Abstract
Neutrophil-specific granule deficiency (SGD) is a rare congenital disorder. The neutrophils of individuals with SGD display atypical bi-lobed nuclei, lack expression of all secondary and tertiary granule proteins, and possess defects in chemotaxis, disaggregation, receptor up-regulation, and bactericidal activity, resulting in frequent and severe bacterial infections. Previously, a homozygous mutation in theCCAAT/enhancer binding protein–ε (C/EBPε) gene was reported for one case of SGD. To substantiate the role of C/EBPε in the development of SGD and elucidate its mechanism of inheritance, the mutational status of the gene was determined in a second individual. An A-nucleotide insertion in the coding region of the C/EBPε gene was detected. This mutation completely abolished the predicted translation of all C/EBPε isoforms. Microsatellite and nucleotide sequence analyses of the C/EBPε locus in the parents of the proband indicated that the disorder may have resulted from homozygous recessive inheritance of the mutant allele from an ancestor shared by both parents. The mutant C/EBPε32 protein localized in the cytoplasm rather than the nucleus and was unable to activate transcription. Consistent with this, a significant decrease in the levels of the messenger RNAs (mRNAs) encoding the secondary granule protein human 18-kd cationic antimicrobial protein (hCAP-18)/LL-37 and the primary granule protein bactericidal/permeability-increasing protein were observed in the patient. The hCAP-18 mRNA was induced by overexpression of C/EBPε32 in the human myeloid leukemia cell line, U937, supporting the hypothesis that C/EBPε is a key regulator of granule gene synthesis. This study strongly implicates mutation of theC/EBPε gene as the primary genetic defect involved in the development of neutrophil SGD and defines its mechanism of inheritance.
Introduction
Neutrophil-specific granule deficiency (SGD) is a rare congenital disorder, possibly inherited in an autosomal recessive fashion. Individuals with SGD (5 reported worldwide) possess atypical bi-lobed nuclei and lack expression of secondary and tertiary granule messenger RNAs (mRNAs) and protein, including lactoferrin, transcobalamin, gelatinase B, and collagenase.1-8Additionally, they display a marked decrease in levels of the primary granule defensins; however, expression of the primary granule genesmyeloperoxidase (MPO) and lysozyme are unaffected.9,10 The neutrophils of SGD patients are defective in chemotaxis, dissaggregation, receptor up-regulation, and bactericidal activity.1-8 More recently, the deficiency in granule gene expression was extended to eosinophils.11These cells from SGD patients lacked eosinophil-specific granule contents, including eosinophil cationic protein, eosinophil-derived neurotoxin, and major basic protein.11 Because of these numerous deficiencies and functional defects, SGD individuals are severely immunocompromised and develop frequent bacterial infections, including Pseudomonas aeruginosa and Staphylococcus aureas.
Since SGD individuals express normal levels of lactoferrin and transcobalamin in their saliva but not in either their plasma or neutrophils, the molecular basis for SGD was hypothesized to involve the mutation of a myeloid-specific transcription factor.8,12-14 A candidate gene encoding such a transcription factor is CCAAT/enhancer binding protein–ε (C/EBPε).15,16 It is expressed primarily during granulocytic differentiation.15-19 Targeted disruption of the gene in mice leads to defects in terminal differentiation of neutrophils with increased numbers of morphologically atypical neutrophils.20 The phenotypic and functional defects of neutrophils from C/EBPε-deficient mice closely parallel those of SGD. They include bi-lobed nuclei, a significantly reduced capacity to produce superoxide when treated with phorbol 12–myristate 13–acetate, and impaired chemotaxis and bactericidal activity.20-22The null mice are susceptible to gram-negative bacterial sepsis, particularly with P aeruginosa, and succumb to systemic infection at 3 to 5 months of age.20 Consistent with SGD, the C/EBPε-deficient mice lack expression of mRNAs encoding the same secondary and tertiary granule proteins. In addition, mRNAs for the cathelicidin B9 and murine cathelin-related antimicrobial peptide (CRAMP) are severely reduced in the bone marrow.21,22These cathelin-like peptides possess potent activity against gram-negative bacteria, including P aeruginosa.23,24 As with SGD, C/EBPε-deficient mice exhibit abnormalities in their eosinophils and lack expression of mRNAs encoding eosinophil peroxidase and major basic protein (our unpublished observations, March 1999).20
Because of the striking similarities between SGD patients and the C/EBPε-deficient mice, the C/EBPε locus was examined for mutations in 1 of the 5 known individuals.25 A homozygous 5–base pair (bp) deletion in the second exon was described.25 In humans, 4 C/EBPε isoforms, referred to as p32, p30, p27, and p14, are generated.16,19 The p32, p27, and p14 forms are encoded by 4 mRNA isoforms generated by transcription from 2 different promoters, Pα and Pβ, combined with differential splicing (see Figure 1B).19 The p30 isoform is synthesized by translation from a downstream start codon at amino acid position 32.16 The predicted 5-bp frameshift resulted in a truncation of the transcriptionally active p32 and p30 isoforms with the loss of the dimerization and DNA-binding domain regions.25 The p27 and p14 isoforms, which are unable to activate transcription, were unaffected (unpublished results, March 1999).19 The truncated C/EBPε32 was unable to activate transcription efficiently.25 These results implicated alteration of the C/EBPε gene in the development of one case of SGD.
The C/EBPε gene contains a frameshift mutation in a patient with SGD.
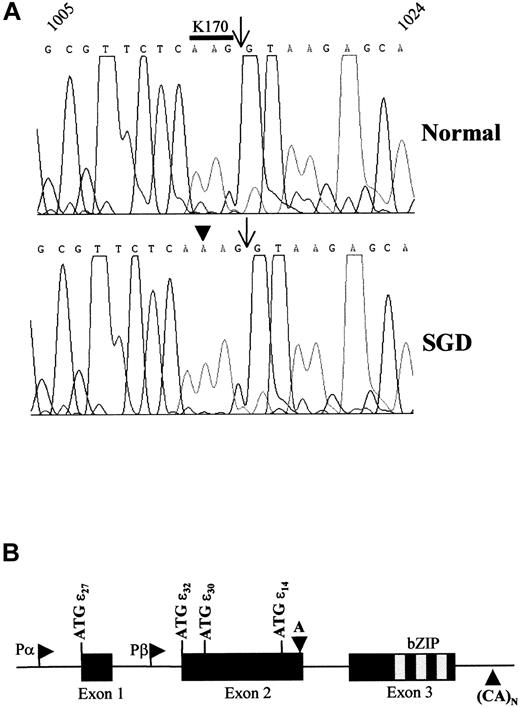
(A) Representative sequence chromatographs from a normal and an SGD patient from nucleotides 1005 through 1024 based on the previously deposited sequence.16 The nucleotide sequence is denoted across the top of the chromatograph. The arrows indicate the boundary between exon 2 and intron 2. The codon encoding amino acid residue lysine 170 (K170) is overlined in the normal sequence. The sequence was determined from 3 separate PCR reactions on the genomic DNA. A total of 12 clones from 3 separate PCR reactions were sequenced from both directions to verify the mutation. (B) Schematic drawing of the C/EBPε genomic locus indicates the 3 exons, translational start codons (ATG) for each isoform, the basic region–leucine zipper (bZIP) domain, and the 2 alternative promoters, Pα and Pβ. The downward arrowhead indicates the location of the A-nucleotide insertion in both panels. This would shift the open reading frame at the K170 codon and result in the lack of the bZIP domain in isoforms p32, p30, p27, and p14. The upward arrowhead indicates the position of CA-repeat microsatellite that is 35 bp 3′ of the polyadenylation site.
The C/EBPε gene contains a frameshift mutation in a patient with SGD.
(A) Representative sequence chromatographs from a normal and an SGD patient from nucleotides 1005 through 1024 based on the previously deposited sequence.16 The nucleotide sequence is denoted across the top of the chromatograph. The arrows indicate the boundary between exon 2 and intron 2. The codon encoding amino acid residue lysine 170 (K170) is overlined in the normal sequence. The sequence was determined from 3 separate PCR reactions on the genomic DNA. A total of 12 clones from 3 separate PCR reactions were sequenced from both directions to verify the mutation. (B) Schematic drawing of the C/EBPε genomic locus indicates the 3 exons, translational start codons (ATG) for each isoform, the basic region–leucine zipper (bZIP) domain, and the 2 alternative promoters, Pα and Pβ. The downward arrowhead indicates the location of the A-nucleotide insertion in both panels. This would shift the open reading frame at the K170 codon and result in the lack of the bZIP domain in isoforms p32, p30, p27, and p14. The upward arrowhead indicates the position of CA-repeat microsatellite that is 35 bp 3′ of the polyadenylation site.
Comparison of the phenotypes of individuals suffering from SGD indicate that it is likely to be a heterogenous disease and suggests the possibility of different underlying genetic defects.4 The purpose of this study is to test the hypothesis that alteration of theC/EBPε gene is the primary defect involved in the development of SGD. We identified a mutation in the C/EBPεlocus of a second individual with SGD and elucidated the mechanism of inheritance by analysis of the locus in the parents of the individual.
Patient, materials, and methods
SGD proband and genomic DNA isolation
The proband is a 27-year old female who has suffered from recurrent pyogenic infections, such as otitis media, skin abscesses, and pneumonia, since infancy. As previously reported,3 8the neutrophils of this individual exhibit unique bi-lobed nuclei and decreased cytoplasmic granularity. These cells display, by histochemical staining, normal peroxidase-positive products and no alkaline phosphatase activity, an absence of specific granules upon electron microscopy, impaired chemotaxis, defective bactericidal activity against S aureus and Escherichia coli, and markedly decreased lactoferrin and transcobalamin content. Peripheral blood was obtained from the proband and her parents after informed consent. The peripheral blood mononuclear cells (PBMCs) were purified by Ficoll gradient centrifugation, and genomic DNA was prepared. For the normal control, genomic DNA was prepared from the bone marrow of healthy volunteers after informed consent. The parents of the proband are healthy with no history of recurrent infections. The number and morphology of the neutrophils of both parents were normal.
RNA isolation and analysis
Total RNA from all nucleated cells present in the peripheral blood was prepared from the proband and her father after informed consent. The RNA was extracted by lysis of cells in TRIzol reagent as described by the manufacturer (Gibco/BRL, Rockville, MD). For reverse-transcription polymerase chain reaction (RT-PCR) analysis, RNA was treated with DNaseI and reversed transcribed by means of random oligonucleotides as described previously.26 Each reverse transcription reaction contained approximately 1 to 2 μg total RNA. Northern blot analysis was performed essentially as described,27 with the use of 10 μg total RNA per lane. Probes (described below) were labeled by means of the StripEZ system (Ambion, Austin, TX), and blots hybridized in ULTRAhyb solution (Ambion) as instructed by the manufacturer. Autoradiographs were exposed to X-OMAT AR film (Kodak, Rochester, NY).
PCR of genomic DNA and complementary DNA
The primers used to amplify the C/EBPεgenomic locus are described in Table 1. The primers were used in the following combinations to amplify overlapping regions of the proband's genomic DNA: (1) Prom-S + Prom-AS; (2) R66 + 440; (3) EX2-S + EX2-AS; (4) R66 + NFM-1; and (5) 404 + NFM-1. PFU (Stratagene, La Jolla, CA) or Advantage Taq (Clontech Laboratories, Palo Alto, CA) polymerases were used to amplify the fragments as described by the manufacturers. The PCR conditions were as follows: 94°C, 3 minutes; then 35 cycles 94°C, 30 seconds; 60°C (for primer sets 1 and 2) or 64°C (for primer sets 3 through 5), 30 seconds; and 72°C, 2 minutes. To amplify the complementary DNA (cDNA) of C/EBPε, primer pair 404 + NFM-2 was used with a 64°C annealing temperature. The products were cloned into either pST-Blue (Novagen, Madison, WI) or pcR2.1 (PE Applied Biosystems, Carlsbad, CA) as described by the manufacturer. Products were sequenced with an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit (Invitrogen, Foster City, CA) by means of primer binding sites available in the plasmids (T7, SP6, and M13R) and the primers described in Table 1 and were analyzed by an ABI 377 sequencing machine.
Primers used for amplification of C/EBPε by polymerase chain reaction
| Primer name . | Nucleotide sequence . | Nucleotide position* . |
|---|---|---|
| Prom-S | 5′-agtcggagggaggaggttgc-3′ | 15-34 |
| Prom-AS | 5′-tggcttcacggcaaagagatc-3′ | 691-671 |
| R66 | 5′-atgtgtgagcatgaggcctccatt-3′ | 602-625 |
| 440 | 5′-cggcagtggccaaaggggcct-3′ | 1790-1770 |
| EX2-S | 5′-cagcctctgcgcgttctcaag-3′ | 995-1015 |
| EX2-AS | 5′-gtccgcagagttaggccgtgc-3′ | 2163-2143 |
| NFM-1 | 5′-ccacaccagcctctccagc-3′ | 1070-1052 |
| 404 | 5′-cagacaggaaggcgctggg-3′ | 795-813 |
| NFM-2 | 5′-gggagggcgccttcaggag-3′ | 1826-1808 |
| Primer name . | Nucleotide sequence . | Nucleotide position* . |
|---|---|---|
| Prom-S | 5′-agtcggagggaggaggttgc-3′ | 15-34 |
| Prom-AS | 5′-tggcttcacggcaaagagatc-3′ | 691-671 |
| R66 | 5′-atgtgtgagcatgaggcctccatt-3′ | 602-625 |
| 440 | 5′-cggcagtggccaaaggggcct-3′ | 1790-1770 |
| EX2-S | 5′-cagcctctgcgcgttctcaag-3′ | 995-1015 |
| EX2-AS | 5′-gtccgcagagttaggccgtgc-3′ | 2163-2143 |
| NFM-1 | 5′-ccacaccagcctctccagc-3′ | 1070-1052 |
| 404 | 5′-cagacaggaaggcgctggg-3′ | 795-813 |
| NFM-2 | 5′-gggagggcgccttcaggag-3′ | 1826-1808 |
Numbering based on published sequence available from EMBL/GenBank/DDBJ under accession No. U80982.
The primers for MPO, lactoferrin,glyceraldehyde-3-phosphate dehydrogenase, and18S ribosomal RNA (rRNA) and Southern blot analysis of PCR products were described previously.17,27,28 The primers for amplification of bactericidal/permeability-increasing (BPI) protein were BPI-S, 5′-cagaagggcctggactac-3′ (nucleotide [nt] 154-171); BPI-AS, 5′-tgctgcagctggagcag-3′ (nt 516-531); and, for hybridization, BPI-Int, 5′-ctgcagaaggagctgaagaggatc (nt 196-219). The primers for amplification of human 18-kd cationic antimicrobial protein (hCAP18) were CAP18-F1, 5′-agctacaaggaagctgtgcttcg-3′ (nt 115-137); CAP18-R1, 5′-tcactgtccccatacaccgc-3′ (nt 333-352); and, for hybridization, CAP18-PR, 5′-caggattgtgacttcaagaaggacg-3′ (nt 299-322). The primers for amplification of human neutrophil peptide 3 (HNP3) were HNP3-S, 5′-gccatgaggaccctcg-3′ (nt 48-63); and HNP3-AS, 5′-gcagcagaatgcccagag-3′ (nt 332-315). These primers also amplified HNP-1 because the genes are nearly identical.29The hCAP18 PCR product was subcloned into pcR2.1 (Invitrogen) and sequenced to verify its identity. The insert was excised with EcoRI and used as a probe in Northern blot analysis.
Microsatellite sequence polymorphism and single-strand conformation polymorphism analyses
The polymorphic CA-repeat microsatellite sequence is located 35-bp 3′ of the polyadenylation site of the C/EBPεgene (Figure 1B). For analysis, the microsatellite was amplified by PCR (as described above) with the use of the primers KO691 (5′-ggcaaagagggcaggacccagc-3′) and KO692 (5′-ggtgcagacctagccacatgc-3′) and an annealing temperature of 55°C.30 The reactions were denatured by heating at 95°C for 5 minutes and electrophoresed through a denaturing 8M-urea, 5% polyacrylamide gel. PCR–single-strand conformation polymorphism (PCR-SSCP) analysis with primers 404 and NFM-1 was performed as described.31 The products were denatured and electrophoresed through a nondenaturing polyacrylamide Mutation Detection Enhancement gel (Biowhittaker Molecular Applications, Rockland, ME) containing 10% glycerol. [33P]–deoxyadenosine triphosphate was added to the reactions in both procedures to facilitate visualization of the products by autoradiography (NEN Lifesciences Products, Boston, MA).
Construction and characterization of mutant C/EBPε
The A-nucleotide insertion was introduced into the wild-type C/EBPε32 cDNA by means of a 2-step PCR approach as described previously.32 Briefly, in one reaction, 100 ng template (pCMV-C/EBPε32) was amplified with the vector-specific primer 32N (5′-tcgccggaattcatgtcccacgggacctactacgagtgtgagccccgg-3′) and the gene-specific primer SGDMUT-AS 5′-ggcctttgagaacgcgcagaggctggccgg-3′). In the second reaction, the vector-specific primer NdelC 5′-agcctggtcgacgtgcccacaatccaccagcca-3′) was mixed with the gene-specific primer SGDMUT-S (5′-gttctcaaaggccccctttggccactgccgc-3′). The products were amplified in a 100-μL reaction by means of Advantage Taq as described by the manufacturer. The resulting products (1 μL each) were mixed and amplified by means of the the 2 vector-specific primers. The full-length product was gel purified, digested with EcoRI and SalI, and ligated with 100 ng pCMV-SPORT (Gibco/BRL) cut with EcoRI and SalI. The resulting transformants were sequenced to verify the presence of the mutation and verify the integrity of the remainder of the insert.
COS-1 and NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with either 10% fetal bovine serum or bovine calf serum, respectively. For cellular localization, COS-1 cells, plated at 70% confluency on a 60-mm dish, were transfected with 3 μg pCMV-SPORT, pCMV-C/EBPε32, or pCMV-SGDε by means of 15 mL GenePorter (Gene Therapy Systems, San Diego, CA), as described by the manufacturer. At 24 hours post-transfection, cells were harvested, and nuclear and cytoplasmic fractions prepared as described previously.16 Whole cell lysates were prepared by lysis in RIPA buffer, and Western blot analysis was performed as described previously.26 The blots were incubated overnight with 0.1 μg anti-C/EBPε antiserum (rabbit) or 0.5 μg anti–β-actin mouse monoclonal antibody (Santa Cruz Biotechology, CA) diluted in phosphate-buffered saline containing 5% powdered milk.16 The primary antibodies were detected with either donkey anti–rabbit horseradish peroxidase (HRP) (1:5000) or anti–mouse-HRP (1:500). The complexes were developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL), as described by the manufacturer and detected by autoradiography.
For transcriptional activation assays, NIH3T3 cells were plated in a 12-well dish at 70% confluency. For each triplicate, the plasmids were prepared as a master mix of 1.0 μg pGCSF-R and 0.1 μg pSV40 Renilla Luciferase (Promega, Madison, WI) plus expression vector plus empty vector for a final total of 3 μg DNA. The combinations and amounts of expression vectors are indicated in the legend of Figure 3. The plasmids in 0.5 mL Opti-MEM (Gibco/BRL) were mixed with 15 μL of GenePorter in 0.5 mL Opti-MEM, incubated 45 minutes, and 0.33 mL aliquoted to each well of the 12-well plate (1 μg DNA per well). The pGCSFR-Luc reporter plasmid was kindly provided by Dan Tenen (Harvard Medical School, Boston, MA). At 24 hours post-transfection, cells were lysed in passive lysis buffer, and luciferase activity was measured by means of a dual luciferase assay (Promega).
Results
Frameshift mutation in coding region of C/EBPεlocus in SGD individual
Sequencing of PCR products from the genomic DNA of the proband revealed a single A-nucleotide insertion at nt 1113 (EMBL/GenBank/DDBJ accession No. U80982) located at the 3′-end of exon 2 (Figure 1A-B). This was not detected in the sequence from a normal individual (Figure 1A). Sequence analysis of this region amplified from the patient's cDNA revealed the presence of the A-nucleotide insertion indicating the mutation was present in the mRNA (data not shown). The mutation predicts a frameshift and premature termination of the encoded C/EBPε isoforms p32, p30, p27, and p14 (Figure 1B). The premature termination would result in the loss of the basic region and leucine zipper domains that are critical for DNA binding and dimerization, respectively.
Sequencing of the cloned PCR products from the genomic DNA of the proband indicated the presence of only the mutant allele. This suggested that the mutation was homozygous. To test this, we used primer pair 404 and NFM-1 to amplify the region of the mutation from the genomic DNA of the father and mother. Sequencing of the cloned products revealed the presence in both parents of wild-type and mutant sequence (data not shown). The mutation in each parent was an A-nucleotide insertion as described above for the proband. To confirm the genotype of the proband and the parents, we performed SSCP analysis using the above primer pair. Two normal subjects (NHBM-1 and NHBM-2) were homozygous for the wild-type (E) allele (Figure2A). The proband was homozygous for the mutant (e) allele, and the parents were heterozygous for both alleles as indicated by the presence of 2 bands in the parents versus 1 in the normal controls and proband (Figure 2A). These results support those observed from sequencing of cloned PCR products.
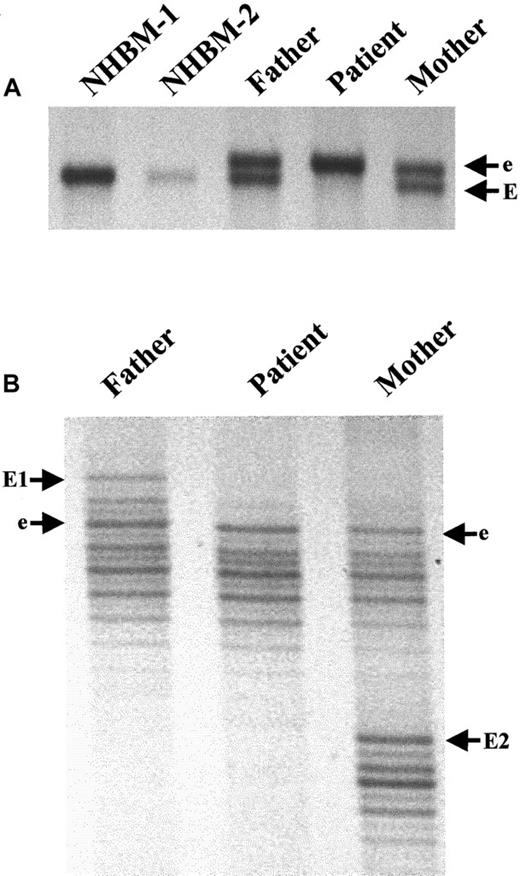
Autosomal recessive inheritance of the mutant allele from a distant relative common to both parents of the proband.
(A) SSCP analysis demonstrates that genomic DNA from NHBM is homozygous for the wild-type allele, EE; the patient is homozygous for the mutant allele containing the A-nucleotide insertion, ee; and both parents are heterozygous, Ee. (B) Microsatellite analysis reveals that the mutant, e, but not the wild-type, E, alleles carried by the parents possesses the same “fingerprint,” indicating they inherited the mutant allele from the same distant relative.
Autosomal recessive inheritance of the mutant allele from a distant relative common to both parents of the proband.
(A) SSCP analysis demonstrates that genomic DNA from NHBM is homozygous for the wild-type allele, EE; the patient is homozygous for the mutant allele containing the A-nucleotide insertion, ee; and both parents are heterozygous, Ee. (B) Microsatellite analysis reveals that the mutant, e, but not the wild-type, E, alleles carried by the parents possesses the same “fingerprint,” indicating they inherited the mutant allele from the same distant relative.
Mechanism of inheritance of mutant allele
The presence of an identical A-nucleotide insertion in one allele from each parent strongly suggested that the parents inherited the same allele from a common distant relative. To test this, we performed microsatellite analysis for a marker that is part of theC/EBPε gene locus. It is located 35 bp downstream of the polyadenylation site of the gene.30 The father and mother possessed 2 patterns (E, wild type; and e, mutant) indicative of their heterozygous status (Figure 2B). Interestingly, they shared one pattern (e) while the other was unique to each parent (E1 and E2). The proband was homozygous for the “e” pattern. The inheritance of an identical microsatellite and nucleotide insertion indicates that the same mutant allele was inherited from each parent (Figure 2B). This most likely occurred because the parents inherited the same mutant allele from a common distant relative. The data support a homozygous recessive inheritance of the mutant allele.
The mutant C/EBPε accumulates in the cytoplasm and is unable to activate transcription
The predicted protein resulting from the mutation would lack the bZIP domain, which is essential for dimerization and DNA binding. In addition, the basic region contains the nuclear localization sequence responsible for targeting the C/EBPs to this compartment.33 We predicted that the mutant C/EBPε would accumulate in the cytoplasm and lack the ability to activate transcription from a promoter containing its binding site. To test this, COS-1 cells were transfected with either empty vector (−), wild-type (W), or mutant (S) C/EBPε32 expression vectors. Whole cell lysates and cytoplasmic (C) and nuclear (N) fractions were analyzed by Western blot (Figure 3A). The empty-vector transfectants did not express the C/EBPε proteins whereas the expected wild-type and mutant forms were expressed at similar levels (Figure 3A). The mutant form did not appear to be unstable; however, it accumulated in the cytoplasmic fraction and not the nuclear fraction, where the wild-type protein localizes (Figure 3A, lanes 4-9).
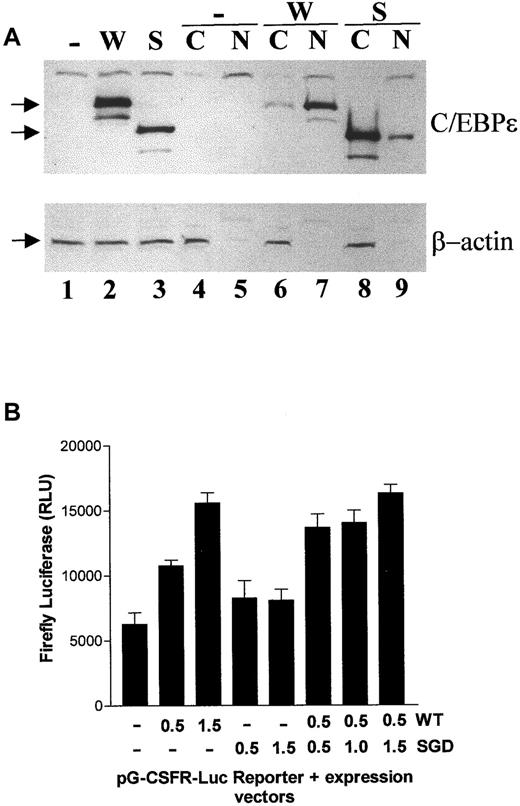
Aberrant cellular localization and transcriptional activation by the mutant C/EBPε.
(A) Western blot analysis of whole cell lysates (lanes 1-3), cytoplasmic (C; lanes 4, 6, and 8), and nuclear (N; lanes 5, 7, and 9) fractions for C/EBPε and β-actin expression. The absence of the cytoplasmic protein β-actin in the nuclear fraction serves as a control for the experiment. The cells were transfected with expression vectors that were either empty (−) or encoded wild-type (W) or mutant (S) C/EBPε32 (32 and 24 kd, respectively). The arrows at the left of the panels indicate the positions of the proteins. (B) NIH3T3 cells were co-transfected with pG-CSFR-luciferase (firefly) and either empty (−), wild-type (WT), or mutant (SGD) C/EBPε32. Luciferase activity was measured and normalized to renilla luciferase used as a control for transfection efficiency. The experiment was performed twice in triplicate with results presented in relative light units (RLUs).
Aberrant cellular localization and transcriptional activation by the mutant C/EBPε.
(A) Western blot analysis of whole cell lysates (lanes 1-3), cytoplasmic (C; lanes 4, 6, and 8), and nuclear (N; lanes 5, 7, and 9) fractions for C/EBPε and β-actin expression. The absence of the cytoplasmic protein β-actin in the nuclear fraction serves as a control for the experiment. The cells were transfected with expression vectors that were either empty (−) or encoded wild-type (W) or mutant (S) C/EBPε32 (32 and 24 kd, respectively). The arrows at the left of the panels indicate the positions of the proteins. (B) NIH3T3 cells were co-transfected with pG-CSFR-luciferase (firefly) and either empty (−), wild-type (WT), or mutant (SGD) C/EBPε32. Luciferase activity was measured and normalized to renilla luciferase used as a control for transfection efficiency. The experiment was performed twice in triplicate with results presented in relative light units (RLUs).
To test the ability of the mutant C/EBPε to activate the transcription of a promoter containing a C/EBP site, the mutant and wild-type forms were co-transfected with a G-CSFR-promoter–luciferase reporter previously shown to be activated by C/EBPε32.19 This reporter was activated in a dose-responsive fashion by the wild-type, but not the mutant, form of C/EBPε (Figure 3B). Additionally, an increasing dose of the mutant form did not demonstrate an ability to interfere with the function of the wild type in a dominant negative fashion (Figure 3B). Together, these data indicate the A-nucleotide insertion would significantly impair the normal function of the C/EBPε protein.
Additional defects in primary and secondary granule gene expression
The C/EBPε-deficient mice have a significant decrease in expression of the cathelin-like genes CRAMP andB9. Because these peptides possess potent bactericidal activity against gram-negative bacteria and are components of the secondary granules, we predicted that the human homologue to murine CRAMP, hCAP18, would be significantly reduced. We examined the expression of hCAP18 mRNA in the proband and the father using RT-PCR analysis and found that its expression was 7-fold less in the proband (Figure 4A). This reduction corresponded with the expected absence of the mRNA encoding the secondary granule protein lactoferrin, which was absent in the proband (Figure 4A). In contrast, the expression of the primary granule gene MPO was unaffected (Figure 4A).
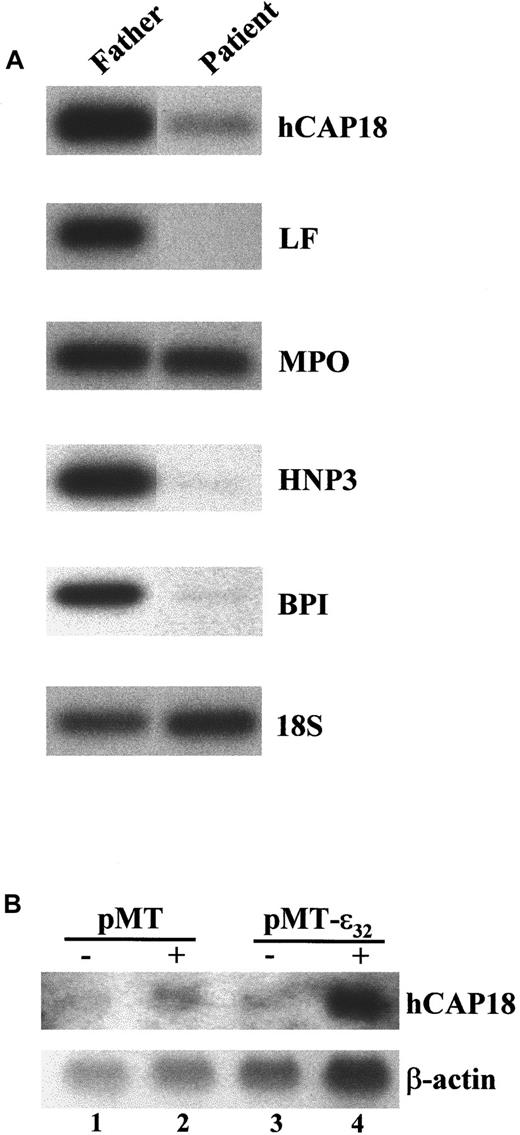
Expression of mRNA encoding the secondary granule protein hCAP18 and primary granule protein BPI is severely reduced in the PBMCs of the SGD patient.
(A) RT-PCR analysis of total RNA prepared from the PBMCs of the patient and father. The cDNAs were analyzed for expression of the primary granule genes MPO, HNP-1 and HNP-3 (HNP1/3), and BPI; the secondary granule geneslactoferrin (LF) and hCAP18; and the control 18S rRNA. The products were Southern blotted and hybridized with either internal oligonucleotide or cDNA probes. (B) Induced expression of C/EBPε32 in U937 activates hCAP18 expression. U937 cells stably transformed with a zinc-inducible empty vector (pMT) or one containing an insert for C/EBPε32 (pMT-ε32) were treated either without (−) or with (+) 100 μM ZnSO4for 24 hours. RNA was harvested and analyzed by Northern blot hybridization with a probe for hCAP18. The blot was stripped and subsequently hybridized for β-actin.
Expression of mRNA encoding the secondary granule protein hCAP18 and primary granule protein BPI is severely reduced in the PBMCs of the SGD patient.
(A) RT-PCR analysis of total RNA prepared from the PBMCs of the patient and father. The cDNAs were analyzed for expression of the primary granule genes MPO, HNP-1 and HNP-3 (HNP1/3), and BPI; the secondary granule geneslactoferrin (LF) and hCAP18; and the control 18S rRNA. The products were Southern blotted and hybridized with either internal oligonucleotide or cDNA probes. (B) Induced expression of C/EBPε32 in U937 activates hCAP18 expression. U937 cells stably transformed with a zinc-inducible empty vector (pMT) or one containing an insert for C/EBPε32 (pMT-ε32) were treated either without (−) or with (+) 100 μM ZnSO4for 24 hours. RNA was harvested and analyzed by Northern blot hybridization with a probe for hCAP18. The blot was stripped and subsequently hybridized for β-actin.
The primary granule protein BPI possesses very potent antimicrobial activity against gram-negative bacteria.34 Since the expression of the neutrophil primary granule defensins is significantly reduced in SGD patients, we hypothesized that BPI gene expression may be similarly affected. RT-PCR analysis revealed an absence of BPI mRNA expression in the proband (Figure 4A). This corresponded with a significant decrease in the mRNA levels of the primary granule neutrophil defensins HNP-1 and HNP-3 (Figure 4A, HNP1/3) and was consistent with the absence of defensins described previously for this patient.10 These results suggest that significantly reduced levels of BPI protein are present in the proband's primary granules.
The lack of expression of secondary and some primary granule proteins in SGD and C/EBPε-deficient mice suggests that the genes are potential targets of C/EBPε. Evidence supporting this hypothesis comes from a myeloid cell line U937 that is stably transformed with a zinc-inducible expression vector for C/EBPε32.27 Previously, we demonstrated that the secondary granule genes encoding lactoferrin and neutrophil collagenase were induced with overexpression of C/EBPε.27 Similarly, strong induction of hCAP18 mRNA expression occurred within 24 hours of zinc treatment (Figure 4B). Taken together, these results reveal additional defects in neutrophil primary and secondary granule gene expression and suggest that C/EBPε directly activates their expression.
Discussion
In this report, we described a 1-bp insertion in the coding region of the C/EBPε gene that results in a frameshift and a truncated protein. This protein is predicted to be nonfunctional, because it would lack the bZIP region that is necessary for subunit dimerization and binding to DNA. This prediction was supported by our functional analysis demonstrating its inabilty to properly localize or activate the G-CSFR promoter. Together with the previous description of a 5-bp deletion in the coding sequence of theC/EBPε gene in another individual,25 our study strongly supports the hypothesis that mutation ofC/EBPε is the primary genetic defect responsible for SGD.
Two other transcription factors known to be important in myeloid cell differentiation are C/EBPα and PU.1. Mice deficient in C/EBPα display an early block in maturation of granulocytes at the myeloblast stage and do not express secondary granules or their proteins.35 In addition, these mice have severe defects in hepatic structure and function, including impaired glycogen storage, and the mice die soon after birth from hypoglycemia.36,37Mice deficient in PU.1 exhibit multiple hematopoietic abnormalities of both myeloid and lymphoid lineages and die either in late embryogenesis or within 2 days of birth.38,39 Those mice that are born soon succumb to an overwhelming systemic bacterial infection.39 With intensive antibiotic treatment, the mice can survive approximately 2 weeks. The neutrophils from these mice are not detected until several days after birth, lack a respiratory burst, are less efficient than normal neutrophils at phagocytosis and bacterial cell killing, and do not express mRNAs for secondary granule proteins, but do contain primary granule proteins.39Although some of the phenotypic features these mice display are similar to those of SGD, a human with a germline mutation in either the C/EBPα or PU.1 gene would probably not survive. The only plausible scenario would involve somatic mutations occurring in the myeloid progenitor cell, thereby resulting in defective granulocyte differentiation.
Our study shows that a germline mutation was inherited by the proband. Characterizing the genotype of the parents revealed that the proband inherited the same A-nucleotide insertion from each parent in a homozygous recessive manner. Inheritance of an identical microsatellite marker that is part of the C/EBPε locus together with the nucleotide insertion indicated that the mutant allele was most likely inherited from a distant relative shared by both parents. The first report of a C/EBPε gene mutation was in an SGD individual whose parents were first cousins once removed.25 In light of our results, we predict that the first patient inherited the mutation in a similar manner.
The mutation of C/EBPε in this SGD individual resulted in additional uncharacterized defects in gene expression. The levels of hCAP18 and BPI mRNAs were severely reduced in the SGD patient, but not in the father. The lack of hCAP18 expression was expected since it is a secondary granule protein, but the absence of BPI, a primary granule protein, was intriguing. Previously, the only primary granule proteins known to be absent in SGD patients were defensins. Both hCAP18 and BPI possess potent activity against gram-negative bacteria. Neutrophils of newborn humans contain 3- to 4-fold less BPI than adult neutrophils and exhibit significantly lower antibacterial activity against gram-negative bacteria.40 This deficiency of BPI may contribute to the increased incidence of gram-negative sepsis among newborns.40 Similarly, in SGD patients, we hypothesize that the lack of the granule proteins hCAP18 and BPI may explain their increased susceptibility to gram-negative bacterial infections.
The striking similarity between the phenotypes of the C/EBPε-null mouse and the SGD patients was critical in suggesting that mutation of C/EBPε in humans was the molecular defect responsible for SGD (Table 2). Interestingly, we have not observed aberrant primary granule gene expression in the mouse. This is partly due to a lack of defensin gene expression in murine neutrophils, and a murine BPI gene has not been described.41 Another interesting observation is the lack of eosinophil peroxidase (EPO) mRNA expression in the bone marrow of the C/EBP-deficient mouse, but the presence of EPO protein in the eosinophils of individuals with SGD.11 Despite these differences, the C/EBPε-null mouse should provide a convenient model system to elucidate the role of C/EBPε in late maturation of granulocytes and the development of specific therapeutic approaches, such as supplemental therapy with antimicrobial peptides to treat SGD. The markedly decreased level of mRNA expression for the primary granule proteins BPI and defensins in SGD patients also suggests a role for C/EBPε in earlier phases of the myeloid differentiation program.9,10 Determining the role of C/EBPε in regulating the expression of granule genes and promoting earlier and later stages of differentiation may be important for understanding other neutropenic disease states, such as acute myeloid leukemia, that show abnormalities in secondary granule gene expression and differentiation. Interestingly, our previous studies suggest thatC/EBPε is a critical downstream target gene that is responsible for retinoic acid–induced granulocytic differentiation of acute promyelocytic leukemia cells.27 In addition, it would be interesting to determine the role of C/EBPε in other models of specific granule deficiciency, including neutrophils from neonates and patients following thermal injury.42
Comparison of SGD and C/EBPε−/− phenotypes
| Phenotype . | SGD . | C/EBPε−/− . |
|---|---|---|
| Neutrophils | ||
| Granule proteins* | ||
| Primary | HNP−, BPI− | Present† |
| Secondary | LF−, NC−, TC−, CAP18− | LF−, NC−, NGAL−, CRAMP−, B9− |
| Tertiary | NG− | NG− |
| Nuclear morphology | Bi-lobed | Bi-lobed |
| Migration | Abnormal | Abnormal |
| Bactericidal activity | Impaired | Impaired |
| Eosinophils | ||
| Granule proteins | MBP−, EDN−, ECP−, EPO+ | MBP−, EPO− |
| Bacterial infection | Staphylococcu aureus, Psuedomonas aeruginosa, Klebsiella | Pseudomonas aeruginosa |
| Phenotype . | SGD . | C/EBPε−/− . |
|---|---|---|
| Neutrophils | ||
| Granule proteins* | ||
| Primary | HNP−, BPI− | Present† |
| Secondary | LF−, NC−, TC−, CAP18− | LF−, NC−, NGAL−, CRAMP−, B9− |
| Tertiary | NG− | NG− |
| Nuclear morphology | Bi-lobed | Bi-lobed |
| Migration | Abnormal | Abnormal |
| Bactericidal activity | Impaired | Impaired |
| Eosinophils | ||
| Granule proteins | MBP−, EDN−, ECP−, EPO+ | MBP−, EPO− |
| Bacterial infection | Staphylococcu aureus, Psuedomonas aeruginosa, Klebsiella | Pseudomonas aeruginosa |
SGD indicates neutrophil-specific granule deficiency (SGD); C/EBPε, CCAAT/enhancer binding protein-ε; HNP, human neutrophil peptide; BPI, bactericidal/permeability-increasing; LF, lactoferrin; NC, neutrophil collagenase; TC, transcobalamin; CAP18, 18-kd cationic antimicrobial protein; NGAL, neutrophil gelatinase-associated lipocalin; CRAMP, cathelin-related antimicrobial peptide; NG, neutrophil gelatinase; MBP, major basic protein; EDN, eosinophil-derived neurotoxin; ECP, eosinophil cationic protein; EPO, eosinophil peroxidase. The (−) denotes severely reduced levels or complete absence of messenger RNA or protein. The (+) indicates presence was detected.
The citations for these data are referred to in the text of this manuscript.
Defensins (HNPs) are not normally expressed in the neutrophils of mice. A murine BPI gene has not been described.
We thank Dr Seiji Kawano for critically reading the manuscript; Drs Dorothy Park, Alexey Chumakov, and Seisho Takeuchi for helpful discussions; Dr Dan Tenen for providing the pG-CSFR-luciferase construct; and the patient and her parents for participating in this study.
Supported by National Institutes of Health grant CA26038-20, the Ko-So Foundation, the Horn Foundation, the Parker Hughes Fund, and the C & H Koeffler Fund. A.F.G. is a recipient of a Lymphoma Research Foundation of America fellowship; H.P.K. is a recipient of the Mark Goodson Chair in Oncology and a member of the Jonsson Cancer Center.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Adrian F. Gombart, Cedars-Sinai Medical Center, Division of Hematology/Oncology, UCLA School of Medicine, Davis Bldg 5065, 8700 Beverly Blvd, Los Angeles, CA 90048; e-mail:gombarta@csmc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal