Von Willebrand disease (VWD) is the commonest congenital bleeding disorder, with a prevalence, estimated from population studies, of about 1%,1,2 although the prevalence of patients requiring treatment is much lower.3 This autosomally inherited disorder is caused by either qualitative (type 2) or quantitative (type 1 and 3) deficiency of von Willebrand factor (VWF). The human VWF gene is located on chromosome 12 and consists of 52 exons.4 A pseudogene on chromosome 22 has also been identified, which is 21-29 kb in length and corresponds to exons 23-34 of the VWF gene. It has 97% homology to the authentic VWF gene, but the presence of multiple stop codons on the pseudogene indicates that this is not a functional gene in humans.5
In recent years, many molecular defects of the VWF gene have been identified in patients with VWD.6,7Molecular defects in types 1 and 3 VWD are not confined to specific regions of the gene, as in type 2 VWD.7-9 The reported molecular defects that have caused autosomal recessive severe type 3 VWD are large gene deletions, frameshift mutations, nonsense mutations, splice-site mutations, defects at the level of mRNA expression, and some candidate missense mutations.6Heterozygotes could present as mild type 1 VWD individuals and may be asymptomatic.10-12
We have investigated a boy who was diagnosed as having severe type 3 VWD at the age of 13 months. His parents are Asian Indians and are first cousins. No previous family history of a bleeding disorder existed. The patient was initially treated for his bleeding episodes with intermediate purity factor VIII concentrate (BPL 8Y). At 6 years of age, he developed a high-titer anti-VWF inhibitor that was identified after a poor response to treatment. This inhibitor caused complete inhibition of von Willebrand factor activity and has persisted at high titer having been unaffected by an immune tolerance treatment regime.
Laboratory investigations of the patient have shown that FVIII:C, VWF:Ag, and VWF:Ricof were all below 0.01 U/mL (normal range, 0.5-1.5 U/mL), and VWF:Ag multimers were absent. The VIII:C, VWF:Ag, and VWF:RiCof values for the patient's mother were 2.04 U/mL, 0.96 U/mL, and 0.60 U/mL, respectively, and for the patient's father were 1.36 U/mL, 0.84 U/mL, and 0.64 U/mL respectively. Both parents have normal plasma VWF:Ag multimers. DNA and RNA were extracted from the propositus and parents to carry out genotypic studies. Haplotypes of parents and propositus were determined by analysis of 2 polymorphic regions of the tetranucleotide (ATCT) simple repeat in intron 40 (VNTR I and II) of the VWF gene, along with 2 restriction fragment length polymorphisms (RFLP) of the VWF gene. These gene-tracking studies showed that the boy had inherited the same affected allele from each parent. The whole VWF gene was screened for mutations using chemical cleavage mismatch detection (CCMD) analysis.13 Any indication of sequence changes was investigated by sequencing.
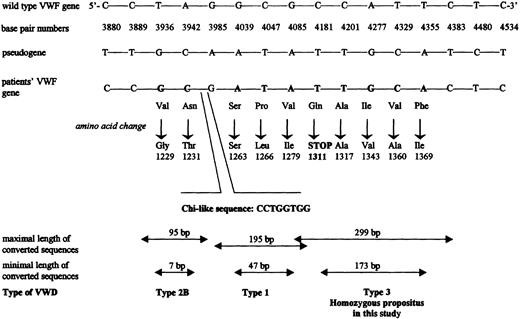
CCMD of exon 28 revealed multiple mismatched bands. The sequencing of this exon revealed a gene conversion to have taken place, for which the boy is homozygous and the parents are heterozygous (see Figure 1). Five base changes corresponding to the pseudogene sequence were identified: 4181C>T, 4201C>T, 4277A>G, 4329T>C, and 4355T>A. The maximal length of the conversion is approximately 299 bp, and the minimal length is 173 bp. The first base change (4181C>T) results in a stop codon, which would result in premature termination of the protein. The nucleotides are numbered from the major transcription cap site (+1), which is located 250 nucleotides upstream of the first nucleotide in the ATG initiation codon. Amino acid residues are numbered from the ATG initiation codon (residue 1) to the carboxy-terminal lysine (residue 2813) of pre-pro-VWF. All changes identified in the VWF gene of the parents and the propositus were confirmed by DNA sequencing and then by restriction enzyme analysis. Two possible factors may contribute to the occurrence of a gene conversion. First, the divergence between the pseudogene and the VWF gene is 3.14%, and the divergence between them for sequences corresponding to exons 23-34 is 2.47%.5 Homology between the 2 genes is greatest in these exonic regions and may contribute to recombinations. Second, it is hypothesized that a chi sequence or sequence similar to a chi sequence can regulate homologous recombination.14 There are 2 consensus chi sequences (5′-GCTGGTGG-3′) around the region of this gene conversion (one in exon 28 and the other in intron 27), which may be responsible for the relatively frequent gene conversion events in exon 28.15Six other cases of homologous gene conversion have been reported previously to be the cause of VWD type 1 or type 2B in 5 unrelated families.15,16,17 A Q1311 mutation has also recently been detected in the homozygous state in 4 Spanish patients from 2 apparently unrelated families of gypsy origin.18
Schematic presentation of the 3′- end of the intron 27 and 5′- end of the exon 28 of the
VWF gene, showing the base pair differences detected in propositus investigated in this study and other reported cases of gene conversion between VWF and pseudogene.Selected nucleotides and their numbers for the wild-type VWFgene are shown together with the corresponding bases of the pseudogene at these positions. Nucleotides in bold in the third line (from the top) shows those that match with bases in the pseudogene in our own and the other reported patients.15-17 Below each changed nucleotide, the resulting amino acid substitution is shown, and finally at the bottom the predicted maximal and minimal length of the putative converted sequences and the reported VWD phenotype are shown. The location and the nucleotides of the chi-like sequence is indicated.19
Schematic presentation of the 3′- end of the intron 27 and 5′- end of the exon 28 of the
VWF gene, showing the base pair differences detected in propositus investigated in this study and other reported cases of gene conversion between VWF and pseudogene.Selected nucleotides and their numbers for the wild-type VWFgene are shown together with the corresponding bases of the pseudogene at these positions. Nucleotides in bold in the third line (from the top) shows those that match with bases in the pseudogene in our own and the other reported patients.15-17 Below each changed nucleotide, the resulting amino acid substitution is shown, and finally at the bottom the predicted maximal and minimal length of the putative converted sequences and the reported VWD phenotype are shown. The location and the nucleotides of the chi-like sequence is indicated.19
The rest of the VWF gene was screened by investigating six overlapping mRNA segments. Another point mutation was found in exon 49, a 8363G>A substitution, which would predict G2705R. This was confirmed in the DNA to be homozygous in the propositus and heterozygous in the parents, suggesting that it be on the same allele as the gene conversion. As the gene conversion is upstream of this mutation and generates a stop codon, this point mutation would not be translated.
In conclusion, we have investigated a boy with severe type 3 VWD who is homozygous for a gene conversion in exon 28 that results in premature termination of the protein. The asymptomatic parents, who are first cousins, are heterozygous for the same mutation. It can be speculated that this mutation produces a truncated dysfunctional protein lacking many of the essential functional sites of VWF. In a patient homozygous for this mutation, the protein cannot be assembled and, therefore, will not be secreted and will be absent from the patient's plasma.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal