.94 |
| DP3-151 | 1.11 | 0.84 -1.48 | 0.46 |
HLA-DQ mismatch defined as disparity at DQA1 or DQB1.
HLA-DP mismatch defined as disparity at DPA1 or DPB1.
The impact of HLA-DRB1 allele disparity on overall survival was examined in the subset of patients in first CP who received transplants from HLA-A, B matched donors. The RR of an HLA-DRB1 mismatch was 1.53 (95% CI, 1.09-2.16; P = .01 No significant effects of disparity for HLA-DQ (RR, 0.88 [95% CI, 0.63-1.21;P = .42]) or HLA-DP (RR, 1.12 [95% CI, 0.76-1.65;P = .56]) were detected.
To better understand the basis for increased mortality in the HLA-DRB1 mismatched population, the primary cause of death as reported by transplant centers was evaluated for the entire study population. Among those patients who died, there were no significant differences in the primary cause of death in the 2 groups. Infection and GVHD were the most commonly reported causes of death in HLA-DRB1 matched and mismatched patients. Among HLA-DRB1 matches, infection accounted for 31% of deaths and GVHD for 18%; among HLA-DRB1 mismatches, infection caused 39% and GVHD caused 17% of deaths.
Discussion
This study confirms and extends previous knowledge of the biologic role of HLA class II gene products in clinical hematopoietic cell transplantation.33-36 We demonstrate that not only is HLA-DRB1 allele disparity detrimental to survival after unrelated stem cell transplantation for CML, the HLA-DRB1 effect is independent of HLA-DQ and HLA-DP. The selection of appropriate stem cell donors should include DNA allele typing of donor and recipient, at least for HLA-DRB1.
The profound adverse effect of HLA-DRB1 disparity on survival was observed in the subset of patients in first CP and in the entire study population. The increased risk for death associated with class II disparity could not be explained by an increased risk for delayed engraftment, chronic GVHD, or hematologic relapse. No effect of HLA-DRB1 disparity on acute GVHD risk could be discerned among the entire study population. However, when the more homogeneous subset of HLA-A, B matched patients who underwent transplantation in first CP was examined, HLA-DRB1 disparity was independently associated with grades III-IV acute GVHD risk. The association of HLA-DRB1 disparity with acute GVHD risk may account in part for the marked decrease in survival of patients after HLA-DRB1–mismatched chronic phase transplantation. Other possible explanations for the poorer outcome associated with HLA-DRB1 disparity may include impaired immune reconstitution or GVHD resistant to therapy. These hypotheses remain to be evaluated.
Hematologic relapse was significantly higher in HLA-DQ mismatches, independent of HLA-DRB1 and HLA-DP mismatch. Although it would be difficult to hypothesize an underlying biologic mechanism for an association between increased risk for relapse and HLA disparity, the analysis demonstrated that HLA-DQ mismatching did not predict for either overall survival or relapse-free survival. There were no imbalances in the study population for factors that could reasonably explain the higher relapse rate. Re-evaluation of relapse and HLA disparity will be warranted in a larger transplant population.
Two previous studies from the Japan Marrow Donor Program (JMDP)37 and the Seattle transplant program38examined the role of HLA class II genes in stem cell transplantation. It is important to note that JMDP, Seattle, and NMDP analyses differed in study design and methodology. Furthermore, the 3 studies are distinguished by the class II loci evaluated and the manner in which a mismatch was defined. The Seattle study examined DRB1, DQB1, and DPB1. In addition to these genes, the NMDP analysis included DQA1 and DPA1, and the JMDP study considered the DRB3, DRB4, and DRB5 genes. The NMDP analysis defined a mismatch as any disparity within A and B genes for HLA-DQ and -DP, whereas the Seattle and JMDP studies defined a mismatch at each gene individually.
Taking these methodological differences into consideration, our NMDP analysis extends observations made by the Seattle transplant program38 in which disparity for HLA-DRB1 or -DQB1 alleles was found to increase acute GVHD risk; however, because the current NMDP analysis used serologic methods to determine HLA class I identity and did not consider allele-level matching for class I genes, potential undetected HLA class I allele disparity among the NMDP-reported HLA-A, -B, -C serologically matched pairs might have obscured a true effect of HLA-DQ and -DP on the clinical endpoints. Such effects have been suggested by others.39 40 However, a definitive understanding of the consequences of HLA class II mismatching on clinical outcome will require not only knowledge of donor-recipient class II identity but also knowledge of the identity of class I genes, other resident genes of the MHC class I and class II regions, and genes that encode minor histocompatibility determinants on donor and recipient cells.
The JMDP, Seattle, and NMDP study populations differ in antigen, allele, and haplotype frequencies.26,41,42 Worldwide, the phenotypes and genotypes of transplant recipients and donors reflect their ethnic and racial backgrounds. Our study included patients of predominantly North American Caucasian background. Extensive analyses of the HLA alleles and haplotypes by the NMDP of its donor population reveal antigen, allele and haplotype frequencies characteristic of persons in North America. For example, the most common HLA-DRB1 alleles represented in our study were DRB1*0701 (15%), *0301 (14%), *1501 (13%), *0401 (10%), *0101 (7%), *1301 (6%), *1101 (5%), and *1302 (5%); similar DRB1 allele frequencies were reported for the Seattle study.42 This is in sharp contrast to the allele frequencies in nonwhite populations.23,41 42 Deduced amino acid sequences of the class II disparities encoded by the Japanese and the North American Caucasian transplants differ by number and nature of amino acid residues. Whether potential differences in GVHD and survival rates across racially diverse transplantation populations reflect differences in the nature of the class II disparities remains to be tested.
In conclusion, we provide strong evidence that donor-recipient differences encoded by HLA-DRB1 alleles are biologically important as transplantation determinants. Current standards for unrelated donor selection rely on testing for HLA-DR phenotypes and subsequent matching for HLA-DRB1 alleles. HLA-DRB1 allele-matched donors should be prioritized over HLA-DRB1 allele-mismatched donors. In some clinical situations, however, proceeding with HLA-DRB1 allele-mismatched transplantation can be successful and may be preferable to the delay imposed by a prolonged search for a DRB1 allele-matched donor. Whether certain HLA-DRB1 allele mismatches are better tolerated than others remains to be determined. Finally, the NMDP experience provides conclusive data on the benefits of unrelated donor transplantation during early-phase CML from genetically well-matched unrelated donors. These results indicate that patients without a sibling donor should be considered early for unrelated donor transplantation in the first chronic phase of disease for optimal transplantation outcome. These long-term results also serve as a standard against which novel, potentially curative approaches to the treatment of CML can be measured.
We thank Janet Hegland, Michelle Setterholm, Rose Fritz, Cynthia McSherry, Tamara Winden, Nancy Morgan, and other NMDP personnel for their assistance with this study. We thank the NMDP Transplant and Donor Center personnel (complete listing available at www.marrow.org) for providing cells and data for this study. We also thank Delene Johnson and the Blood Centers of the Pacific, Irwin Center of the NMDP Research Sample Repository for storing and preparing the samples, and the personnel at the NMDP DNA-based high-resolution HLA typing laboratories, as follows, for providing the retrospective allele-level HLA typing results: Zuheir Awdeh, CBR Laboratories, Boston, MA; Lee Ann Baxter-Lowe, University of South Carolina, Columbia; Ann Begovich, Roche Molecular Systems, Alameda, CA; Michael Chopek, American Red Cross–National Histocompatibility Laboratory, Rockville, MD; Marcelo Fernández-Viña, American Red Cross–National Histocompatibility Laboratory, Baltimore, MD; Harriet Noreen, University of Minnesota, Minneapolis; Marcela Salazar, ARC–New England Region, Dedham, MA; Barbara Schmeckpeper, Johns Hopkins University, Baltimore, MD; Thomas M. Williams, University of New Mexico, Albuquerque; and Edmond Yunis, Dana-Farber Cancer Institute, Boston, MA.
Supported by grants from the Office of Naval Research (N0014-93-1-0658 and N0014-95-1-0005) to the National Marrow Donor Program.
The views expressed in this article are those of the authors and do not reflect the official policy or the position of the Department of the Navy, the Department of Defense, or the United States Government.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Philip McGlave, Division of Hematology, Oncology and Transplantation, University of Minnesota Medical School, MMC 480, 420 Delaware St SE, Minneapolis, MN 55455; e-mail:mcgla001@maroon.tc.umn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal