Abstract
Evi-1 is a zinc finger nuclear protein whose inappropriate expression leads to leukemic transformation of hematopoietic cells in mice and humans. This was previously shown to block the antiproliferative effect of transforming growth factor β (TGF-β). Evi-1 represses TGF-β signaling by direct interaction with Smad3 through its first zinc finger motif. Here, it is demonstrated that Evi-1 represses Smad-induced transcription by recruiting C-terminal binding protein (CtBP) as a corepressor. Evi-1 associates with CtBP1 through one of the consensus binding motifs, and this association is required for efficient inhibition of TGF-β signaling. A specific inhibitor for histone deacetylase (HDAc) alleviates Evi-1–mediated repression of TGF-β signaling, suggesting that HDAc is involved in the transcriptional repression by Evi-1. This identifies a novel function of Evi-1 as a member of corepressor complexes and suggests that aberrant recruitment of corepressors is one of the mechanisms for Evi-1–induced leukemogenesis.
Introduction
The Evi-1 gene was first identified as a common locus of retroviral integration in myeloid tumors in AKXD mice.1,2 It encodes a transcriptional regulator with 2 zinc finger domains. Evi-1 is shown to be highly expressed in human myeloid leukemias and myelodysplastic syndromes by chromosomal rearrangements involving 3q26, to which Evi-1 is mapped,3-6 although it is expressed at a very low level in a limited stage of normal myeloid cell differentiation. The most frequent rearrangements involving 3q26 are the t(3;3)(q21;q26) and the inv(3)(q21;q26). Cases of myelodysplastic syndrome with these anomalies are characterized by the increase in the platelet count and the dysplastic features of megakaryocytes and are designated as 3q21q26 syndrome.7 Aberrant expression of Evi-1 as a fusion transcript with AML1 (AML1/Evi-1) leads to blastic transformation in patients with chronic myelogenous leukemia.8 Even in the absence of cytogenetically evident abnormalities at chromosome 3q26, overexpression of Evi-1gene has been shown in a variety of myelogenous leukemias.9 These facts strongly suggest a critical role for Evi-1 in human leukemogenesis.
Our previous studies revealed that Evi-1 possesses diverse functions as an oncoprotein.10-12 Among these studies is our demonstration that Evi-1 and AML1/Evi-1 repress transforming growth factor β (TGF-β) signaling.13,14 TGF-β is a multifunctional peptide hormone that regulates various biological processes according to cellular contexts. In many types of cells, TGF-β acts as a negative regulator for cellular proliferation by inhibiting cell-cycle progression, leading to differentiation or apoptosis. In hematopoietic cells, TGF-β has also been shown to play an important role as a regulator of cellular growth and differentiation. Many studies show that early myeloid progenitors are sensitive to growth inhibition by TGF-β.15 Several cell lines derived from leukemic cells were reported to be resistant to the growth-inhibitory effect of TGF-β, and loss of responsiveness to TGF-β is supposed to contribute, at least in part, to the leukemogenesis.16
Intracellular mechanisms that transmit TGF-β signaling have been elucidated in detail. Upon binding of TGF-β to its receptors, Smad2 and Smad3, also called receptor-activated Smads (R-Smads), are phosphorylated by the activated TGF-β receptors and oligomerize with Smad4, called common Smads (Co-Smads). Then, the complexes of R-Smads and Co-Smads accumulate in the nucleus, interact with DNA, and activate transcription of TGF-β–responsive genes. This process is apparently simple, but many proteins, including inhibitory Smads, participate in regulating the process, and modifiy cellular responses to the stimuli.17,18 Genetic impairments in genes encoded for the Smad proteins that strongly associate with carcinogenesis. Smad4 is deleted in about 50% of pancreatic cancers.19 Germline mutations of humanSmad4 contribute to familial juvenile polyposis, an autosomal dominant disorder characterized by predisposition to gastrointestinal cancer.20Smad2 is also frequently inactivated by somatic mutations in sporadic cases of human colorectal cancer.21 From these observations, the Smad proteins are envisioned as acting as tumor suppressors. On the other hand, the proteins that inhibit TGF-β–induced signaling might have oncogenic property. To date, several proteins, including TGIF, Ski, and Sno, are shown to block Smad-mediated signaling.22-25 However, the implication of these Smad-inhibitory proteins in human cancer has not been well determined.
Evi-1 and AML1/Evi-1 antagonize the growth-inhibitory effect of TGF-β by interacting with Smad3 through its first zinc finger motif.13,14 This inhibitory effect on TGF-β signaling may be one of the critical mechanisms for leukemogenesis by Evi-1 and AML1/Evi-1. MOLM-1 cells, a human megakaryoblastoid cell line carrying the inv(3)(q21;q26), uniquely express the truncated form of the Evi-1 protein in which the C-terminal 44 amino acids of wild-type Evi-1 were replaced by 5 amino acids.3 MOLM-1 cells are relatively resistant to the growth-inhibitory effect of TGF-β. However, suppression of endogenous Evi-1 expression in these cells by means of antisense oligonucleotides restores TGF-β responsiveness.13 Thus, loss of responsiveness to TGF-β signaling may play an important role in leukemogenesis in these Evi-1–expressing cells.
C-terminal binding protein (CtBP) was originally identified as a protein that interacts with the C-terminal region of adenoviral oncoprotein E1A, which results in the reduced ability of E1A to transform cells.26,27 To date, 2 highly related homologues, CtBP1 and CtBP2, have been identified in vertebrates. It is known that CtBP2 is expressed primarily during embryogenesis in mice, whereas CtBP1 is widely expressed throughout the developmental stages. Differences in function between them, however, remain elusive.28 CtBP has been shown to have a transcriptional repression activity when fused to the heterologous DNA-binding motif.29 The Drosophila homologue of CtBP was shown to mediate transcriptional repression by Knirps, Krüppel, and Snail in the early embryo.29,30 In vertebrates, CtBP was also shown to act as a cofactor of certain transcriptional repressors, including basic Krüppel-like factor (BKLF), friend of GATA (FOG), and T-cell factor (TCF).31-33 These facts suggest a critical role for CtBP in transcriptional repression of genes in a wide variety of cells. CtBP was shown to interact with E1A of human adenoviruses 12 and 2 through the 5–amino acid sequences PLDLS and PVDLS (single-letter amino acid codes), respectively. These sequences are highly conserved among the E1A proteins.26 Repressor proteins that are described as interacting with CtBP so far also contain similar 5–amino acid sequences that are called CtBP-binding motifs.31-33 We have reported a repressor domain of Evi-1 in the region between amino acids 608 and 732.14 In this study, we identified another region of Evi-1, spanning amino acids 544 to 607, that is required for full repressor activity. Recently, Turner and Crossley31identified 2 CtBP-binding-motif–like sequences in this region of Evi-1 and showed physical interaction between mCtBP2 and a portion of Evi-1, including this region by the yeast 2-hybrid system. However, it remains to be determined whether Evi-1 interacts with CtBP in vivo and whether Evi-1–mediated repression depends on the interaction with CtBP. Furthermore, the most important point is to elucidate the biological phenomenon and its signaling pathway, where the physical interaction between Evi-1 and CtBP occurs.
To address these issues, we further investigated the mechanism for Smad-inhibition by Evi-1 and demonstrated that CtBP acts as an essential cofactor for Evi-1–mediated repression of TGF-β signaling.
Materials and methods
Cell culture and establishment of stable clones
COS7, HepG2, and Mv1Lu cells were grown in Dulbecco modified Eagle medium supplemented with penicillin and streptomycin and 10% fetal calf serum at 37°C in a 5% CO2 incubator. To generate stable Mv1Lu clones overexpressing Evi-1 and its mutant, the constructs subcloned into pME-18Sneo vector were transfected by means of Lipofectamine (Gibco BRL, Gaithersburg, MD) according to the manufacturer's instructions. These cells were selected in medium containing G418 (800 μg/mL). G418-resistant clones were screened for expression of Evi-1 by Western blotting. For each construct, 2 independent clones with comparable expression were used in further assays.
Plasmids
The human Evi-1 complementary DNA was inserted into theEcoRI site of plasmid pME18S or pME18Sneo.34The deletion mutants of Evi-1, Δ544- 607, Δ608-732, and Δ544-732, were constructed by means of a polymerase chain reaction (PCR) method. Mutant forms of Evi-1, AS/DL, DL/AS, and AS/AS, were generated by the site-directed mutagenesis method.35 Glutathione S-transferase (GST) fusion proteins of Evi-1 (544-607) were obtained by inserting in frame the corresponding fragments into pGEX-2TK (Amersham Pharmacia Biotech, Uppsala, Sweden). Smad3 and Smad4 were subcloned into pcDNA3 (Invitrogen, Carlsbad, CA). To construct fragments of CtBP fused to GST, corresponding CtBP fragments obtained by means of PCR were inserted in frame into pEBG, the eukaryotic GST fusion protein expression vector.
Luciferase assay
For analysis of luciferase activities, HepG2 cells were seeded in 12-well culture plates at a density of 4 × 104 per well. At 12 hours after seeding, the cells were transfected with 1 μg p3TP-Lux or p800neoLUC reporter plasmid along with the effector plasmids (400 ng for pME-Evi-1 or the equivalent molar for their derivatives, and 400 ng for Smads in pcDNA3) with SuperFect (Qiagen, Valencia, CA) according to the manufacturer's instructions. For analysis of the luciferase activity derived from cotransfection with several expression plasmids, the total amount of DNA in terms of weight was adjusted to be equal by adding the plasmid pUC13. As an internal control of transfection efficiency, a plasmid expressing β-galactosidase was cotransfected. The cells were harvested 48 hours after transfection and assayed for the luciferase activity by means of the luciferase assay system (Promega, Madison, WI) and a luminometer (Lumat, Berthold, Badwildbod, Germany). The data were normalized to the β-galactosidase activity. Cells were treated with 200 pM TGF-β (Roche Diagnostics, Mannheim, Germany) for 24 hours before harvesting. For the luciferase assay using the histone deacetylase (HDAc) inhibitor, 50 ng/mL trichostatin A (Waco, Osaka, Japan), dissolved in ethanol, was added to culture medium for 8 hours before harvesting. The same amounts of ethanol were added to the control cells.
Immunoprecipitation and Western blotting
COS7 cells were transfected by the diethylaminoethyl-dextran method as described previously.34 The cells were cultured for 48 to 72 hours after transfection and were lysed in the TNE buffer.13 For immunoprecipitation, cell lysates were incubated with the anti-T7 monoclonal antibody (Novagen, Madison, WI) for 3 hours at 4°C. Then the samples were incubated with protein A-Sepharose (Sigma, St Louis, MO) for 1 hour at 4°C. Immunoprecipitates were washed 5 times with the TNE buffer and were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analysis by Western blotting. Immunoblotting was performed with anti–Evi-1 serum10 and anti-T7 and was detected with the enhanced chemiluminescence system (Amersham Pharmacia Biotech).
GST pull-down assay
[35S]methionine-labeled CtBP1 was synthesized in vitro by means of TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. We collected 5 μg bacterially produced GST fusion proteins on Glutathione Sepharose 4B beads (Amersham Pharmacia Biotech) and incubated them with [35S]methionine-labeled CtBP1 for 3 hours at 4°C in the TNE buffer. Then, the beads were washed 5 times with the TNE buffer and subjected to SDS-PAGE followed by detection with autoradiography.
Growth-inhibition assay
The stable clones derived from Mv1Lu cells were seeded in duplicate at a density of 1 × 104 per well in 24-well culture plates. At 12 hours later, different doses of TGF-β were added, and the cells were incubated for 24 hours. During the last 4 hours, the cells were labeled with 1 μCi/mL [3H]thymidine (Amersham Pharmacia Biotech). The cells were washed with phosphate-buffered saline, fixed with 10% trichroloacetate, and solubilized with 0.5 M NaOH. The cell extracts were neutralized with HCl, and [3H] radioactivity was measured in a liquid scintillation β-counter (Aloka, Mitaka, Tokyo, Japan).
Results
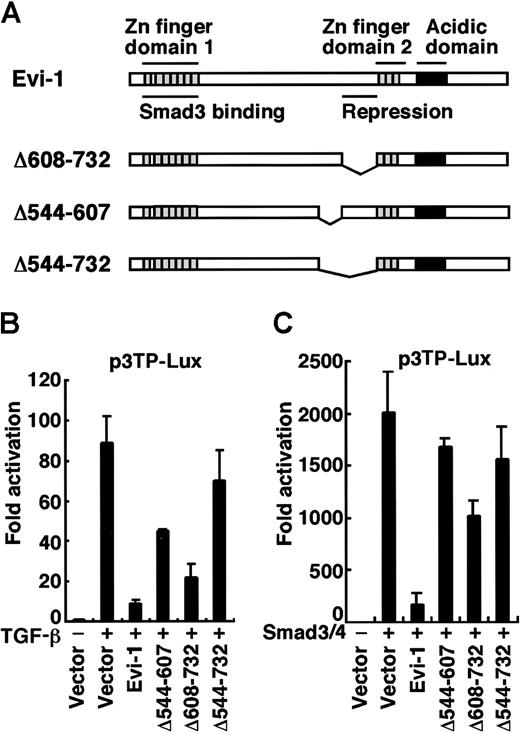
The region between amino acids 544 and 607 of Evi-1 is required to block TGF-β signaling
We previously identified 2 domains of Evi-1 that are indispensable for repression of TGF-β signaling.14 One is the first zinc finger domain, through which Evi-1 binds to Smad3 directly (Figure1A). The Evi-1 mutant that lacks this domain has lost the ability to suppress TGF-β signaling. Using C-terminal–truncated mutants of Evi-1, we found that the other domain, the region between amino acids 608 and 732 of Evi-1, is also required for efficient repression of TGF-β signaling, although it does not contribute to binding to Smad3. We have therefore termed this region the repression domain.
Identification of a domain responsible for repression in Evi-1.
(A) Structures of wild-type Evi-1 and its deletion mutants. The Smad3-binding domain and the repression domain are shown. (B) HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the absence or the presence of 200 pM TGF-β as indicated. (C) HepG2 cells were transfected with Smad3, Smad4, and p3TP-Lux together with wild-type or each mutant of Evi-1 as indicated in the absence of TGF-β. Luciferase activities were measured, and the values relative to the basal activity of the reporter are presented. The representative data of 3 independent experiments in duplicate are shown. Values and error bars depict the mean and the SD, respectively.
Identification of a domain responsible for repression in Evi-1.
(A) Structures of wild-type Evi-1 and its deletion mutants. The Smad3-binding domain and the repression domain are shown. (B) HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the absence or the presence of 200 pM TGF-β as indicated. (C) HepG2 cells were transfected with Smad3, Smad4, and p3TP-Lux together with wild-type or each mutant of Evi-1 as indicated in the absence of TGF-β. Luciferase activities were measured, and the values relative to the basal activity of the reporter are presented. The representative data of 3 independent experiments in duplicate are shown. Values and error bars depict the mean and the SD, respectively.
To define the full picture of the Evi-1 domains that contribute to inhibition of TGF-β signaling, we employed a series of deletion mutants of Evi-1 for specific regions and tested them for their repression activities. As shown in Figure 1, effector plasmids for Evi-1 were transfected with the reporter plasmid p3TP-Lux, a synthetic TGF-β–responsive reporter that contains the region of the plasminogen activator inhibitor 1 (PAI-1) promoter.36 As we previously showed, wild-type Evi-1 effectively repressed transcriptional activation induced by TGF-β (Figure 1B). There was no effect of Evi-1 on the basal activity of the promoter, indicating that the Evi-1 effect is specific for TGF-β–induced transcriptional activation (data not shown).14 We found that Δ608-732, which lacks the repression domain, had diminished repression activity. Remarkably, deletion of the region between amino acids 544 and 607 in Δ544-607 also severely impaired the repression activity. Deletion of both the regions (Δ544-732) completely abolished the activity of repression (Figure 1A-B). As overexpression of Smad3 mimics the effect of TGF-β on activation of the PAI-1 promoter,37 we performed the transcriptional response experiments by overexpressing Smad3 and Smad4 instead of exposing cells to TGF-β and obtained similar results (Figure 1C). We performed these experiments using p800neoLUC containing −799 to +81 of the 5′ end of the human PAI-1 gene38,39 as a reporter, and similar results were again obtained (data not shown). Although Evi-1 has been shown to be a DNA-binding protein,40 41there are no consensus sequences for Evi-1 binding in both of the promoters we used. Taken together, the region between amino acids 544 and 607 of Evi-1 (Evi-1 [544-607]) also participates in efficient inhibition of TGF-β–mediated transcriptional responses.
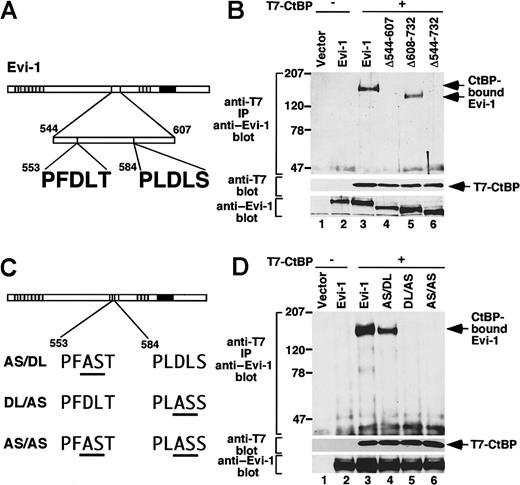
Evi-1 interacts with CtBP1 through its consensus motif
Recently, it was reported that Evi-1 contains 2 amino acid sequences, PFDLT (single-letter amino acid codes) and PLDLS, that fit to the CtBP-binding motif (Figure2A).31 These sequences are included in the region between amino acids 544 and 607 of Evi-1, which we showed as participating in efficient inhibition of TGF-β signaling, and they are completely conserved between mouse and human Evi-1 proteins.42 These findings allowed us to perform a coprecipitation assay to confirm whether Evi-1 interacts with CtBP in mammalian cells. We introduced T7-tagged CtBP1 with wild-type or the deletion mutants of Evi-1 into COS7 cells. Cell lysates were subjected to immunoprecipitation with anti-T7, followed by immunoblotting with anti–Evi-1. We observed that wild-type Evi-1 was coprecipitated with T7-CtBP1. However, Δ544-607 or Δ544-732, which lacks putative CtBP-binding motifs, was not. In contrast, Δ608-732, which retains the domain including the motifs, was coprecipitated with T7-CtBP1 (Figure 2B). From these results, we concluded that Evi-1 interacts with CtBP1 in vivo and that the region between amino acids 544 and 607 is responsible for binding to CtBP1. Next we examined the relative contribution of the 2 putative CtBP-binding motifs to the interaction with CtBP. For this purpose, we constructed Evi-1 mutants that harbor specific amino acid substitutions. The previous report demonstrated that substituting the AS residues for the DL residues in the CtBP-binding motif of E1A completely eliminates binding to CtBP1.26 Therefore, we introduced the same substitution into Evi-1 at the PFDLT (AS/DL), the PLDLS (DL/AS), or both (AS/AS) (Figure 2C). We transfected these mutants with T7-CtBP1 into COS7 cells and performed immunoprecipitation experiments. Each mutant is expressed comparably at the anticipated size. Both wild-type Evi-1 and the mutant for PFDLT (AS/DL) interacted with T7-CtBP1. However, neither of the 2 mutants for PLDLS (DL/AS and AS/AS) retained the ability to interact with CtBP (Figure 2D). Thus, of the 2 CtBP-binding-motif–like sequences, PLDLS at amino acid 584 is responsible for the interaction between Evi-1 and CtBP1, and PFDLT at amino acid 553 is not.
Evi-1 associates with CtBP1 through PLDLS.
(A) Evi-1 contains 2 putative CtBP-binding motifs. (B) The pME18S empty vector (lane 1), Evi-1 (lanes 2, 3), Δ544-607 (lane 4), Δ608-732 (lane 5), or Δ544-732 (lane 6) in pME18S was transfected into COS7 cells (2 × 106) with pRc/CMV empty vector (lanes 1, 2) or T7-CtBP in pRc/CMV (lanes 3 to 6). After 48 hours, cells were lysed and subjected to immunoprecipitation with anti-T7. T7-CtBP–bound Evi-1 was detected by Western blot by means of anti–Evi-1 (top). Positions of size markers (in kilodaltons) are indicated on the left. Expression of T7-CtBP and Evi-1 is monitored with anti–T7 (middle) and anti–Evi-1 (bottom), respectively. (C) Structures of the mutants of Evi-1. Mutations from DL to AS within the 5 amino acid motifs are underlined. (D) The pME18S empty vector (lane 1), Evi-1 (lanes 2, 3), AS/DL (lane 4), DL/AS (lane 5), or AS/AS (lane 6) was transfected into COS7 cells (2 ×106) together with pRc/CMV empty vector (lanes 1, 2) or T7-CtBP (lanes 3 to 6) in pRc/CMV. The immunoprecipitation was performed, and the results are shown as in Figure 2B.
Evi-1 associates with CtBP1 through PLDLS.
(A) Evi-1 contains 2 putative CtBP-binding motifs. (B) The pME18S empty vector (lane 1), Evi-1 (lanes 2, 3), Δ544-607 (lane 4), Δ608-732 (lane 5), or Δ544-732 (lane 6) in pME18S was transfected into COS7 cells (2 × 106) with pRc/CMV empty vector (lanes 1, 2) or T7-CtBP in pRc/CMV (lanes 3 to 6). After 48 hours, cells were lysed and subjected to immunoprecipitation with anti-T7. T7-CtBP–bound Evi-1 was detected by Western blot by means of anti–Evi-1 (top). Positions of size markers (in kilodaltons) are indicated on the left. Expression of T7-CtBP and Evi-1 is monitored with anti–T7 (middle) and anti–Evi-1 (bottom), respectively. (C) Structures of the mutants of Evi-1. Mutations from DL to AS within the 5 amino acid motifs are underlined. (D) The pME18S empty vector (lane 1), Evi-1 (lanes 2, 3), AS/DL (lane 4), DL/AS (lane 5), or AS/AS (lane 6) was transfected into COS7 cells (2 ×106) together with pRc/CMV empty vector (lanes 1, 2) or T7-CtBP (lanes 3 to 6) in pRc/CMV. The immunoprecipitation was performed, and the results are shown as in Figure 2B.
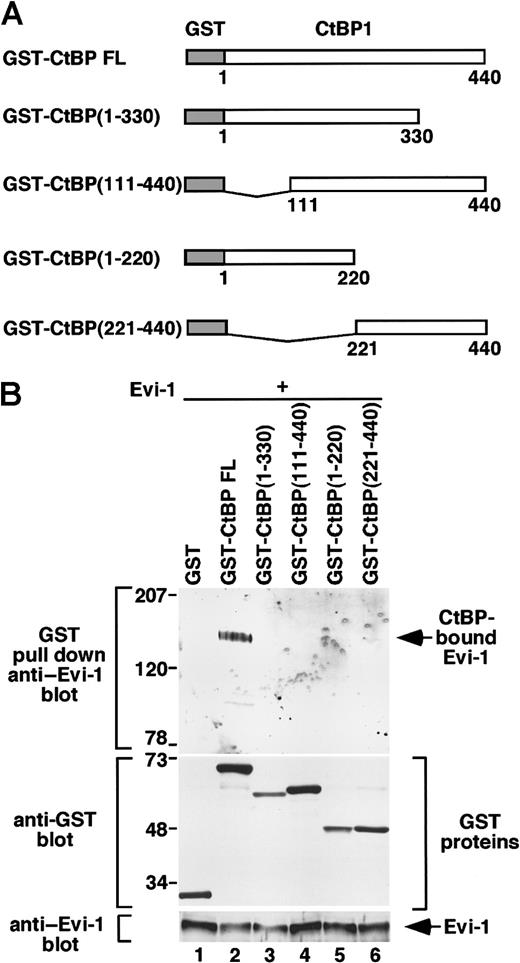
To determine the region of CtBP1 that interacts with Evi-1, we overexpressed wild-type Evi-1 with full-length or truncated forms of CtBP1 fused to GST in COS7 cells (Figure3A). Whereas a full-length CtBP1 strongly binds to Evi-1, none of the truncated forms of CtBP1 interacted with Evi-1 (Figure 3B). These results suggest that the integrity of the CtBP protein is required for interacting with Evi-1.
The whole structure of CtBP is required for interacting with Evi-1.
(A) Schematic presentation of GST-CtBP FL, GST-CtBP (1-330), GST-CtBP (111-440), GST-CtBP (1-220), and GST-CtBP (221-440). (B) pEBG (lane 1), CtBP FL (lane 2), CtBP (1-330) (lane 3), CtBP (111-440) (lane 4), CtBP (1-220) (lane 5), or CtBP (221-440) (lane 6) in pEBG was transfected into COS7 cells (4 × 106) together with Evi-1 in pME18S. Cell extracts were incubated with Glutathione Sepharose beads and washed extensively, and GST-CtBP–bound Evi-1 was detected by Western blot by means of anti–Evi-1 (top). Expression of GST-CtBP fusion constructs and Evi-1 is monitored with anti-GST (middle) and anti–Evi-1 (bottom), respectively. Positions of size markers (in kilodaltons) are indicated on the left.
The whole structure of CtBP is required for interacting with Evi-1.
(A) Schematic presentation of GST-CtBP FL, GST-CtBP (1-330), GST-CtBP (111-440), GST-CtBP (1-220), and GST-CtBP (221-440). (B) pEBG (lane 1), CtBP FL (lane 2), CtBP (1-330) (lane 3), CtBP (111-440) (lane 4), CtBP (1-220) (lane 5), or CtBP (221-440) (lane 6) in pEBG was transfected into COS7 cells (4 × 106) together with Evi-1 in pME18S. Cell extracts were incubated with Glutathione Sepharose beads and washed extensively, and GST-CtBP–bound Evi-1 was detected by Western blot by means of anti–Evi-1 (top). Expression of GST-CtBP fusion constructs and Evi-1 is monitored with anti-GST (middle) and anti–Evi-1 (bottom), respectively. Positions of size markers (in kilodaltons) are indicated on the left.
To determine whether Evi-1 binds to CtBP in vitro, we performed a pull-down assay using GST fusion proteins. Evi-1 [544-607] and its mutants for the CtBP-binding motifs were fused to GST (Figure4A) and expressed in bacteria. These proteins are immobilized onto Glutathione Sepharose 4B beads and incubated with [35S]methionine-labeled, in vitro–translated CtBP1. GST-DL/AS and GST-AS/AS failed to bind to CtBP, whereas GST-DL/DL and GST-AS/DL interacted with in vitro–translated CtBP1 (Figure 4B). These data indicate that Evi-1 interacts with CtBP1 in vitro.
Evi-1 interacts with CtBP1 in vitro.
(A) Construction of GST fusion proteins of the wild-type or mutant forms of Evi-1 (544-607). (B) GST pull-down experiments. Here, 5 μg bacterially expressed GST alone (lane 1), GST-DL/DL (lane 2), GST-AS/DL (lane 3), GST-DL/AS (lane 4), and GST-AS/AS (lane 5) were incubated with in vitro–translated [35S]methionine-labeled CtBP1. After extensive washing, the samples were subjected to SDS-PAGE followed by autoradiography (top). Half of the input of in vitro–translated [35S]methionine-labeled CtBP was presented in lane 6. Positions of size markers (in kilodaltons) are indicated on the left. Coomassie stains of GST-fusion proteins are indicated (bottom).
Evi-1 interacts with CtBP1 in vitro.
(A) Construction of GST fusion proteins of the wild-type or mutant forms of Evi-1 (544-607). (B) GST pull-down experiments. Here, 5 μg bacterially expressed GST alone (lane 1), GST-DL/DL (lane 2), GST-AS/DL (lane 3), GST-DL/AS (lane 4), and GST-AS/AS (lane 5) were incubated with in vitro–translated [35S]methionine-labeled CtBP1. After extensive washing, the samples were subjected to SDS-PAGE followed by autoradiography (top). Half of the input of in vitro–translated [35S]methionine-labeled CtBP was presented in lane 6. Positions of size markers (in kilodaltons) are indicated on the left. Coomassie stains of GST-fusion proteins are indicated (bottom).
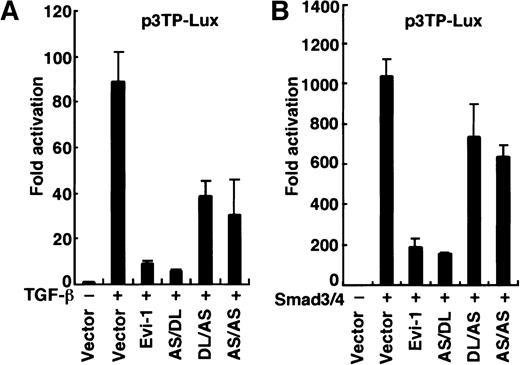
Evi-1 requires CtBP as a corepressor in inhibition of Smad3
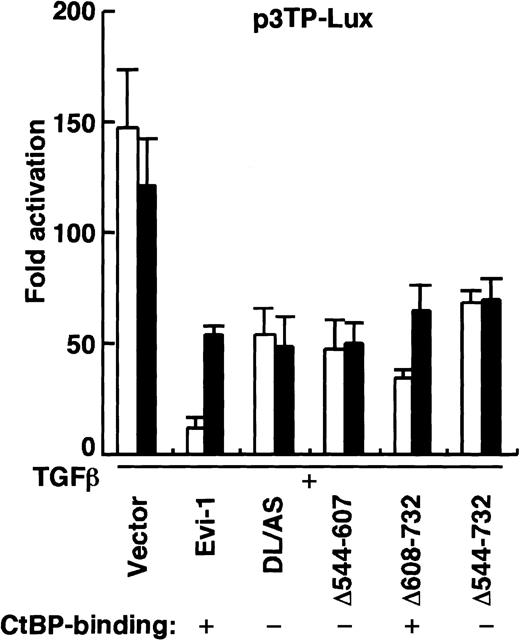
CtBP is shown to act as a transcriptional corepressor inDrosophila and in vertebrates.28,29 43 To investigate a role for CtBP in Evi-1–mediated repression, we tested whether the AS mutation, which abrogates the binding of CtBP1 to Evi-1, would impair the ability of Evi-1 to repress TGF-β signaling. We transfected p3TP-Lux into HepG2 cells together with the effector plasmids as indicated in Figure 5. The AS/DL mutant, which retains the ability to interact with CtBP1, repressed TGF-β–mediated transactivation as efficiently as wild-type Evi-1 (Figure 5A). In contrast, DL/AS or AS/AS mutant, in which the interaction with CtBP1 is specifically abrogated, showed a severely reduced ability of repression. We also performed these experiments using overexpression of Smad3 and Smad4 instead of exposure to TGF-β, and similar results were obtained (Figure 5B). The experiments using p800neoLUC as a reporter plasmid demonstrated similar results (data not shown). Taken together, these data indicate that recruitment of CtBP plays an important role in full repression of TGF-β signaling by Evi-1.
Evi-1 requires CtBP as a corepressor for inhibition of TGF-β signaling.
(A) HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the absence or the presence of 200 pM TGF-β as indicated. (B) Smad3, Smad4, and p3TP-Lux were transfected into HepG2 cells together with wild-type or each mutant of Evi-1 in the absence of TGF-β. Luciferase activities were assayed and presented as described in the legend for Figure 1.
Evi-1 requires CtBP as a corepressor for inhibition of TGF-β signaling.
(A) HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the absence or the presence of 200 pM TGF-β as indicated. (B) Smad3, Smad4, and p3TP-Lux were transfected into HepG2 cells together with wild-type or each mutant of Evi-1 in the absence of TGF-β. Luciferase activities were assayed and presented as described in the legend for Figure 1.
An HDAc inhibitor alleviates Evi-1–mediated Smad repression
An HDAc is known to mediate transcriptional repression by rendering the nearby chromatin inaccessible to transcriptional activators through deacetylation of histone proteins.44,45HDAc forms a large multiprotein complex with corepressor proteins, including the nuclear hormone corepressor (N-CoR), the silencing mediator for retinoid and thyroid-hormone receptors, and the mammalian homologue of yeast transcriptional repressor SIN3 (mSin3).46,47 The recent study demonstrates that CtBP interacts with HDAc1, a member of HDAc family, both in vitro and in vivo.48 We also performed the immunoprecipitation study by coexpressing CtBP1 and HDAc1 in COS7 cells and confirmed the interaction between them (data not shown). These observations suggest that CtBP might play a role as a corepressor by recruiting HDAcs. To investigate whether Evi-1–mediated repression involves histone deacetylation, we performed a luciferase assay using a specific HDAc inhibitor, trichostatin A.49 Trichostatin A has been shown to relieve transcriptional repression by Mad-Max complex or by other repressor proteins, which are mediated by HDAc.50 51 We cultured HepG2 cells with 50 ng/mL trichostatin A for 8 hours before harvesting, and the treatment had little effect on the basal or TGF-β–induced activity of p3TP-Lux (Figure 6). In contrast, repression of TGF-β signaling elicited by Evi-1 was significantly attenuated by the treatment with trichostatin A. To determine whether the effect of trichostatin A is dependent on recruitment of CtBP, we tested Evi-1 mutants with altered CtBP-binding properties for trichostatin A–induced derepression in this assay. DL/AS mutant and Δ544-607, both of which do not interact with CtBP, were little affected by trichostatin A treatment. However, Δ608- 732 mutant, which lacks the previously identified repression domain but preserves binding to CtBP, was sensitive to trichostatin A treatment (Figure 6). These results suggest that an HDAc activity is involved in repression of TGF-β signaling by Evi-1 and that this is mediated chiefly through the interaction with CtBP.
Effect of trichostatin A on Evi-1–mediated repression.
HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the presence of 200 pM TGF-β as indicated. Luciferase activities were measured 48 hours after transfection following an 8-hour treatment with (▪) or without (■) 50 ng/mL trichostatin A. Luciferase activities relative to the basal activity of the reporter are presented. Values and error bars depict the mean and the SD, respectively.
Effect of trichostatin A on Evi-1–mediated repression.
HepG2 cells were transfected with p3TP-Lux together with wild-type or each mutant of Evi-1 in the presence of 200 pM TGF-β as indicated. Luciferase activities were measured 48 hours after transfection following an 8-hour treatment with (▪) or without (■) 50 ng/mL trichostatin A. Luciferase activities relative to the basal activity of the reporter are presented. Values and error bars depict the mean and the SD, respectively.
Association with CtBP is required for Evi-1 to inhibit TGF-β–induced growth inhibition
Mv1Lu cell is a mink lung endothelial cell line that is highly responsive to TGF-β–mediated growth inhibitory signals. Overexpression of Evi-1 in Mv1Lu cells abrogates the antiproliferative effect of TGF-β.14 To investigate a role for CtBP in the Evi-1–mediated block of antiproliferative effect of TGF-β, we established Mv1Lu clones that stably express either wild-type Evi-1 or the DL/AS mutant. We introduced the expression vector for wild-type Evi-1 or the DL/AS mutant that enables concomitant expression of the neomycin-resistant gene into Mv1Lu cells and selected them for neomycin resistance in the presence of G418. Of the resultant G418-resistant clones, Western blot analyses using anti–Evi-1 revealed 4 representative clones that express high levels of wild-type Evi-1 (W-55 and W-60) or the DL/AS mutant (A-72 and A-82) (Figure7A). Two independent Mv1Lu clones, M-2 and M-4, which were transfected with the empty vector, were used as controls. In these experiments, intrinsic Evi-1 was below the detectable level in the naive Mv1Lu cells. When cultured in complete medium without TGF-β, all of these clones showed comparable viabilities and proliferative abilities (data not shown). We then evaluated growth-inhibitory effects of TGF-β on these Mv1Lu clones using [3H]thymidine-incorporation assays. Shown in Figure7B are the effects on [3H]thymidine uptake when these clones were exposed to increasing amounts of TGF-β. The Mv1Lu clones, which overexpress Evi-1 (W-55 and W-60), showed reduced responsiveness to TGF-β, in comparison with mock-transfected clones (M-2 and M-4). In contrast, the growth of A-72 and A-82 clones, which overexpress the DL/AS mutant, was inhibited in response to TGF-β as efficiently as that of mock-transfected clones. These results suggest that recruitment of CtBP is required for Evi-1 to release cells from the growth-inhibitory effect of TGF-β in vivo.
Association with CtBP is required for Evi-1 to block TGF-β–induced growth inhibition.
(A) Expression of wild-type Evi-1 and its mutant form, DL/AS, in the stable Mv1Lu transfectant clones. Clones W-55 (lane 3) and W-60 (lane 4) were established from cells transfected with wild-type Evi-1, while A-72 (lane 5) and A-82 (lane 6) were from cells transfected with the DL/AS mutant of Evi-1. M-2 (lane 1) and M-4 (lane 2) are mock clones transfected with pME18Sneo empty vector. The results of immunoblotting with anti–Evi-1 are shown. (B) The Mv1Lu clones stably transfected with wild-type Evi-1 (W-55, ▵, and W-60, ▴), DL/AS (A-72, ■, and A-82, ▪), and control clones (M-2, ○, and M-4, ●) were subjected to a [3H]thymidine-incorporation assay in the presence of the indicated concentrations of TGF-β. Results are expressed as percentages relative to values obtained from control cultures that did not receive TGF-β. Values and error bars depict the mean and the SD, respectively.
Association with CtBP is required for Evi-1 to block TGF-β–induced growth inhibition.
(A) Expression of wild-type Evi-1 and its mutant form, DL/AS, in the stable Mv1Lu transfectant clones. Clones W-55 (lane 3) and W-60 (lane 4) were established from cells transfected with wild-type Evi-1, while A-72 (lane 5) and A-82 (lane 6) were from cells transfected with the DL/AS mutant of Evi-1. M-2 (lane 1) and M-4 (lane 2) are mock clones transfected with pME18Sneo empty vector. The results of immunoblotting with anti–Evi-1 are shown. (B) The Mv1Lu clones stably transfected with wild-type Evi-1 (W-55, ▵, and W-60, ▴), DL/AS (A-72, ■, and A-82, ▪), and control clones (M-2, ○, and M-4, ●) were subjected to a [3H]thymidine-incorporation assay in the presence of the indicated concentrations of TGF-β. Results are expressed as percentages relative to values obtained from control cultures that did not receive TGF-β. Values and error bars depict the mean and the SD, respectively.
Discussion
In this study, we investigated the mechanism through which Evi-1 blocks TGF-β signaling and found that the CtBP protein acts as a corepressor of Evi-1. Furthermore, we demonstrated that one of the mechanisms for Smad3 inhibition by Evi-1 should be recruitment of an HDAc complex through the CtBP protein. Recently, several proteins were also shown to negatively regulate the TGF-β–induced signals.17,18,22-25 Among them, TGIF, Ski, and Sno are shown to act as members of transcriptional corepressor complexes, including HDAc.22-25
Involvement of corepressor proteins in human leukemogenesis has been proposed with the demonstration that PML/RARα, the fusion protein of promyelocytic leukemia (PML) and retinoic acid α (RARα), recruits a corepressor complex, including N-CoR and HDAc, to RARα target genes, thereby blocking myeloid cell differentiation in acute promyelocytic leukemia.51 The AML1/ETO chimeric protein, which is generated in acute myelogenous leukemia with t(8;21), has also been shown to recruit corepressor complexes.52 53 Our study, for the first time, suggests a role for a distinct type of corepressor, CtBP, in human leukemogenesis.
In the previous study, we showed that Evi-1 directly binds to the MH2 domain of Smad3 in the nucleus through its first zinc finger domain and affects the interaction of the complex of Smad3 and Smad4 with its DNA-binding sequence. A C-terminal portion of Evi-1 spanning amino acids 608 to 732 (Evi-1 [608-732]), which is not involved in the binding to Smad3, also contributes to the repression.14 In the present study, we identified another region of Evi-1, spanning amino acids 544 to 607, that is also required for repressing TGF-β signaling. This region overlaps with the region between amino acids 514 and 633, which, on the basis of results of the artificial transcriptional response analysis, has been proposed as the transcriptional repression domain.54 This region contains 2 amino acid sequences reminiscent of CtBP-binding motifs, which are completely conserved between mouse and human Evi-1 proteins. Recently, another group showed that the region between amino acids 514 and 726 of Evi-1 physically interacts with mCtBP2 in the yeast 2-hybrid system.31 However, a role for the CtBP protein in Evi-1–mediated repression remained to be determined because the interaction with CtBP does not immediately indicate CtBP-dependent repression. For example, FOG-2, which also physically interacts with CtBP, was shown to repress GATA4-mediated transcription by a CtBP-independent mechanism.55 In this study, we showed that Evi-1 interacts with CtBP in vivo and that, of these 2 CtBP-binding-motif–like sequences, PLDLS at amino acid 584 is required for Evi-1 to interact with CtBP. Furthermore, we found that the interaction with CtBP is crucial for the efficient repression of TGF-β signaling by Evi-1. These results strongly suggest that CtBP acts as a corepressor of Evi-1 in repressing TGF-β signaling. In support of this is our demonstration that disruption of the CtBP-binding motifs of Evi-1 completely abrogated its ability to block the growth-inhibitory effect of TGF-β in vivo. CtBP proteins have been shown to act as a corepressor of growing members of transcriptional repressors in Drosophila and vertebrates; among those are Hairy, Knirps, Krüppel, and Snail inDrosophila,29 and CtBP-interacting protein (CtIP),56,57 BKLF,31 FOG,32Net,43ZEB/δEF-1,28,58 TCF,33 and Ikaros59 in vertebrates. These findings propose another mechanism for Evi-1–mediated repression of TGF-β signaling. In addition to inhibiting Smad3 from binding to DNA, Evi-1 may, as a corepressor, recruit CtBP to Smad complexes to fully repress the expression of target genes. Given the fact that mutant forms of Evi-1, which are unable to associate with CtBP, retained the partial transcriptional repression activity in the reporter assay, CtBP-independent mechanisms should be active in the repression of TGF-β signaling. In these lines, transcriptional repressors, such as BKLF, Hairy, FOG, and δEF1, require CtBP binding for their repression activity, but also contain CtBP-independent repression domains.28,31,32 60 The repression domain that we had previously identified (Evi-1 [608-732]) might be one of these domains because it does not contribute to the interaction between Evi-1 and CtBP1. One possibility is that another factor yet to be identified might bind to this region and act in cooperation with CtBP. Further studies to elucidate the precise mechanism of repression are in progress.
It remains to be determined how CtBP works as a transcriptional corepressor. One possibility is that CtBP recruits an HDAc complex. CtBP has been shown to interact with HDAc1 both in vitro and in vivo.48 We demonstrated that trichostatin A, a specific inhibitor of HDAc, significantly alleviates the Evi-1–mediated repression. Moreover, Evi-1 mutants that cannot bind to CtBP are not vulnerable to trichostatin A, which suggests that derepression by trichostatin A is dependent on CtBP. Ski, Sno, and TGIF are transcriptional corepressor proteins that associate with Smad proteins and inhibit their transcriptional activation with dependence on an HDAc activity.22-25 Although these corepressors for Smad proteins are known to be endogenously expressed in a variety of cells, the present study is hardly affected by them, if at all, because trichostatin A treatment does not change the basal transcriptional activity or the activity induced by TGF-β in our transcriptional response assay. These findings strongly support the model for repression in which Evi-1 recruits CtBP, and this in turn conducts an HDAc complex, which exerts the transcriptional repression activity. Recent reports, however, propose another model for CtBP-mediated repression through polycomb complexes.59,61 In this model, CtBP, as a dimer, acts as a bridge between specific repressor proteins and the polycomb complex, which is supposed to package regions of DNA into heterochromatin-like structures without the help of HDAcs.62 63 To date, however, there is no experimental demonstration that polycomb proteins play a central role in CtBP-mediated transcriptional repression.
Our study provides a novel function of Evi-1 as a transcriptional corepressor that interacts with DNA-binding transcription factors. Thus far, Evi-1 is proposed as a DNA-binding protein because of the observation that it directly interacts with specific sequences of DNA in vitro.40 41 Although these sequences overlap with the binding site for the GATA-family transcription factors, target genes that Evi-1 directly regulates have not been identified so far. Thus, Evi-1 may, in addition to binding to DNA directly, regulate transcription by interacting with DNA-binding proteins, as in the case of Smad3, and by recruiting corepressor complexes, including CtBP. Identification of those target DNA-binding proteins may greatly contribute to understanding Evi-1–induced leukemogenesis.
We thank G. Chinnadurai (St Louis University, MO) for providing T7-hCtBP1, J. Massagué (Memorial Sloan-Kettering Cancer Center) for Smad4, R. Derynck (University of California) for Smad3, K. Miyazono (The Cancer Institute of the Japanese Foundation for Cancer Research) for p3TP-Lux, M. Abe (Institute of Development, Aging and Cancer, Tohoku University, Sendai, Japan) for p800neoLUC, B. J. Mayer (Howard Hughes Medical Institute, Harvard Medical School) for pEBG, S. L. Schreiber (Howard Hughes Medical Institute) for HDAc1, and K. Arai (The Institute of Medical Science, University of Tokyo, Japan) for pME18Sneo.
Supported in part by The Mitsubishi Foundation and grants-in-aid for cancer research from the Ministry of Health and Welfare and the Ministry of Education, Science and Culture of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hisamaru Hirai, Dept of Hematology and Oncology, Graduate School of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, Japan; e-mail: hhirai-tky@umin.ac.jp.

![Fig. 4. Evi-1 interacts with CtBP1 in vitro. / (A) Construction of GST fusion proteins of the wild-type or mutant forms of Evi-1 (544-607). (B) GST pull-down experiments. Here, 5 μg bacterially expressed GST alone (lane 1), GST-DL/DL (lane 2), GST-AS/DL (lane 3), GST-DL/AS (lane 4), and GST-AS/AS (lane 5) were incubated with in vitro–translated [35S]methionine-labeled CtBP1. After extensive washing, the samples were subjected to SDS-PAGE followed by autoradiography (top). Half of the input of in vitro–translated [35S]methionine-labeled CtBP was presented in lane 6. Positions of size markers (in kilodaltons) are indicated on the left. Coomassie stains of GST-fusion proteins are indicated (bottom).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2815/6/m_h80910983004.jpeg?Expires=1767698633&Signature=bWLJ0hzuJ0vAIfaApc3exLJ6lF6c9AYkAQzIZUep9GLiSXnucG6ax6vOA46rG-XEIMYOs2zI9TSSU0afE4szqeXB~uMocaJz4~m44S5t1hKiZ~ePNquEu4kMlFC3hjX-GNd1uNCPeFzHB1hNpVp0bti2NC3EVfk2Cb1coHMvUfkGKwRDcarWd62z2qF6dbo3S75goc84e8~dDVjNt5hMMUl8-qclhBjPjjLA5UofEP1PAmOSlrlqWJ36tWIm9-H5335OvlV6D00HcAweZHUdxns-O~OMoeCkRzxe3sOkZA47WI4J2mXJ-TLd3PGlc87pStqkDVBAx8BOYVSUplQ3LA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Association with CtBP is required for Evi-1 to block TGF-β–induced growth inhibition. / (A) Expression of wild-type Evi-1 and its mutant form, DL/AS, in the stable Mv1Lu transfectant clones. Clones W-55 (lane 3) and W-60 (lane 4) were established from cells transfected with wild-type Evi-1, while A-72 (lane 5) and A-82 (lane 6) were from cells transfected with the DL/AS mutant of Evi-1. M-2 (lane 1) and M-4 (lane 2) are mock clones transfected with pME18Sneo empty vector. The results of immunoblotting with anti–Evi-1 are shown. (B) The Mv1Lu clones stably transfected with wild-type Evi-1 (W-55, ▵, and W-60, ▴), DL/AS (A-72, ■, and A-82, ▪), and control clones (M-2, ○, and M-4, ●) were subjected to a [3H]thymidine-incorporation assay in the presence of the indicated concentrations of TGF-β. Results are expressed as percentages relative to values obtained from control cultures that did not receive TGF-β. Values and error bars depict the mean and the SD, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2815/6/m_h80910983007.jpeg?Expires=1767698633&Signature=UFK5k2~rGqhuHwYXiNX4sxLRfGkHYUIQ0mTaSNfWO5NF6F1Au8n2XacJ-YQmfY~QfNglCuwYgaBiDMcnCxEJwtmkoQRBOWsr33-AHohr8TZiqIfe19eACBo95Nu-HgYA1P~jxqRZKwWgyRTdGyfDeI2TorUtIIfs0yBFPl2UVVLpgXlKLPX8iDXlcMrmKdoVkfVTfkIoNu2VlXFJ2wza9V8jJzhYuqV919acJLvnCGiUtrzu047S~yzSt5oZID881T63X3eITe7Tl0bl5~OkIoK7AOWmC-MohNH9yb0esR~HznwiWv~PqsJGuJqAEth297UzZH73Uh1JTysV5WUj~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal