The molecular genetics of the α-thalassemias has been comprehensively reviewed.1 Of the numerous mutations that have been described, deletions at the α-globin gene locus account for the vast majority of α-thalassemia alleles.2 The most widely occurring of these are the -α3.7 and -α4.2 single α-globin gene deletions, while double α-globin gene deletions in cis, such as the - -SEA, - -FIL, and - -THAIalleles are very common within Southeast Asia, and the - -MED and -(α)20.5 double-gene deletions occur more frequently in the Mediterranean area.
Since the publication of the entire sequence of the human α-globin gene cluster,3 we and others have developed multiplex polymerase chain reaction (PCR) methods to diagnose different subsets of α-thalassemia deletional determinants.4 5 Generally, however, the high G+C nucleotide content and high degree of homology between the genes and pseudogenes at this locus have made it technically challenging to develop a multiplex PCR assay capable of detecting all 7 of the above mutations [-α3.7, -α4.2, - -SEA, - -FIL, - -MED, -(α)20.5, and - -THAI] in a single reaction. We have now successfully developed an improved single-tube multiplex PCR assay that can detect heterozygosity, homozygosity, and compound heterozygosity of these 7 α-globin gene deletions.
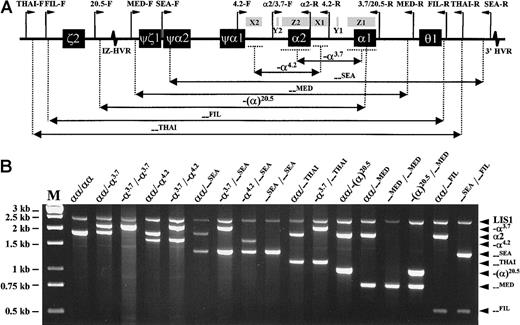
To achieve this, several aspects of our original multiplex PCR assay were modified. These included the redesign of several primers, inclusion of additional new primers, reoptimization of primer concentrations, and use of a different, chemically modified automatic hot-start DNA polymerase. Each 50-μL reaction contained 200 μM of each dNTP, 1.5 mM MgCl2, 1 × Q-solution (Qiagen, Hilden, Germany), 2.5 U HotStarTaq DNA polymerase in supplied reaction buffer (Qiagen), 100-200 ng of genomic DNA, and 16 different primers at various concentrations (Table 1). Reactions were conducted in a T3 thermal cycler (Biometra, Göttingen, Germany), with an initial 15-minute denaturation at 96°C, followed by 30 cycles of 98°C denaturation for 45 seconds, 60°C annealing for 90 seconds, and 72°C extension for 135 seconds. A final 5-minute extension at 72°C completed the reaction. Ten microliters of each amplified product was analyzed by electrophoresis through a 1% agarose gel in 1 × Tris-Borate-EDTA buffer at 10 volts/cm for an hour. The expected amplicon sizes for each of the deletion junction fragments and the controlα2 globin gene and LIS1 gene 3′ untranslated region (UTR) fragments are listed in Table 1. Because any of the 7 deletions either partially or completely removes theα2 globin gene, its positive amplification serves to indicate heterozygosity when a deletion allele is also present. The LIS1 gene 3′ UTR fragment serves as a separate control for general amplification success. Multiplex PCR results from representative DNA samples with various α-thalassemia genotypes are shown in Figure 1.
Primer sequences for α-thalassemia multiplex PCR and expected amplicon sizes
| Name . | 5′→3′ sequence . | GenBank ID: nucleotides . | Concentration . | Amplicon (size) . |
|---|---|---|---|---|
| LIS1-F | ATACCATGGTTACCCCATTGAGC | HSLIS10:510→532 | 0.5 μM } | LIS1 3′UTR fragment (2350 bp) |
| LIS1-R | AGGGCTCATTACATGTGGACCC | HSLIS10:2859→2838 | 0.5 μM | |
| α2/3.7-F | CCCCTCGCCAAGTCCACCC | HUMHBA4:5676→5694 | 0.2 μM } | -α3.7jxna fragment (2022/2029 bp) |
| 3.7/20.5-R | AAAGCACTCTAGGGTCCAGCG | HUMHBA4:11514→11494 | 0.2 μM | |
| α2/3.7-F | As above | As above | — } | α2gene (1800 bp) |
| α2-R | AGACCAGGAAGGGCCGGTG | HUMHBA4:7475→7457 | 0.2 μM | |
| 4.2-F | GGTTTACCCATGTGGTGCCTC | HUMHBA4:3064→3084 | 0.5 μM } | -α4.2 jxn fragment (1628 bp) |
| 4.2-R | CCCGTTGGATCTTCTCATTTCCC | HUMHBA4:8942→8920 | 0.5 μM | |
| SEA-F | CGATCTGGGCTCTGTGTTCTC | HSGG1:26120→26140 | 0.2 μM } | --SEA jxn fragment (1349 bp) |
| SEA-R | AGCCCACGTTGTGTTCATGGC | HSCOS12:3817→3797 | 0.2 μM | |
| THAI-F | GACCATTCCTCAGCGTGGGTG | HSGG1:9592→9612 | 0.3 μM } | --THAI jxn fragment (1153 bp) |
| THAI-R | CAAGTGGGCTGAGCCCTTGAG | HSCOS12:1241→1221 | 0.3 μM | |
| 20.5-F | GCCCAACATCCGGAGTACATG | HSGG1:17904→17924 | 0.2 μM } | -(α)20.5 jxn fragment (1007 bp) |
| 3.7/20.5-R | As above | As above | — | |
| MED-F | TACCCTTTGCAAGCACACGTAC | HSGG1:23123→23144 | 0.2 μM } | --MED jxn fragment (807 bp) |
| MED-R | TCAATCTCCGACAGCTCCGAC | HSGG1:41203→41183 | 0.2 μM | |
| FIL-F | TTTAAATGGGCAAAACAGGCCAGG | HSGG1:12304→12327 | 1.0 μM } | --FIL jxn fragment (546 bp) |
| FIL-R | ATAACCTTTATCTGCCACATGTAGC | HSCOS12:570→546 | 1.0 μM |
| Name . | 5′→3′ sequence . | GenBank ID: nucleotides . | Concentration . | Amplicon (size) . |
|---|---|---|---|---|
| LIS1-F | ATACCATGGTTACCCCATTGAGC | HSLIS10:510→532 | 0.5 μM } | LIS1 3′UTR fragment (2350 bp) |
| LIS1-R | AGGGCTCATTACATGTGGACCC | HSLIS10:2859→2838 | 0.5 μM | |
| α2/3.7-F | CCCCTCGCCAAGTCCACCC | HUMHBA4:5676→5694 | 0.2 μM } | -α3.7jxna fragment (2022/2029 bp) |
| 3.7/20.5-R | AAAGCACTCTAGGGTCCAGCG | HUMHBA4:11514→11494 | 0.2 μM | |
| α2/3.7-F | As above | As above | — } | α2gene (1800 bp) |
| α2-R | AGACCAGGAAGGGCCGGTG | HUMHBA4:7475→7457 | 0.2 μM | |
| 4.2-F | GGTTTACCCATGTGGTGCCTC | HUMHBA4:3064→3084 | 0.5 μM } | -α4.2 jxn fragment (1628 bp) |
| 4.2-R | CCCGTTGGATCTTCTCATTTCCC | HUMHBA4:8942→8920 | 0.5 μM | |
| SEA-F | CGATCTGGGCTCTGTGTTCTC | HSGG1:26120→26140 | 0.2 μM } | --SEA jxn fragment (1349 bp) |
| SEA-R | AGCCCACGTTGTGTTCATGGC | HSCOS12:3817→3797 | 0.2 μM | |
| THAI-F | GACCATTCCTCAGCGTGGGTG | HSGG1:9592→9612 | 0.3 μM } | --THAI jxn fragment (1153 bp) |
| THAI-R | CAAGTGGGCTGAGCCCTTGAG | HSCOS12:1241→1221 | 0.3 μM | |
| 20.5-F | GCCCAACATCCGGAGTACATG | HSGG1:17904→17924 | 0.2 μM } | -(α)20.5 jxn fragment (1007 bp) |
| 3.7/20.5-R | As above | As above | — | |
| MED-F | TACCCTTTGCAAGCACACGTAC | HSGG1:23123→23144 | 0.2 μM } | --MED jxn fragment (807 bp) |
| MED-R | TCAATCTCCGACAGCTCCGAC | HSGG1:41203→41183 | 0.2 μM | |
| FIL-F | TTTAAATGGGCAAAACAGGCCAGG | HSGG1:12304→12327 | 1.0 μM } | --FIL jxn fragment (546 bp) |
| FIL-R | ATAACCTTTATCTGCCACATGTAGC | HSCOS12:570→546 | 1.0 μM |
jxn, junction.
Strategy and results of α-thalassemia multiplex polymerase chain reaction analysis.
(A) Schematic representation of the α-globin gene cluster, indicating extents of the 7 deletions and relative positions of the primers (except for the control LIS1-F and LIS1-R primers, which are located on a different chromosome). Locations of X, Y, and Z sequence homology boxes and hypervariable regions (HVRs) are also shown. (B) Multiplex PCR results from genomic DNA samples with various α-globin genotypes. M indicates Generuler 1kb DNA ladder (Fermentas, St Leon-Rot, Germany).
Strategy and results of α-thalassemia multiplex polymerase chain reaction analysis.
(A) Schematic representation of the α-globin gene cluster, indicating extents of the 7 deletions and relative positions of the primers (except for the control LIS1-F and LIS1-R primers, which are located on a different chromosome). Locations of X, Y, and Z sequence homology boxes and hypervariable regions (HVRs) are also shown. (B) Multiplex PCR results from genomic DNA samples with various α-globin genotypes. M indicates Generuler 1kb DNA ladder (Fermentas, St Leon-Rot, Germany).
This simple assay has been validated on over two hundred DNA samples from α-thalassemia-1 and HbH disease individuals, and represents a rapid and reliable method for detecting 7 of the most common mutations of α-thalassemia.
Supported by Singapore grant NMRC/0365/1999 to S.S.C.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal