Abstract
Plasminogen activator inhibitor-1 (PAI-1) expression is induced by hypoxia (8% O2) via the PAI-1 promoter region −175/−159 containing a hypoxia response element (HRE-2) binding the hypoxia-inducible factor-1 (HIF-1) and an adjacent response element (HRE-1) binding a so far unknown factor. The aim of the present study was to identify this factor and to investigate its role in the regulation of PAI-1 expression. It was found by supershift assays that the upstream stimulatory factor-2a (USF-2a) bound mainly to the HRE-1 of the PAI-1 promoter and to a lesser extent to HRE-2. Overexpression of USF-2a inhibited PAI-1 messenger RNA and protein expression and activated L-type pyruvate kinase expression in primary rat hepatocytes under normoxia and hypoxia. Luciferase (Luc) gene constructs driven by 766 and 276 base pairs of the 5′-flanking region of the PAI-1 gene were transfected into primary hepatocytes together with expression vectors encoding wild-type USF-2a and a USF-2a mutant lacking DNA binding and dimerization activity (ΔHU2a). Cotransfection of the wild-type USF-2a vector reduced Luc activity by about 8-fold, whereas cotransfection of ΔHU2a did not influence Luc activity. Mutation of the HRE-1 (−175/−168) in the PAI-1 promoter Luc constructs decreased USF-dependent inhibition of Luc activity. Mutation of the HRE-2 (−165/−158) was less effective. Cotransfection of a HIF-1α vector could compete for the binding of USF at HRE-2. These results indicated that the balance between 2 transcriptional factors, HIF-1 and USF-2a, which can bind adjacent HRE sites, appears to be involved in the regulation of PAI-1 expression in many clinical conditions.
Introduction
The tissue-type and the urokinase-type plasminogen activators (tPA and uPA) are serine proteases converting the inactive zymogen plasminogen to the active endopeptidase plasmin.1The tPA and uPA activity is regulated, in part, by plasminogen activator inhibitors (PAIs).2 Among 2 identified inhibitors, PAI-1 and PAI-2, PAI-1 is the primary physiologic inhibitor of both tPA and uPA.3 PAI-1 is a 50-kd glycoprotein from the serpin superfamily.4 It can be produced by platelets, vascular endothelial cells,5 vascular smooth muscle cells,6 and several nonvascular cell types,7,8 including hepatocytes.9 PAI-1 has also been identified as a component of the extracellular matrix.10
The plasminogen activator inhibitors are involved in many functions, both under normal and pathological conditions, including fibrinolysis, extracellular matrix turnover, and fibrosis.11 PAI-1 also participates in wound healing and cancer metastasis.12 13
Certain pathophysiologic processes in which PAI-1 levels increase (eg, prethrombotic events, hemorrhage, and thrombus formation) are associated with hypoxia. PAI-1 gene expression was induced by mild hypoxia (8% O2) via an O2-responsive promoter sequence (−175/−158) containing hypoxia response element-1 (HRE-1, −175/−168) and hypoxia response element-2 (HRE-2, −165/−158) in primary cultured rat hepatocytes.14 HRE-2 was shown to bind the hypoxia-inducible factor-1 (HIF-1) and thus to be most critical for the increase in PAI-1 gene expression under hypoxia. The HRE-1 sequence was shown to bind a factor other than HIF-1.
The HRE-1 and also HRE-2 include a CACGTG-like sequence that can be recognized by transcription factors containing basic helix-loop-helix (bHLH) domains. Besides HIF-1 consisting of the bHLH-PAS (Per-ARNT-Sim) domain proteins HIF-1α and HIF-1β (ARNT, or arylhdrocarbon receptor nuclear translocator),15 the bHLH leucine zipper (bHLH-zip) proteins, upstream stimulatory factors (USF),16 or the Myc/Max 16,17 transcription factors also can bind to the CACGTG sequence. USF was initially identified in HeLa cells as a protein binding the CACGTG sequence located immediately upstream of the TATA sequence of the adenovirus major late promoter,16 thereby activating transcription.18
Two different ubiquituously expressed forms of USF (USF-1 and USF-2) with different molecular weights have been identified; however, their relative abundance varies in different cell types.19,20The USF family members are quite different in their N-terminal amino acid sequences, whereas their DNA binding and dimerization domains were highly identical.19 Thus, it may be likely that proteins of the bHLH-zip family can bind to the PAI-1 HRE sites. Therefore, it was the aim of the present study to identify the HRE-1 binding factor and to investigate its role in the regulation of PAI-1 expression using primary cultured rat hepatocytes as a model system. By using electrophoretic mobility shift assays (EMSAs) and supershift assays, it was shown that USF proteins bound to the hypoxia-responsive elements of the rat PAI-1 gene promoter, mainly HRE-1, and that overexpression of USF-2a inhibited PAI-1 expression under normoxia and hypoxia to the same extent. Mutations of both HREs abolished the inhibition of PAI-1 with a more prominent effect on HRE-1. Furthermore, the inhibitory action of USF-2 at HRE-2 could be antagonized by transfection of a HIF-1α expression vector. Thus, USF-2a appears to play a modulatory role in the PAI-1 expression under different conditions such as hypoxia and proliferation.
Materials and methods
All biochemicals and enzymes were of analytical grade and were purchased from commercial suppliers.
Animals
Male Wistar rats (200-260 g) were kept on a 12-hour day-night rhythm with free access to water and food. Rats were anesthetized with pentobarbital (60 mg/kg body weight) prior to preparation of hepatocytes.
Cell culture experiments
Hepatocytes were isolated by collagenase perfusion. Cells (1 × 106 per dish) were in a normoxic atmosphere of 16% O2, 79% N2, and 5% CO2 (by volume) in medium M-199 containing 0.5 nM insulin, 100 nM dexamethasone as permissive hormones, and 4% fetal calf serum for the initial 4 hours of culture. Cells were then cultured in serum-free medium from 4 to 24 hours at normal arterial 16% O2. At 24 hours, medium was changed and culture was continued at normoxia 16% O2or at mild hypoxia 8% O2 (87% N2, 5% CO2 by volume).
Plasmid constructs
The pGl3PAI-766 plasmid, containing the rat PAI-1 promoter 5′-flanking region21 from −766 to +31, as well as pGl3PAI-766M1 and pGl3PAI-766M2 were previously described.14 Plasmids pGl3PAI-276, pGl3PAI-276M1, and pGl3PAI-276M2 were constructed from pGl3PAI-766, pGl3PAI-766M1, and pGl3PAI-766M2 by excision of a 490–base pair (bp) KpnI fragment and subsequent relegation of the remaining vector. The construct L-type pyruvate kinase–183 luciferase (PKL-183 Luc) was constructed by excision of the PKL-183 promoter sequence from PKL-183 CAT,22 withSacI and subsequent ligation into the SacI sites of pGl3 basic (Promega, Heidelberg, Germany). The human USF-2a, ΔHU2a, and ΔTDU2 plasmids were a kind gift from Dr A. Kahn and Dr M. Raymondjean and, as well as the HIF-1α expression vector, they have been already described.14 23
RNA preparation and Northern analysis
Isolation of total RNA and Northern analysis were performed as described.24 Digoxigenin (DIG)-labeled antisense RNAs served as hybridization probes; they were generated by in vitro transcription from pBS–PAI-1 and pBS-PKL using T3 RNA polymerase or from pBS–β-actin using T7 RNA polymerase and RNA labeling mixture containing 3.5 mM 11-DIG–UTP, 6.5 mM UTP, 10 mM GTP, 10 mM CTP, and 10 mM ATP. Hybridizations and detections were carried out essentially as previously described.24 Blots were quantified with a videodensitometer (Biotech Fischer, Reiskirchen, Germany).
Western blot analysis
Western blot analysis was carried out as described.25 In brief, media from primary cultured hepatocytes were collected, and the protein content was determined using the Bradford method. A total of 50 μg protein was loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel and after electrophoresis blotted onto nitrocellulose membranes. The primary rabbit antibody against rat PAI-1 (American Diagnostics, Greenwich, CT) was used in a 1:200 dilution. The secondary antibody was a goat antirabbit immunoglobulin G (Santa Cruz Biotechnology, Santa Cruz, CA) used in a 1:2000 dilution. The PKL Western analysis was performed as described 26 except that the primary antibody was a mouse monoclonal antibody against rat PKL,27 which was used in a 1:100 dilution. The secondary antibody was an antimouse IgG horseradish peroxidase (Santa Cruz Biotechnology) used in a 1:2000 dilution. The enhanced chemiluminescence Western blotting system (Amersham, Freiburg, Germany) was then used for detection. Under these conditions PAI-1 was seen as a double band, the major 49-kd band, and the minor 46-kd band,25 and PKL was visible as a 60-kd band.
Cell transfection and Luc assay
Freshly isolated rat hepatocytes (about 1 × 106cells per dish) were transfected as described.26 In brief, 2 μg of the appropriate PAI-1 or PKL promoterFirefly Luc construct was transfected together with 500 ng USF-2a, ΔHU2a, or ΔTDU2 expression vectors or in the controls with 500 ng USF-2a backbone plasmid pCMV. In competition experiments, 2 μg of the appropriate PAI-1 promoter Luc construct was transfected together with 500 ng USF-2a and 500 ng HIF-1α expression vector or with 500 ng USF-2a or HIF-1α vector plus 500 ng pCMV vector. In the controls, 1 μg pCMV vector was used with 2 μg of the appropriate PAI-1 construct. After 5 hours, the medium was changed and the cells were cultured under normoxia for 19 hours. Then, medium was changed again and the cells were further cultured for 24 hours under normoxia or mild hypoxia.
Preparation of nuclear extracts and EMSA
Nuclear extracts were prepared by modification of a standard protocol,28,29 with buffers A and C containing 0.5 mM dithioerythritol (Sigma, Deisenhofen, Germany), 0.4 mM phenylmethylsulfonyl fluoride (Serva), 2 μg leupeptin per milliliter (Roche, Mannheim, Germany), 2 μg pepstatin per milliliter (Roche), 2 μg aprotinin per milliliter (Bayer, Leverkusen, Germany), 1 mM sodium vanadate (Sigma), and the “complete” protease inhibitor cocktail tablets (Roche) essentially as described.14 The sequences of the PAI-1 oligonucleotides used for the EMSA are shown in Figure1A. Equal amounts of complementary oligonucleotides were annealed and labeled by 5′-end labeling with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (MBI, St Leon-Rot, Germany). They were purified with the Nucleotide Removal Kit (Qiagen, Hilden, Germany). Binding reactions were carried out in a total volume of 20 μL containing 50 mM KCl, 1 mM MgCl2, 1 mM ethylenediaminetetraacetic acid, 5% glycerol, 10 μg nuclear extract, 250 ng poly(dIdC), and 5 mM dithioerythritol. For competition analyses, a 1-, 5-, 10-, or 50-fold molar excess of unlabeled annealed oligonucleotide was added. After preincubation for 5 minutes at room temperature, 1 μL of the labeled probe (104 cpm) was added, and the incubation was continued for an additional 10 minutes. For supershift analysis, 1 μL USF-1 (C20), USF-2 (N18), Myc (C33), Max (C17), or SP-1 (PEP2-G) antibodies (Santa Cruz Biotechnology) as well as a rabbit preimmune serum14were added to the EMSA reaction and then incubated at 4°C for 2 hours. The electrophoresis was then performed with a 5% nondenaturing polyacrylamide gel in TBE buffer (89 mM Tris, 89 mM boric acid, 5 mM ethylenediaminetetraacetic acid) at 200 V. After electrophoresis the gels were dried and exposed to a phosphorimager screen.
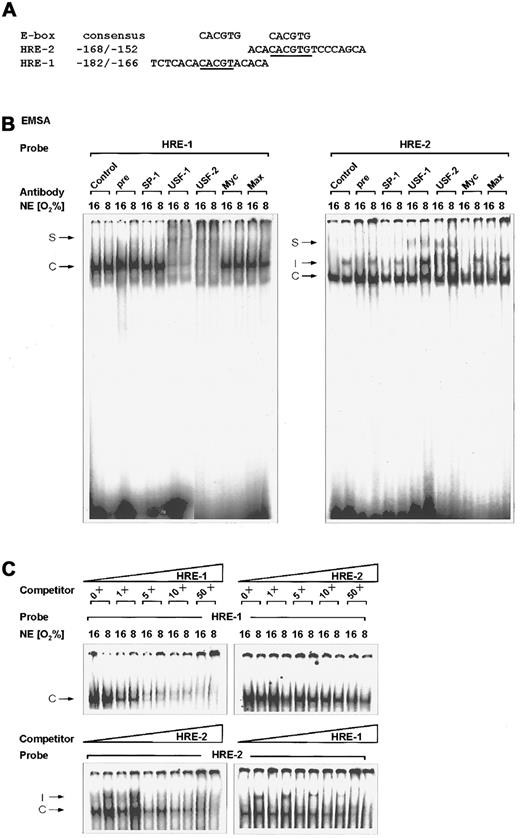
Binding of USF-2a to the rat PAI-1 HREs.
(A) Oligonucleotides. The E-box consensus sequence CACGTG and the sense strands of the rat PAI-1 promoter sequence oligonucleotides containing HRE-2 and HRE-1 are shown. Bases matching the consensus sequences are underlined. (B) EMSA. The 32P-labeled PAI-1 HRE-1 (−182/−166) (left panel) and HRE-2 (−168/−152) (right panel) oligonucleotides were incubated with either 10 μg protein of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells as indicated (see “Materials and methods”). In EMSA with antibodies the nuclear extracts were preincubated with 1 μL preimmune serum, the USF-1, USF-2, Myc, Max, or SP-1 antibodies for 2 hours at 4°C before adding the labeled probe. The DNA protein binding was analyzed by electrophoresis on 5% native polyacrylamide gels. (C) EMSA competition assays. The 32P-labeled HRE-1 or HRE-2 oligonucleotide was incubated with nuclear extracts as in panel B. For competition, increasing concentrations, as indicated, of nonlabeled HRE-1 or HRE-2 oligonucleotides were added. C indicates constitutive complex; I, induced HIF-1 complex; S, supershifted USF complex.
Binding of USF-2a to the rat PAI-1 HREs.
(A) Oligonucleotides. The E-box consensus sequence CACGTG and the sense strands of the rat PAI-1 promoter sequence oligonucleotides containing HRE-2 and HRE-1 are shown. Bases matching the consensus sequences are underlined. (B) EMSA. The 32P-labeled PAI-1 HRE-1 (−182/−166) (left panel) and HRE-2 (−168/−152) (right panel) oligonucleotides were incubated with either 10 μg protein of nuclear extracts from normoxic (16% O2) or hypoxic (8% O2) cells as indicated (see “Materials and methods”). In EMSA with antibodies the nuclear extracts were preincubated with 1 μL preimmune serum, the USF-1, USF-2, Myc, Max, or SP-1 antibodies for 2 hours at 4°C before adding the labeled probe. The DNA protein binding was analyzed by electrophoresis on 5% native polyacrylamide gels. (C) EMSA competition assays. The 32P-labeled HRE-1 or HRE-2 oligonucleotide was incubated with nuclear extracts as in panel B. For competition, increasing concentrations, as indicated, of nonlabeled HRE-1 or HRE-2 oligonucleotides were added. C indicates constitutive complex; I, induced HIF-1 complex; S, supershifted USF complex.
Results
Binding of USF-2a to HRE sequences of the rat PAI-1 promoter
There are 2 E-box–like sequences, E1 (−175/−170) and E2 (−165/−160), inside the “C” site of the rat PAI-1 promoter, which was identified by footprint analysis with liver nuclear extracts.30 Both E-boxes acted as HREs and were thus designated HRE-1 and HRE-2. Besides HRE-2 (−165/−160), which was found to bind the HIF-1, HRE-1 (−175/−170) bound a so far unknown factor.14 Because the CACGTG motif, which includes the canonical E-box sequence CANNTG, appears to be the common recognition site for bHLH transcription factors such as USF,16 it was supposed that the HRE-1 or even both HREs can be bound by USF. The first potential USF binding element, HRE-1 5′-CACGTA-3′, matches the consensus sequence in 5 of 6 bp. The second potential USF binding element, HRE-2 5′-CACGTG-3′, contains the canonical USF binding sequence CACGTG (Figure 1).
The binding of nuclear proteins to oligonucleotide probes spanning the HRE-1 and HRE-2 sites of the rat PAI-1 promoter was examined by EMSA. The oligonucleotide −182/−166 containing HRE-1 was able to bind a very strong complex, which was independent from the pO2 (Figure 1B, left panel). In contrast, with the PAI-1 promoter oligonucleotide −168/−152 containing HRE-2 beside a constitutive protein complex, a hypoxia-inducible nuclear protein complex was detectable (Figure 1B, right panel). This hypoxia-induced DNA protein complex was shown to contain HIF-1.14 The formation of specific complexes was no longer detectable when mutations were introduced in the HRE-2 and HRE-1 sites.14
To investigate the presence of USF in these complexes, antibodies against USF-1 and USF-2 were included in the binding reaction. The USF-1 antibody was raised against a C-terminal 18–amino acid polypeptide of human origin, whereas the USF-2 antibody was raised against an N-terminal 18–amino acid peptide of mouse origin. Addition of the USF-1 antibody to the EMSA reaction strongly reduced, whereas the USF-2 antibody inhibited, the formation of the DNA complex with the HRE-1 oligonucleotide, and both led to a supershifted complex. With the HRE-2 oligonucleotide, the USF antibodies slightly produced a supershifted complex, which appeared stronger with the USF-2 antibody. Furthermore, it appeared that the addition of the USF antibodies favored binding of the HIF-1 complex to the HRE-2. To ensure specificity of the supershifts mediated by the USF antibody, the EMSA was also performed in the presence of a rabbit preimmune serum and an antibody against the GC-box binding factor SP-1. Neither the preimmune serum nor the SP-1 antibody influenced the complex formation with the HRE-1 and the HRE-2 oligonucleotides. To test whether other members of the bHLH family such as Myc or Max may participate in binding to the HRE-1 and HRE-2, supershifts with Myc and Max antibodies were performed. Addition of antibodies against Myc or Max did not result in a supershift or inhibition of complex formation (Figure 1B). Thus, it appears that USF-2 could play the more important role in PAI-1 gene expression.
To investigate whether HRE-1 could bind the USF complex with higher affinity, competition experiments were performed. The formation of the USF complex with the HRE-1 oligonucleotide could be clearly reduced by unlabeled HRE-1 starting with a 5-fold molar excess of cold HRE-1 oligonucleotides (Figure 1C, upper part, left). At this point it appeared that only 2 faint complexes were visible, consistent with the finding that both USF-1 and USF-2 can bind to the HRE-1. In contrast, the competition of HRE-1 binding with the cold HRE-2 oligonucleotide was not as prominent as with the unlabeled HRE-1 oligonucleotide; clear reduction of binding activity was visible at a 10- to 50-fold molar excess of HRE-2 oligonucleotide (Figure 1C, upper part, right). Vice versa it could be shown by competition of the HRE-2 binding complexes that, again, unlabeled HRE-2 competed with higher affinity for the hypoxia-inducible as well as the constitutive complex than the HRE-1 (Figure 1C, lower part). These findings together indicated that the PAI-1 HRE sites could be bound by USF proteins and that the HRE-1 site is the major USF binding site.
Inhibition of PAI-1 expression and induction of PKLexpression by overexpression of USF-2a under both normoxia and mild hypoxia
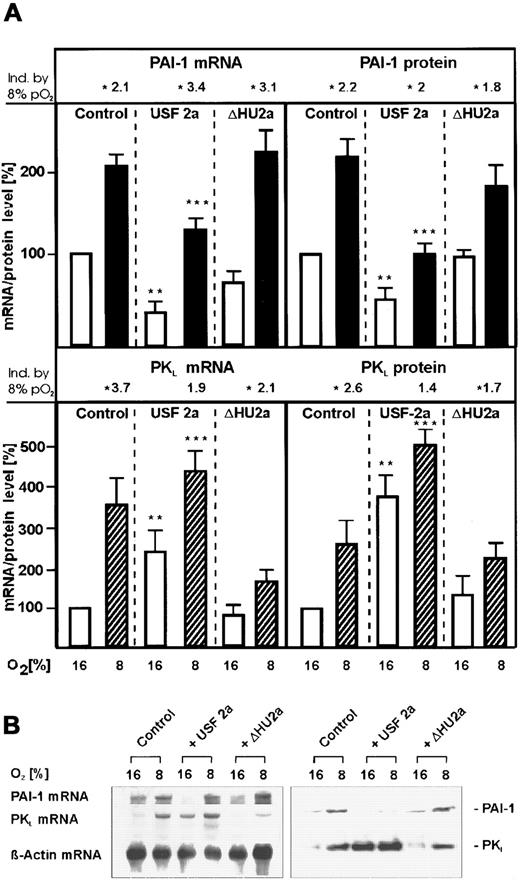
To investigate the role of USF-2a in the regulation of PAI-1 gene expression, primary cultured rat hepatocytes were transfected with expression vectors encoding wild-type human USF-2a or the mutant USF-2a protein ΔHU2a lacking the second helix of the HLH domain and thus unable to bind DNA and to dimerize.23 Hepatocytes cultured under mild hypoxia and transfected with the empty control vector displayed an about 2-fold–enhanced PAI-1 messenger RNA (mRNA) level, in line with a previous study.14 In the cells transfected with the vector encoding wild-type USF-2a and cultured under normoxia, PAI-1 mRNA expression was inhibited by about 70% (Figure2A). Transfection of the USF-2a expression vector reduced the hypoxia-dependent PAI-1 mRNA induction by about 60%. However, the mRNA levels were still about 2-fold higher than in the USF-2a–transfected cells cultured under normoxia. This inhibition of PAI-1 mRNA expression was not observed in cells transfected with the ΔHU2a vector.
Inhibition of PAI-1 and induction of PKLmRNA and protein expression by overexpression of USF-2a under both normoxia and mild hypoxia.
Hepatocytes transfected either with 8 μg USF-2a or ΔHU2a expression vectors or the control vector pCMV were cultured for 24 hours under arterial pO2. At 24 hours the medium was changed and cells were further cultured for the next 24 hours under normoxic (16% O2) or hypoxic (8% O2) conditions. (A, left panels) The PAI-1 and PKL mRNA levels were measured by Northern blotting (see panel B). The mRNA level under normoxia (16% O2) was set equal to 100%. (A, right panels) The PAI-1 and PKL protein levels were measured by Western blotting (see panel B). The protein level under normoxia (16% O2) was set equal to 100%. The fold induction by 8% PO2 was calculated in each series to the corresponding 16% values of PAI-1 and PKL mRNA and protein and is indicated above. (B) Representative Northern and Western blot. For Northern analysis, 15 μg total RNA prepared from the cultured hepatocytes was hybridized to DIG-labeled PAI-1, PKL, and β-actin antisense RNA probes (see “Materials and methods”). A total of 50 μg of protein from the medium or of the cultured hepatocytes was subjected to Western analysis with an antibody against rat PAI-1 or rat PKL (see “Materials and methods”). Autoradiographic signals were obtained by chemiluminescence and scanned by videodensitometry. Values are means ± SEM of 3 independent culture experiments. Statistics, Student t test for paired values: *, significant difference 16% O2 versus 8% O2; **, significant difference 16% O2 + USF-2a versus 16% O2 + pCMV (control); ***, significant difference 8% O2 + USF-2a versus 8% O2 + pCMV (control); P < .05. ΔHU2a, USF-2 mutant lacking the second helix of the HLH domain.23
Inhibition of PAI-1 and induction of PKLmRNA and protein expression by overexpression of USF-2a under both normoxia and mild hypoxia.
Hepatocytes transfected either with 8 μg USF-2a or ΔHU2a expression vectors or the control vector pCMV were cultured for 24 hours under arterial pO2. At 24 hours the medium was changed and cells were further cultured for the next 24 hours under normoxic (16% O2) or hypoxic (8% O2) conditions. (A, left panels) The PAI-1 and PKL mRNA levels were measured by Northern blotting (see panel B). The mRNA level under normoxia (16% O2) was set equal to 100%. (A, right panels) The PAI-1 and PKL protein levels were measured by Western blotting (see panel B). The protein level under normoxia (16% O2) was set equal to 100%. The fold induction by 8% PO2 was calculated in each series to the corresponding 16% values of PAI-1 and PKL mRNA and protein and is indicated above. (B) Representative Northern and Western blot. For Northern analysis, 15 μg total RNA prepared from the cultured hepatocytes was hybridized to DIG-labeled PAI-1, PKL, and β-actin antisense RNA probes (see “Materials and methods”). A total of 50 μg of protein from the medium or of the cultured hepatocytes was subjected to Western analysis with an antibody against rat PAI-1 or rat PKL (see “Materials and methods”). Autoradiographic signals were obtained by chemiluminescence and scanned by videodensitometry. Values are means ± SEM of 3 independent culture experiments. Statistics, Student t test for paired values: *, significant difference 16% O2 versus 8% O2; **, significant difference 16% O2 + USF-2a versus 16% O2 + pCMV (control); ***, significant difference 8% O2 + USF-2a versus 8% O2 + pCMV (control); P < .05. ΔHU2a, USF-2 mutant lacking the second helix of the HLH domain.23
The USF-mediated decrease of PAI-1 mRNA was followed by a decrease of PAI-1 protein. It was found that USF-2a–transfected hepatocytes cultured under normoxia or hypoxia secreted about 50% less PAI-1 protein into the medium than the control cells (Figure 2A). However, as observed with the PAI-1 mRNA, the amount of PAI-1 protein in the USF-transfected cells measured under hypoxia was still enhanced by about 2-fold compared with the PAI-1 levels under normoxia. Again, the amount of PAI-1 protein in the medium of the cells transfected with the ΔHU2a vector remained almost the same as in the controls.
To investigate that the inhibitory action of USF-2a is not due to the effect of the protein to form inactive homodimers, the PKLexpression, which should be activated by USF,23 was examined. In the cells cultured under mild hypoxia and transfected with the empty control vector, PKL mRNA levels were about 3.5 times higher than under normoxia. The same was found for the PKL protein (Figure 2A, lower panel). In the cells transfected with the USF-2a expression vector, the PKL mRNA levels were increased by USF-2a by about 2.5 fold under normoxia and by about 4.5 fold under hypoxia, and the PKL protein level was enhanced by about 3.5 fold under normoxia and by about 5-fold under hypoxia (Figure 2).
When the ΔHU2a vector was transfected into hepatocytes, the USF-dependent induction of PKL expression was no longer present but the hypoxia-induced PKL expression was still observed (Figure 2).
Inhibition of PAI-1 promoter–controlled Luc expression and induction of PKL promoter–controlled Luc expression by overexpression of USF-2a under both normoxia and mild hypoxia
To further substantiate that the binding of USF-2a to the HREs, as revealed by EMSA (Figure 1), may be involved in the regulation of PAI-1 expression not only under normoxic but also under hypoxic conditions, primary hepatocytes were cotransfected with the hypoxia responsive Luc gene construct pGl3PAI-76614 and either control pCMV plasmid, USF-2a, or ΔHU2a vectors. In hepatocytes transfected with pGl3PAI-766, Luc activity was about 2.5-fold higher at 8% O2 than at 16% O2, in line with a previous study.14 In hepatocytes cotransfected with pGI3PAI-766 and USF-2a, the Luc activity was decreased by about 8-fold under both normoxia and hypoxia (Figure 3). Thus, USF-2a cotransfections did not abolish induction of Luc activity under hypoxia. The Luc activity in pGl3PAI-766– and ΔHU2a-cotransfected hepatocytes did not differ significantly from the control values.
Inhibition of PAI-1 and induction of PKLpromoter–controlled Luc expression by overexpression of USF-2a under both normoxia and mild hypoxia.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with the pGl3PAI-766 Luc or pGl3PKL-183 Luc constructs and the USF-2a, ΔHU2a, or ΔTDU2 expression vectors or the control pCMV vector. After 24 hours the transfected cells were cultured for the next 24 hours under hypoxic (8% O2) or normoxic (16% O2) conditions, as indicated. In each experiment the percentage of Luc activity was determined relative to the normoxic pGl3PAI-766 Luc or pGl3PKL-183 Luc controls, which were set equal to 100%. The fold induction by 8% O2was calculated in each set of transfections relative to the corresponding Luc activity obtained under 16% O2. Values are means ± SEM of 3 independent culture experiments. Statistics, Student t test for paired values: *, significant difference 16% O2 versus 8%O2; **, significant difference 16% O2 + USF-2a or ΔTDU2 versus 16% O2 + pCMV (control); ***, significant difference 8% O2 + USF-2a or ΔTDU2 versus 8% O2 + pCMV (control); P < .05. ΔHU2a, USF-2 mutant lacking the second helix of the HLH domain; ΔTDU2, USF-2 mutant lacking the first 198 amino acids of the transactivation domain.23 L1-L4: L indicates liver-specific; L1, binding site for hepatocyte nuclear factor 1, L2, binding site for nuclear factor 1; L3, binding site for hepatocyte nuclear factor 4; L4, binding site for USF.31 32
Inhibition of PAI-1 and induction of PKLpromoter–controlled Luc expression by overexpression of USF-2a under both normoxia and mild hypoxia.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with the pGl3PAI-766 Luc or pGl3PKL-183 Luc constructs and the USF-2a, ΔHU2a, or ΔTDU2 expression vectors or the control pCMV vector. After 24 hours the transfected cells were cultured for the next 24 hours under hypoxic (8% O2) or normoxic (16% O2) conditions, as indicated. In each experiment the percentage of Luc activity was determined relative to the normoxic pGl3PAI-766 Luc or pGl3PKL-183 Luc controls, which were set equal to 100%. The fold induction by 8% O2was calculated in each set of transfections relative to the corresponding Luc activity obtained under 16% O2. Values are means ± SEM of 3 independent culture experiments. Statistics, Student t test for paired values: *, significant difference 16% O2 versus 8%O2; **, significant difference 16% O2 + USF-2a or ΔTDU2 versus 16% O2 + pCMV (control); ***, significant difference 8% O2 + USF-2a or ΔTDU2 versus 8% O2 + pCMV (control); P < .05. ΔHU2a, USF-2 mutant lacking the second helix of the HLH domain; ΔTDU2, USF-2 mutant lacking the first 198 amino acids of the transactivation domain.23 L1-L4: L indicates liver-specific; L1, binding site for hepatocyte nuclear factor 1, L2, binding site for nuclear factor 1; L3, binding site for hepatocyte nuclear factor 4; L4, binding site for USF.31 32
Again, to further corroborate the positive effect of USF-2a on PKL expression, hepatocytes were cotransfected with the USF expression vectors and a PKL promoter Luc construct (pGl3PKL-183) that was shown to be activated by USF.23 The cells were then cultured under normoxia or mild hypoxia. In hepatocytes transfected with pGl3PKL-183 and the pCMV control vector, hypoxia elicited only a marginal induction of Luc activity. In the USF-2a– and pGl3PKL-183–cotransfected cells, Luc activity was increased by about 3-fold under both normoxia and hypoxia. By contrast, ΔHU2a cotransfection did not result in activation of Luc activity compared with the controls (Figure 3).
Thus, PAI-1 mRNA protein as well as PAI-1–promoter controlled Luc expression in primary rat hepatocytes cultured under normoxia and hypoxia were inhibited by overexpression of USF-2a.
Inhibition of transfected rat PAI-1 promoter Luc gene constructs by wild-type and mutant USF-2a
To determine what domains of USF-2a were involved in the regulation of PAI-1 gene expression, the wild-type rat PAI-1 promoter Luc construct pGl3PAI-766 and plasmids expressing wild-type USF-2a, the mutant protein ΔHU2a, and the mutant ΔTDU2, which lacks the first 198 amino acids of the transactivation domain, were cotransfected. In hepatocytes cotransfected with pGI3PAI-766 and USF-2a, the Luc activity was decreased (Figure 3). The Luc activity in pGl3PAI-766– and ΔHU2a-cotransfected hepatocytes was about the same as in the control. In hepatocytes cotransfected with pGI3PAI-766 and the ΔTDU2 vector, which contains the intact DNA-binding domain,23 the Luc activity was decreased by about 5-fold (Figure 3). In contrast, after cotransfection of the ΔTDU2 vector and the pGl3-183PKLconstruct, the Luc activity was in the range of the controls (Figure3). These data demonstrated that only the bHLH-zip domain but not the N-terminal transactivation domain was necessary for the USF-2a–dependent inhibition of PAI-1 expression in primary hepatocytes, whereas PKL promoter activation by USF-2 required the transactivation domain.
Cotransfection of the plasmid pGl3PAI-276 containing a 276-bp fragment of the rat PAI-1 promoter in front of the Luc gene with the USF-2a plasmid resulted in the same inhibition of Luc activity as with the pGl3PAI-766 Luc construct. (Figure 4). The inhibition was not observed when the pGl3PAI-276 construct was cotransfected with the ΔHU2a vector. These results also indicated that the deletion of about 490 bp of the PAI-1 promoter did not abolish the inhibitory effect of USF-2a on the PAI-1 expression probably exerted via HRE-1 and HRE-2.
Inhibition of Luc activity by USF-2a is conferred by HRE-1 and HRE-2 in the rat PAI-1 promoter.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with either USF-2a or ΔHU2a expression plasmids and Luc gene constructs driven by a wild-type 276-bp rat PAI-1 promoter (pGl3PAI-276) or the 276 bp promoter mutated at either the HRE-1 (pGl3PAI-276M1) or HRE-2 (pGl3PAI-276M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-276 + pCMV control, which was set equal to 100%. In pGl3PAI-276M1 and pGl3PAI-276M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-276 + USF-2a versus pGl3PAI-276 + pCMV control; pGl3PAI-276M1 + USF-2a versus pGl3PAI-276M1 + pCMV control; pGl3PAI-276M2 + USF-2a versus pGl3PAI-276M2 + pCMV control; P < .05.
Inhibition of Luc activity by USF-2a is conferred by HRE-1 and HRE-2 in the rat PAI-1 promoter.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with either USF-2a or ΔHU2a expression plasmids and Luc gene constructs driven by a wild-type 276-bp rat PAI-1 promoter (pGl3PAI-276) or the 276 bp promoter mutated at either the HRE-1 (pGl3PAI-276M1) or HRE-2 (pGl3PAI-276M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-276 + pCMV control, which was set equal to 100%. In pGl3PAI-276M1 and pGl3PAI-276M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-276 + USF-2a versus pGl3PAI-276 + pCMV control; pGl3PAI-276M1 + USF-2a versus pGl3PAI-276M1 + pCMV control; pGl3PAI-276M2 + USF-2a versus pGl3PAI-276M2 + pCMV control; P < .05.
Mutations of the HRE sequences in PAI-1 Luc gene constructs abolished the inhibition of Luc activity
To investigate the role of the HRE-1 and HRE-2 sequences in the USF-2a–dependent inhibition of PAI-1 expression, the wild-type PAI-276 promoter Luc construct and PAI-276 promoter Luc constructs with either a mutated HRE-1 or a HRE-2 were cotransfected with the expression vectors for USF-2a, ΔHU2a, or the control pCMV into primary hepatocytes (Figure 4). Mutation of the nucleotides −175/−170 encompassing the HRE-1 site in the construct pGl3PAI-276M1 resulted in a strong (about 12-fold) induction of Luc activity compared with pGl3PAI-276–transfected hepatocytes. In cotransfection experiments, USF-2a acted as inhibitor and reduced the Luc activity by about 7-fold, but the overall Luc activity was about 5-fold higher than the wild-type pGl3PAI-276 control. Cotransfection of pGl3PAI-276M1 and ΔHU2a led to the maximal observed (about 15-fold) induction of Luc activity (Figure 4).
When the nucleotides −165/−160 inside HRE-2 were mutated in the construct pGl3PAI-276M2, the Luc activity was also enhanced, but this increase of Luc activity was much less then the induction observed with pGl3PAI-276M1 (Figure 4). In hepatocytes transfected with pGl3PAI-276M2, Luc activity was about 4.5-fold higher than with pGl3PAI-276. Cotransfection of the USF-2a vector resulted in an about 2.5-fold inhibition of Luc activity, which was then still about 1.8-fold higher than the Luc activity of wild-type pGl3PAI-267. Cotransfection of pGl3PAI-276M2 with ΔHU2a resulted in an about 6-fold induction of Luc activity. These data supported the idea that both HREs in the PAI-1 promoter contributed to the USF-2a–dependent inhibition of PAI-1 gene transcription and that the HRE-1 is the main USF-2a binding site.
Competition by HIF-1α via HRE-2 of the USF-2a–dependent inhibition of pGl3PAI-766 Luc expression
The results from the supershift assays and the transfection experiments suggested that HRE-2 might be involved in the competition for the binding of 2 different transcription factors, namely USF and HIF-1. To test whether HIF-1 could counteract the USF-dependent inhibition of PAI-1 expression, cotransfections with the USF-2a and HIF-1α expression vectors together with the PAI-1 promoter Luc constructs pGl3PAI-766, pGl3PAI-766M1, and pGl3PAI-766M2 were performed.
Cotransfection of pGl3PAI-766 with the USF-2a vector again reduced Luc activity by about 8-fold, whereas cotransfection of the HIF-1α vector enhanced Luc activity by about 4-fold. In the cotransfection experiments of both the USF-2a and HIF-1α expression vectors with pGl3PAI-766, the HIF-1α vector counteracted the USF-2a–dependent inhibition of Luc activity and restored the control values (Figure5). When the cells were cotransfected with the construct pGl3PAI-766M1 in which the major USF binding site HRE-1 was mutated and the expression vector for USF-2a Luc activity was still inhibited by about 4-fold compared with the pGl3PAI-766M1 control. When the HIF-1α vector was cotransfected together with the HRE-1–mutated plasmid pGl3PAI-766M1, Luc activity was induced by about 2-fold, in line with a previous study.14 After cotransfection of both the USF-2a and the HIF-1α vector, the negative effect of USF-2a again could be attenuated by HIF-1α; Luc activities were now about 2-fold higher than in the presence of USF-2a alone (Figure 5).
Competition by HIF-1α via HRE-2 of the USF-2a–dependent inhibition of pGl3PAI-766 Luc expression in hepatocytes.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the USF and HIF-1 binding element14 matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with expression plasmids for either USF-2a, HIF-1α, or both and Luc gene constructs driven by a wild-type 766-bp rat PAI-1 promoter (pGl3PAI-766) or the 766-bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-766 + pCMV control, which was set equal to 100%. In pGl3PAI-766M1 and pGl3PAI-766M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-766 + USF-2a versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + USF-2a versus pGl3PAI-766M1 + pCMV control; pGl3PAI-766M2 + USF-2a versus pGl3PAI-766M2 + pCMV control; **, significant difference pGl3PAI-766 + HIF-1α versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + HIF-1α versus pGl3PAI-766M1 + pCMV control; ***, significant difference pGl3PAI-766 + USF-2a + HIF-1α versus pGl3PAI-766 + USF-2a; pGl3PAI-766M1 + USF-2a + HIF-1α versus pGl3PAI-766M1 + USF2a; P < .05.
Competition by HIF-1α via HRE-2 of the USF-2a–dependent inhibition of pGl3PAI-766 Luc expression in hepatocytes.
(A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the USF and HIF-1 binding element14 matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with expression plasmids for either USF-2a, HIF-1α, or both and Luc gene constructs driven by a wild-type 766-bp rat PAI-1 promoter (pGl3PAI-766) or the 766-bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-766 + pCMV control, which was set equal to 100%. In pGl3PAI-766M1 and pGl3PAI-766M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-766 + USF-2a versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + USF-2a versus pGl3PAI-766M1 + pCMV control; pGl3PAI-766M2 + USF-2a versus pGl3PAI-766M2 + pCMV control; **, significant difference pGl3PAI-766 + HIF-1α versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + HIF-1α versus pGl3PAI-766M1 + pCMV control; ***, significant difference pGl3PAI-766 + USF-2a + HIF-1α versus pGl3PAI-766 + USF-2a; pGl3PAI-766M1 + USF-2a + HIF-1α versus pGl3PAI-766M1 + USF2a; P < .05.
In hepatocytes transfected with the HRE-2–mutated construct pGl3PAI-766M2 and the USF-2a vector, Luc activity was diminished by about 4-fold. As shown,14 cotransfection of pGl3PAI-766M2 and the HIF-1α vector had no effect on Luc activity and also did not compete for the USF-2a–dependent inhibition of Luc activity (Figure5). These results strengthen the proposal that the HRE-2 site can bind both USF and HIF-1 and might therefore be involved in the fine-tuning of PAI-1 gene expression.
Discussion
In this study it was demonstrated that the transcription factor USF-2a acted as inhibitor of the rat PAI-1 gene expression via binding to the noncanonical E-box sequence HRE-1. The inhibitory effect of USF-2a on the PAI-1 gene expression was due to the DNA-binding domain of USF-2a and not exerted by the transactivation domain. The HRE-1 is separated by only 4 bp from the other E-box–like sequence in the rat PAI-1 promoter, HRE-2, which could bind HIF-1 acting as the activator of the rat PAI-1 expression under mild hypoxia. Overexpression of USF-2a in general inhibited the expression of PAI-1 but did not interfere with the induction of PAI-1 expression by mild hypoxia per se.
The upstream stimulatory factors as inhibitors and activators
The finding that USF inhibited PAI-1 production in hepatocytes was at a first glance unexpected because USF was originally identified from HeLa cell nuclei as an activator of the adenovirus major late promoter.16 However, as in this study, USF was able to act as an inhibitor of gene expression. Using chromatin cross-linking and immunoprecipitation protocols, it was demonstrated that both c-Myc and USF bound to exactly the same E-box site on the cad (carbamoyl-phosphate synthase/aspartate carbamoyltransferase/dihydroorotase) promoter in cultured B5-4 cells.33,34 Binding of USF and competition with TFE3 at the microE3 box within the immunoglobulin heavy chain enhancer lead to inhibition of enhancer activity in NIH3T3 cells.35Furthermore, USF was also shown to compete with the arylhdrocarbon receptor/arylhdrocarbon receptor nuclear translocator (AHR-ARNT) heterodimer for binding to the xenobiotic-responsive element (XRE) in the rabbit CYP1A1 gene,36 thereby attenuating the AHR-ARNT mediated activation of XRE-TK/Luc gene constructs in RK13 cells. Moreover, USF repressed XMyoD binding to the XMyoDa promoter, thereby preventing XMyoDa autoactivation in 10T[½] cells.37 Thus, the findings of this study with the PAI-1 gene are another example for the role of USF as an inhibitor of gene expression.
Despite the fact that in vivo most of the USF proteins exist as heterodimers,19 one might expect that the USF-2 homodimers formed after overexpression of USF-2 alone could result in a squelching mechanism, thereby mediating the suppression via an indirect effect. However, homodimers formed after overexpression of USF-1 or USF-2 in HeLa or hepatoma cells were shown to activate a promoter containing 4 copies of an E-box or the −183-bp PKL gene promoter.20 Meanwhile, USF consisting of either homodimers and heterodimers of USF-1 and USF-2a are known to activate the expression of genes, encoding regulators involved in cellular proliferation such as p53,38 cyclin B1,39 and transforming growth factor β240 as well as glucose-controlled genes such as fatty acid synthetase41and PKL.23 In line with this, in the present study USF-2a overexpression enhanced PKL mRNA and protein levels as well as Luc activity driven by the −183 PKL gene promoter (Figures 2, 3). This PKL promoter contains the L4 binding site (ie, the glucose response element), which was shown to be able to bind USF-2 dimers, which appear to be the main functional activators of the glucose responsive complex of the PKLgene.42 Thus, the down-regulation of PAI-1 gene expression by overexpression of USF-2a in this study appears not to be the result of a dominant negative effect of USF-2 homodimers.
The PAI-1 HREs as USF binding sites: competition between USF and HIF
In a previous study it was shown that the PAI-1 promoter region −177/−152 contains 2 E-box sequences 5′-CACGTA-3′ (−175/−170) and 5′-CACGTG-3′ (−165/−160), which were named HRE-1 and HRE-2, respectively. The induction of PAI-1 gene expression by hypoxia was shown to depend on both the HRE-1 and HRE-2. Gel shift analysis indicated that HRE-2 bound the HIF-1. In contrast, the oligonucleotide spanning the HRE-1 was able to bind a complex, formation of which was independent from the pO2.14 Thus, it was proposed that other partners of the bHLH-protein family such as USF or Myc may interact at the HRE-1 of the PAI-1 promoter. Indeed, this study showed binding of USF-1 and USF-2 to both HREs of the PAI-1 promoter. In line with the present findings, it was shown during this study that with nuclear extracts obtained from serum-stimulated renal epithelial cells (NRK-52E, clone EC-1) the HRE-2 can be bound by USF-1, whereas the HRE-1 sequence was not investigated; but, again, incubation of NRK-52E nuclear extracts with antibodies to Myc or Max failed to produce a supershift with the HRE-2 oligonucleotide.43
The gel shift and supershift assays demonstrated that USF-2 bound with high affinity to the HRE-1 and to a lesser extent to the HRE-2. Because the preferred E-box core sequence for USF is 5′-CACGTG-3′,16,44 one would have expected that the HRE-2 functions as the predominant USF binding site within the PAI-1 promoter. Indeed, supershift assays and the cotransfection experiments of pGl3PAI-276M1 in which the HRE-2, but not the HRE-1, was intact with an expression vector for USF-2a caused inhibition of Luc activity (Figures 1, 4). However, this inhibition was not as strong as with the wild-type pGl3PAI-276 construct or with the HRE-2–defective plasmid pGl3PAIM2, indicating that HRE-1 appeared to be the primary USF binding site. Thus, the finding that USF bound the imperfect E-box HRE-1 5′-CACGTA-3′ in the PAI-1 promoter is also in good agreement with previous reports in which USF has been shown to bind the imperfect E-boxes 5′-CCCGTG-3′ as in the rat γ-fibrinogen promoter,45 the 5′-CGCGTG-3′ box as in the mouse metallothionine I promoter,46 and the 5′-CACCTG-3′ as in the human apolipoprotein A-II gene promoter.47
In the cotransfection experiments with the vectors encoding a USF-2a protein lacking DNA-binding activity (ΔHU2a) or lacking the transactivation domain (TDU2) and pGl3PAI-276, it was shown that the inhibitory action of USF-2 on the PAI-1 expression was mediated by the DNA-binding domain (Figure 3). This finding is in line with the observation that the inhibition of the rat ribosomal RNA gene transcription by USF-2 in Chinese hamster ovary cells48 and the inhibitory action of USF-2 on ras-mediated transformation in FR3T3 and 293 cells was also dependent on the DNA-binding domain of USF-2.49
The DNA-binding domain–dependent inhibitory action of USF-2 on the PAI-1 gene expression might play a role in the regulation of PAI-1 expression in response to diverse stimuli. Because the HRE-2 bound USF as well as HIF-1, it makes this site attractive to a competitive regulation by the DNA binding of both transcription factors. This was indeed shown with the cotransfection experiments with the USF-2a and HIF-1α expression vectors together with the PAI-1 promoter Luc gene constructs pGl3PAI-766, pGl3PAI-766M1, and pGl3PAI-766M2 (Figure 5). Cotransfection of the wild-type PAI-1 promoter Luc construct pGl3PAI-766 or the construct pGl3PAI-766M1, in which the major USF binding site HRE-1 was mutated with the USF-2a vector, reduced the Luc activity; the HIF-1α vector alone induced Luc activity and, after cotransfection of both, the HIF-1α vector attenuated the negative effect of USF-2a (Figure 5). In hepatocytes transfected with the HIF-1 binding site (HRE-2) mutated construct pGl3PAI-766M2, the HIF-1α vector had no effect on Luc activity and thus could not compete for the USF-2a–dependent inhibition of Luc activity (Figure 5). These results strengthen the proposal that the HRE-2 site can bind both USF and HIF-1 and might therefore be involved in the fine-tuning of PAI-1 gene expression. Thus, under normoxia the PAI-1 HREs might be occupied to some extent by the transcription factor USF, leading to a low level of PAI-1 expression. Under hypoxic conditions the HIF-1α protein50 is stabilized and transported into the nucleus,51 leading to accumulation of the active HIF-1α/HIF-1β heterodimer. The active HIF-1 can then outcompete USF from binding to the HRE sites, leading to the induction by hypoxia of PAI-1 gene expression.
Role of the PAI-1 inhibition by USF in growth progression and carcinogenesis
The finding of this study that USF inhibited PAI-1 gene expression may have an important role during growth and regeneration processes. It was suggested that when cells progress from quiescence into the S phase, the transcription factor Myc/Max heterodimer competed with USF proteins for binding to the E-box.33 Accordingly, USF-1 and USF-2 mediated antiproliferative properties; when overexpressed in REF cells they inhibited c-Myc–induced cellular transformation.52 The Myc-dependent cellular transformation was inhibited only when wild-type USF-1 and USF-2 or vectors for USF mutants lacking the N-terminal transactivation domain were transfected. The DNA-binding–deficient forms of USF-1 and USF-2 did not affect the Myc-dependent foci formation in REF cells.52 This is in line with the results of the present study in which the cotransfection experiments with the USF-2a mutants lacking the DNA-binding domain (ΔHU2a) and the PAI-1 Luc constructs demonstrated the necessity of the DNA-binding domain for the inhibitory action of USF (Figure 3). On one hand, the results support that the role of USF-2 does not appear to be cell-specific because USF-2 also exerted a strong inhibition of E1A-mediated transformation of REF cells and a strong suppression of HeLa cell colony formation52; however, on the other hand, in the Saos-2 osteosarcoma cell line, USF proteins had no effect on cell proliferation, suggesting a model involving a cell type–specialized coactivator.53
The antiproliferative activities of USF suggested that inactive USF could promote carcinogenesis. Indeed, in several cell lines from breast tumors USF was completely inactive.54 Thus, the loss of the repression by USF with the PAI-1 promoter gene constructs with the mutated USF site and the subsequent higher PAI-1 Luc expression as shown in Figure 4 would also coincide with the observation that PAI-1 was found to be expressed at higher levels in breast and other cancer cells55; it may be necessary for optimal metastasis formation as shown for cultured lung cancer cells.56Moreover, in a number of clinical studies it was demonstrated that high PAI-1 levels in patients suffering from various types of cancer indicated a poor prognosis.57-59 Thus, it appears that USF controls the production of metabolic enzymes such as fatty acid synthetase,41 PKL60 and, as shown in this study, inhibitors of proteolysis such as PAI-1, which contributes to the control of cell proliferation and tumor growth.
We thank Dr A. Kahn and Dr M. Raymondjean (Institut Cochin de Genetique Moleculaire, Universite Rene Descartes, Paris) for the kind gift of the −183PKLCAT construct and the human USF-2a, ΔHU2a, and ΔTDU2 plasmids and Dr T. D. Gelehrter (Department of Human Genetics, University Michigan Medical School, Ann Arbor, MI) for the kind gift of PAI-1 complementary DNA.
Supported by the Deutsche Forschungsgemeinschaft SFB 402 Teilprojekt A1 and GRU 335.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
T. Kietzmann, Institut für Biochemie und Molekulare Zellbiologie, Humboldtallee 23, D-37073 Göttingen, Germany; e-mail: tkietzm@gwdg.de.

![Fig. 3. Inhibition of PAI-1 and induction of PKLpromoter–controlled Luc expression by overexpression of USF-2a under both normoxia and mild hypoxia. / (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with the pGl3PAI-766 Luc or pGl3PKL-183 Luc constructs and the USF-2a, ΔHU2a, or ΔTDU2 expression vectors or the control pCMV vector. After 24 hours the transfected cells were cultured for the next 24 hours under hypoxic (8% O2) or normoxic (16% O2) conditions, as indicated. In each experiment the percentage of Luc activity was determined relative to the normoxic pGl3PAI-766 Luc or pGl3PKL-183 Luc controls, which were set equal to 100%. The fold induction by 8% O2was calculated in each set of transfections relative to the corresponding Luc activity obtained under 16% O2. Values are means ± SEM of 3 independent culture experiments. Statistics, Student t test for paired values: *, significant difference 16% O2 versus 8%O2; **, significant difference 16% O2 + USF-2a or ΔTDU2 versus 16% O2 + pCMV (control); ***, significant difference 8% O2 + USF-2a or ΔTDU2 versus 8% O2 + pCMV (control); P < .05. ΔHU2a, USF-2 mutant lacking the second helix of the HLH domain; ΔTDU2, USF-2 mutant lacking the first 198 amino acids of the transactivation domain.23 L1-L4: L indicates liver-specific; L1, binding site for hepatocyte nuclear factor 1, L2, binding site for nuclear factor 1; L3, binding site for hepatocyte nuclear factor 4; L4, binding site for USF.3132](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2657/6/m_h80910984003.jpeg?Expires=1770994616&Signature=l1I5M0JNOeKwYFJu4o0J3IrLN7bI3f8CuuMkHLeSYZz8Cf4gaNVIQXVzJcvrK9WQv-X33ruFkf8PBVGlYax9jnbD0f6M4694EOYUbRgPLQgNbIxo3eBEtOCGoBv9OvKH82dRHt-yEuW-1N-n6Yy0UYrYidPaTOSFQRtpLJfn8qcab~WZByrNzjQwnV31qru6geX3SWz6hf7ML0eRM6kXSzitRPLF0WDS~nNPUa7BeM4A2EtRywrkwL669wL4rTU1Q3T1sG3-O4-ACs65-vqw5cctYBy34sakRPw7pc54hRZRW3ssLO2EQ45mMesjT9xWk-y8F92I3IpRvaZC4WfHTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Inhibition of Luc activity by USF-2a is conferred by HRE-1 and HRE-2 in the rat PAI-1 promoter. / (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the 2 potential USF-2a binding elements (HRE) matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with either USF-2a or ΔHU2a expression plasmids and Luc gene constructs driven by a wild-type 276-bp rat PAI-1 promoter (pGl3PAI-276) or the 276 bp promoter mutated at either the HRE-1 (pGl3PAI-276M1) or HRE-2 (pGl3PAI-276M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-276 + pCMV control, which was set equal to 100%. In pGl3PAI-276M1 and pGl3PAI-276M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-276 + USF-2a versus pGl3PAI-276 + pCMV control; pGl3PAI-276M1 + USF-2a versus pGl3PAI-276M1 + pCMV control; pGl3PAI-276M2 + USF-2a versus pGl3PAI-276M2 + pCMV control; P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2657/6/m_h80910984004.jpeg?Expires=1770994616&Signature=PMiZglTo4bY1d8arxn5WBTrKS9L~abIekm2TlSEs2BNER157Yj7GgvTZz7fNDFzAxLBSiBg-D~qT-Rz7osx-PBGs8xAFbNiROzFBdE9kp49A5V59-rwCpE8sgPlEOcjVcVv9DJpDWZ~IaIY8NFtTSgy0xyn1qCJ8NRyoNr~6u90s0R9jCCVIFbwBwp8SnHpNPNeeE4k37OTtk7YSG2fZG0w~YiYG0KYm7UZwWIDucb37EuSp4a8hQhV3eMLDPdUwAY1zFMeEei99Cz9oDwDVaU9-nL8~1uCO~rc53A3q2GuwHTZlbV4VFjZQH5~whaOIWHZw-Fu321BdyruB4ZuQcA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Competition by HIF-1α via HRE-2 of the USF-2a–dependent inhibition of pGl3PAI-766 Luc expression in hepatocytes. / (A) The 5′-flanking region of the rat PAI-1 gene with its footprinted sites [A-G]. In the “C-site” the USF and HIF-1 binding element14 matching the E-box consensus sequence are underlined. S (G/C) is shown because the actual PAI-1 sequence does not match bases allowed by the consensus. (B) Hepatocytes were transiently cotransfected with expression plasmids for either USF-2a, HIF-1α, or both and Luc gene constructs driven by a wild-type 766-bp rat PAI-1 promoter (pGl3PAI-766) or the 766-bp promoter mutated at either the HRE-1 (pGl3PAI-766M1) or HRE-2 (pGl3PAI-766M2) site. In control experiments Luc constructs were cotransfected with pCMV plasmid. In each experiment the percentage of Luc activity was determined relative to the pGl3PAI-766 + pCMV control, which was set equal to 100%. In pGl3PAI-766M1 and pGl3PAI-766M2 the wild-type PAI-1 sequence is shown on the upper strand, and mutated bases are indicated by an asterisk and are shown in lowercase letters. The values represent means ± SEM of 3 independent experiments. Statistics, Student t test for paired values: *, significant difference pGl3PAI-766 + USF-2a versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + USF-2a versus pGl3PAI-766M1 + pCMV control; pGl3PAI-766M2 + USF-2a versus pGl3PAI-766M2 + pCMV control; **, significant difference pGl3PAI-766 + HIF-1α versus pGl3PAI-766 + pCMV control; pGl3PAI-766M1 + HIF-1α versus pGl3PAI-766M1 + pCMV control; ***, significant difference pGl3PAI-766 + USF-2a + HIF-1α versus pGl3PAI-766 + USF-2a; pGl3PAI-766M1 + USF-2a + HIF-1α versus pGl3PAI-766M1 + USF2a; P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2657/6/m_h80910984005.jpeg?Expires=1770994616&Signature=sptuJU3aFDQwcicBqXe4YS329d7SF8p-GhttWhcVd9CxbY5YbFzrWX91AQnTtF06TJox~dNPGPsm08EE~mZoe55ARRfR77Cmzs5ay0i3EUo51lBmGGqM0KqGXwLQFYV4bhGZyeeN6-ZdSAwfKprKDQGdUsJnsmi~5YtgGRhOxzUDiakSfgKoi~-sOwNWXG8AAlwK4tAUnsaUzpla8s2q4vCVKEkOespOSjwyNs5A9e8Rk7b5P67ObS07cPp0SgGGIrWxmB6cpn4fX96MU-dpy3jUwej-dg2DrM4BjDbs9MCelyn2bIrzKPGvJoymWGPf-czK6POyf8SlZYiAZTqj2A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal