Abstract
Hereditary hemochromatosis usually results from C282Y homozygosity in the HFE gene on chromosome 6p. Recently, a new type of hemochromatosis (HFE3) has been characterized in 2 unrelated Italian families with a disorder linked to 7q. Patients with HFE3 have transferrin receptor 2 (TFR2) inactivated by a homozygous nonsense mutation (Y250X). Here the identification of 2 newTFR2 mutations is reported. In a large inbred family from Campania, a frameshift mutation (84-88 insC) in exon 2 that causes a premature stop codon (E60X) is identified. In a single patient with nonfamilial hemochromatosis, a T→A transversion (T515A), which causes a Methionine→Lysine substitution at position 172 of the protein (M172K), has been characterized. TFR2 gene gives origin to 2 alternatively spliced transcripts—the α-transcript, which may encode a transmembrane protein, and the β-transcript, a shorter, possibly intracellular variant. Based on their positions, the effects of the identified mutations on the 2 TFR2 forms are expected to differ. Y250X inactivates both transcripts, whereas E60X inactivates only the α-form. M172K has a complex effect: it causes a missense in the α-form, but it may also prevent the β-form production because it affects its putative initiation codon. Analysis of the clinical phenotype of 13 HFE3 homozygotes characterized at the molecular level has shown a variable severity, from nonexpressing patients to severe clinical complications. The identification of new mutations of TFR2 confirms that this gene is associated with iron overload and offers a tool for molecular diagnosis in patients without HFE mutations.
Introduction
Inherited disorders of iron metabolism that cause iron overload are heterogeneous. Hereditary hemochromatosis, the most prevalent form in whites, is due to inappropriately high intestinal iron absorption and may cause several clinical complications in middle age, including liver cirrhosis, diabetes, heart failure, hypogonadism, and arthritis. Hemochromatosis is associated with C282Y mutation in theHFE gene.1 This gene encodes a protein that interacts with transferrin receptor (TFRC) and negatively affects cellular iron uptake from transferrin.2,3 Most patients are C282Y homozygous; a minority are C282Y/H63D compound heterozygous.1,4-10 Other HFE mutations are rare and usually private.11-13 Few patients with severe, early-onset disease14,15 have wild-type HFE and a distinct disorder,16 which is linked to the long arm of chromosome 117 (juvenile hemochromatosis, hemochromatosis type 2, or HFE2). A new type of hemochromatosis (hemochromatosis type 3 or HFE3) has been recently characterized in 6 patients from 2 Italian families with a disorder linked to 7q22. These patients are homozygous for a nonsense mutation (Y250X) of TFR2,18 a recently identified member of the transferrin receptor family, that shows moderate homology to TFRC and is presumed to mediate cellular iron uptake.19 At variance with TFRC,the regulation of TFR2 is not iron-dependent,19 suggesting that TFR2 has a distinct function in iron metabolism.20 The lack of interaction between TFR2 and HFE, recently demonstrated using soluble proteins, suggests that TFR2 has an independent role in iron regulation compared to theHFE/TFRC pathway.21 Two alternatively spliced forms (α and β) of TFR2 have been isolated. The α-transcript is mostly expressed in the liver, and the predicted α-protein could be a transmembrane protein.19The β-transcript is widely expressed at low level; it lacks cytoplasmic domain, transmembrane domain, and part of the extracellular domain, resulting in a possible intracellular protein.19The Y250X mutation that occurs in exon 6 inactivates both α and βTFR2.
In this paper we report the identification of 2 new mutations that disrupt TFR2 in patients with HFE3 with different effects on the 2 transcripts. In addition, we describe the clinical phenotypes of all the patients thus far characterized with TFR2 mutations.
Patients, materials, and methods
Patients and families
This study includes 4 different families with hemochromatosis unrelated to HFE. Six patients belong to the 2 previously described Sicilian families (families 1 and 2) and are Y250X homozygous.22 Four patients belong to a large inbred family (family 3) originating from Campania, a region in southern Italy. The description of the 2 index cases of family 3 and the exclusion of chromosome 6p linkage were previously reported.22 Recently, we recruited other members of the same family for a total of 14 subjects. The pedigree of family 3 is shown in Figure 1A. A 43-year-old man with heavy iron overload, several clinical complications, and wild-type HFE genotype was already reported.23 His family (family 4, pedigree in Figure 1B) originated from Lazio, in central Italy.
Pedigree of families 3 and 4.
(A) Family 3 harbors the 84-88 insC mutation. Genotypically affected patients are represented by filled symbols, unaffected relatives by open symbols. NE, not examined. (B) Pedigree of family 4. The genotypically affected patient is represented by a filled symbol, unaffected relatives by open symbols. The coinheritance of β-thalassemia is indicated by the asterisk.
Pedigree of families 3 and 4.
(A) Family 3 harbors the 84-88 insC mutation. Genotypically affected patients are represented by filled symbols, unaffected relatives by open symbols. NE, not examined. (B) Pedigree of family 4. The genotypically affected patient is represented by a filled symbol, unaffected relatives by open symbols. The coinheritance of β-thalassemia is indicated by the asterisk.
Healthy subjects were 50 blood donors, originating from southern Italy, with normal iron parameters. Informed consent was obtained for molecular studies according to the guidelines of the different institutions.
Molecular studies
DNA was prepared from peripheral blood buffy coats or the patient's lymphoblastoid cell lines by standard phenol-chloroform extraction.24 C282Y and H63D mutations in HFE were studied on genomic DNA using polymerase chain reaction (PCR)–based tests and restriction enzyme digestion with RsaI and MboI (New England Biolabs, Berkeley, MA), respectively.5Linkage to the HFE3 locus was established in family 3 analyzing microsatellites D7S651, D7S2498, D7S662, D7S477, and D7S1588 allele segregation and 2 TFR2 intragenic repeats (R1 and R2).18 Pairwise linkage analysis was performed using the MLINK program (J. Ott, Rockefeller University, New York, NY) from the LINKAGE computer package, as previously described.17
PCR was performed in a Thermal Cycler using 10 pmol each primer, with an average protocol of 32 cycles (denaturation, 94°C for 30 seconds; annealing, 56°C for 45 seconds; extension, 72°C for 45 seconds) and 1 U AmpliTaq DNA polymerase (Perkin Elmer). PCR primers forTFR2 exon amplification were obtained from databases and are reported in Table 1. MaeI enzyme was used to detect Y250X mutant on the amplification product of exon 6. Restriction enzyme analysis of the PCR products was performed according to the manufacturer's recommendations.
Sequences of TFR2 primers used in the polymerase chain reaction reactions
| Exons . | . | Sequence . | Fragment length (bp) . |
|---|---|---|---|
| 1 | F | 5'-CCTGCCAGGACTGATAAGGG-3' | 250 |
| R | 5'-GGTGGGAAGAAGCGAGGTCA-3' | ||
| 2 | F | 5'-TCACTGACCTCATTATTGCC-3' | 440 |
| R | 5'-AAGGCTGGCGGGTGGCAAGA-3' | ||
| 3 | F | 5'-GATTTCTGGGGTCCAGAGGT-3' | 370 |
| R | 5'-AGGCAGATGGGAGGACTCAG-3' | ||
| 4 | F | 5'-ACGTCTCTGGCATCCTTCCCT-3' | 276 |
| R | 5'-GGTGAGCGCCCCGAGCCGCG-3' | ||
| 5/6 | F | 5'-CGCGGCTCGGGGCGCTCACC-3' | 450 |
| R | 5'-CCTGAACGATTCTCACTGGC-3' | ||
| 7/8 | F | 5'-CTCCTGCCCTTATGAATCGGG-3' | 447 |
| R | 5'-GCGGGAGGCAATGTCTGCAC-3' | ||
| 9 | F | 5'-AGCGATCTGGAGCCAGAAAG-3' | 350 |
| R | 5'-CTATCTTGCCAGGGTGTAT-3' | ||
| 10 | F | 5'-GGGCTGAGAGACACAGGCAG-3' | 340 |
| R | 5'-CAGGTACAATGTGGATGCCG-3' | ||
| 11/12 | F | 5'-CCTCGCTAAGCCTGTCCTCC-3' | 325 |
| R | 5'-CCCAGAGGCAAGGGTGGACC-3' | ||
| 13/14/15 | F | 5'-TGAGCCCTGAGCCTG-3' | 522 |
| R | 5'-GCCAGGACAGGGTGGACGCTGG-3' | ||
| 16 | F | 5'-CCCAGCGTCCACCCTGTCCTGGC-3' | 368 |
| R | 5'-CTGGATTGCCAGAGAGGACC-3' | ||
| 17 | F | 5'-GGGTCCTGGCCCCTGCCGCCAG-3' | 258 |
| R | 5'-AGGTCCAGGAGGTGGAG-3' | ||
| 18 | F | 5'-CCTTTGCTCCCGAGCCAGAGCC-3' | 375 |
| R | 5'-AGCAGAGGAGCTCTTGACTG-3' |
| Exons . | . | Sequence . | Fragment length (bp) . |
|---|---|---|---|
| 1 | F | 5'-CCTGCCAGGACTGATAAGGG-3' | 250 |
| R | 5'-GGTGGGAAGAAGCGAGGTCA-3' | ||
| 2 | F | 5'-TCACTGACCTCATTATTGCC-3' | 440 |
| R | 5'-AAGGCTGGCGGGTGGCAAGA-3' | ||
| 3 | F | 5'-GATTTCTGGGGTCCAGAGGT-3' | 370 |
| R | 5'-AGGCAGATGGGAGGACTCAG-3' | ||
| 4 | F | 5'-ACGTCTCTGGCATCCTTCCCT-3' | 276 |
| R | 5'-GGTGAGCGCCCCGAGCCGCG-3' | ||
| 5/6 | F | 5'-CGCGGCTCGGGGCGCTCACC-3' | 450 |
| R | 5'-CCTGAACGATTCTCACTGGC-3' | ||
| 7/8 | F | 5'-CTCCTGCCCTTATGAATCGGG-3' | 447 |
| R | 5'-GCGGGAGGCAATGTCTGCAC-3' | ||
| 9 | F | 5'-AGCGATCTGGAGCCAGAAAG-3' | 350 |
| R | 5'-CTATCTTGCCAGGGTGTAT-3' | ||
| 10 | F | 5'-GGGCTGAGAGACACAGGCAG-3' | 340 |
| R | 5'-CAGGTACAATGTGGATGCCG-3' | ||
| 11/12 | F | 5'-CCTCGCTAAGCCTGTCCTCC-3' | 325 |
| R | 5'-CCCAGAGGCAAGGGTGGACC-3' | ||
| 13/14/15 | F | 5'-TGAGCCCTGAGCCTG-3' | 522 |
| R | 5'-GCCAGGACAGGGTGGACGCTGG-3' | ||
| 16 | F | 5'-CCCAGCGTCCACCCTGTCCTGGC-3' | 368 |
| R | 5'-CTGGATTGCCAGAGAGGACC-3' | ||
| 17 | F | 5'-GGGTCCTGGCCCCTGCCGCCAG-3' | 258 |
| R | 5'-AGGTCCAGGAGGTGGAG-3' | ||
| 18 | F | 5'-CCTTTGCTCCCGAGCCAGAGCC-3' | 375 |
| R | 5'-AGCAGAGGAGCTCTTGACTG-3' |
RNA-SSCP was performed according to previously described protocols.24-26 After PCR reaction, transcription was carried out with 10 U T7 RNA polymerase in a final volume of 10 μL containing 10 mM dithiothreitol, 40 mM Tris, pH 7.5, 6 mM MgCl, 2 mM spermidine, 10 mM NaCl, 5 nmol each ribonucleoside, 10 U RNase, and 0.2 μL S35 UTP. After electrophoresis, gels were dried and subjected to autoradiography. Bands showing electrophoretically altered mobility were directly sequenced. For direct sequencing, PCR products were run on 1% agarose gel, purified using QIAquick PCR purification kit (QIAGEN, Valencia, CA), and sequenced using Thermo Sequenase Cy5.5 dye terminator cycle sequencing kit. After purification from unincorporated dye with Autoseq G-50 columns, sequencing products were electrophoresed in an automatic sequencer (model 373A; Applied Biosystem) according to the manufacturer's protocols.
Total RNA was prepared by standard methods from peripheral blood buffy coats and retrotranscribed using the RNA GeneAmp kit (Roche Molecular System, Branchburg, NJ). Primers used to discriminate α and β transcripts were as reported.19
Results
Molecular studies
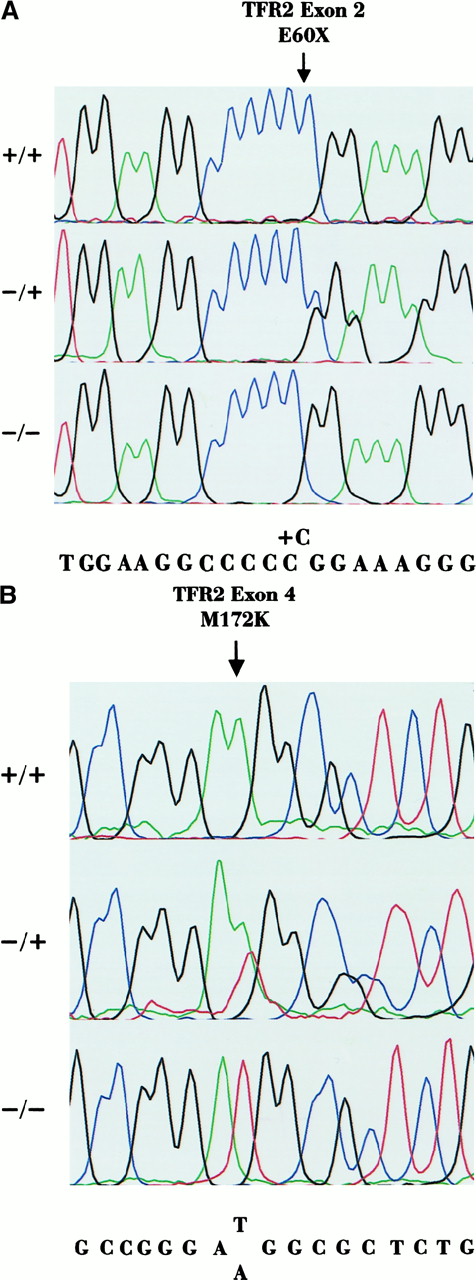
As expected, because of consanguinity, patients VI-4, VI-5, VI-7, and VI-8 of family 3 were homozygous at TFR2 intragenic repeats and at the other microsatellite markers of 7q (not shown). Based on the principle of homozygosity mapping, this finding was suggestive of linkage to this genomic area. When DNA of other family members became available, pairwise linkage analysis was calculated. A LOD score of 3.19 at θ = 0.0 was obtained for the intragenic repeat R1. The whole coding sequence and intron-exon boundaries ofTFR2 were scanned for mutations using direct sequencing in subjects VI-7 and VI-8. An insertion of a cytosine residue at the homozygous state was identified in exon 2 in a polyC tract (84-88 insC). This mutation resulted in a frameshift followed by a premature stop codon at amino acid 60 (E60X) in the protein (Figure2A). The C insertion was investigated by sequencing the DNA of all family members available. Subjects V-2, VI-3, VI-4, and VI-5 had the mutation at the homozygous state (Table2) and subjects V-4, VI-1, VI-6, VII-3, and VII-4 had the mutation at the heterozygous state (Table3). The same mutation was not found in 50 normal DNA samples analyzed by RNA-SSCP.
Sequencing chromatographs.
(A) Sequencing chromatographs of the forward sequence of exon 2 spanning the C insertion, which originates the E60X mutation. Subjects VI-6 (+/+) and VI-2 (+/−) are shown compared to a healthy control (−/−). Position of the C insertion is indicated by the arrow. (B) Sequencing chromatographs of the forward sequence of exon 4 spanning the T515A (M172K) mutation. Subjects II-2 (+/+) and I-2 (+/−) are shown compared to a healthy control (−/−). Mutation is indicated by the arrow.
Sequencing chromatographs.
(A) Sequencing chromatographs of the forward sequence of exon 2 spanning the C insertion, which originates the E60X mutation. Subjects VI-6 (+/+) and VI-2 (+/−) are shown compared to a healthy control (−/−). Position of the C insertion is indicated by the arrow. (B) Sequencing chromatographs of the forward sequence of exon 4 spanning the T515A (M172K) mutation. Subjects II-2 (+/+) and I-2 (+/−) are shown compared to a healthy control (−/−). Mutation is indicated by the arrow.
Clinical data of patients with hemochromatosis type 3 with different TFR2 mutations
| . | Sex . | Age at diagnosis . | TS (%) . | SF (μg/L) . | Liver function (HII) . | Additional clinical findings . | IR (g) . | HFEmutations . | TFR2 mutations . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C282Y . | H63D . | Y250X . | E60X . | M172K . | ||||||||
| Family 1 | ||||||||||||
| V-11 | M | 47 | 100 | 3200 | Cirrhosis | Arthritis | * | −/− | −/− | +/+ | −/− | −/− |
| V-12 | M | 36 | 100 | 2600 | Abnormal function findings | * | −/− | −/− | +/+ | −/− | −/− | |
| V-13 | F | 45 | 78 | 329 | Abnormal function findings | * | −/− | −/− | +/+ | −/− | −/− | |
| VII-1 | F | 33 | 100 | 2310 | Normal | Hypogonadism | 9.2 | −/− | −/− | +/+ | −/− | −/− |
| VII-2 | M | 27 | 100 | 2930 | Normal | Hypogonadism | 15 | −/− | −/− | +/+ | −/− | −/− |
| Family 2 | ||||||||||||
| II-3 | F | 45 | 100 | 4300 | Cirrhosis | Diabetes arthritis | * | −/− | +/+ | +/+ | −/− | −/− |
| Family 3 | ||||||||||||
| V-2 | M | 82 | ND | 2550 | Abnormal function findings | Arthritis | −/− | −/− | −/− | +/+ | −/− | |
| VI-3 | F | 45 | 100 | 400 | Normal | −/− | −/− | −/− | +/+ | −/− | ||
| VI-4† | F | 43 | 6 | 6 | Normal | −/− | −/− | −/− | +/+ | −/− | ||
| VI-5 | M | 39 | 100 | 938 | Fibrosis (1.99) | 4.9 | −/− | −/− | −/− | +/+ | −/− | |
| VI-7 | M | 44 | 70 | 1100 | Cirrhosis (2.85) | 7.2 | −/− | −/− | −/− | +/+ | −/− | |
| VI-8 | M | 29 | 78 | 1800 | Cirrhosis | 24 | −/− | −/− | −/− | +/+ | −/− | |
| Family 4 | ||||||||||||
| II-2‡ | M | 43 | 96 | 2350 | Cirrhosis (11.6) | Hypogonadism | 26 | −/− | −/− | −/− | −/− | +/+ |
| Cardiopathy arthritis | ||||||||||||
| . | Sex . | Age at diagnosis . | TS (%) . | SF (μg/L) . | Liver function (HII) . | Additional clinical findings . | IR (g) . | HFEmutations . | TFR2 mutations . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C282Y . | H63D . | Y250X . | E60X . | M172K . | ||||||||
| Family 1 | ||||||||||||
| V-11 | M | 47 | 100 | 3200 | Cirrhosis | Arthritis | * | −/− | −/− | +/+ | −/− | −/− |
| V-12 | M | 36 | 100 | 2600 | Abnormal function findings | * | −/− | −/− | +/+ | −/− | −/− | |
| V-13 | F | 45 | 78 | 329 | Abnormal function findings | * | −/− | −/− | +/+ | −/− | −/− | |
| VII-1 | F | 33 | 100 | 2310 | Normal | Hypogonadism | 9.2 | −/− | −/− | +/+ | −/− | −/− |
| VII-2 | M | 27 | 100 | 2930 | Normal | Hypogonadism | 15 | −/− | −/− | +/+ | −/− | −/− |
| Family 2 | ||||||||||||
| II-3 | F | 45 | 100 | 4300 | Cirrhosis | Diabetes arthritis | * | −/− | +/+ | +/+ | −/− | −/− |
| Family 3 | ||||||||||||
| V-2 | M | 82 | ND | 2550 | Abnormal function findings | Arthritis | −/− | −/− | −/− | +/+ | −/− | |
| VI-3 | F | 45 | 100 | 400 | Normal | −/− | −/− | −/− | +/+ | −/− | ||
| VI-4† | F | 43 | 6 | 6 | Normal | −/− | −/− | −/− | +/+ | −/− | ||
| VI-5 | M | 39 | 100 | 938 | Fibrosis (1.99) | 4.9 | −/− | −/− | −/− | +/+ | −/− | |
| VI-7 | M | 44 | 70 | 1100 | Cirrhosis (2.85) | 7.2 | −/− | −/− | −/− | +/+ | −/− | |
| VI-8 | M | 29 | 78 | 1800 | Cirrhosis | 24 | −/− | −/− | −/− | +/+ | −/− | |
| Family 4 | ||||||||||||
| II-2‡ | M | 43 | 96 | 2350 | Cirrhosis (11.6) | Hypogonadism | 26 | −/− | −/− | −/− | −/− | +/+ |
| Cardiopathy arthritis | ||||||||||||
TS indicates transferrin saturation; SF, serum ferritin; HII, hepatic iron index; IR, iron removed; ND, not determined.
Long-term phlebotomies (IR not determined).
Excessive bleeding through menses.
β-thalassemia trait.
Hematologic data and iron parameters of hemochromatosis type 3 carriers with different TFR2 mutations
| . | Sex . | Age . | TS (%) . | SF (μg/L) . | Hb (g/L) . | HFEmutations . | TFR2 mutations . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| C282Y . | H63D . | Y250X . | E60X . | M172K . | ||||||
| Family 1 | ||||||||||
| V-2 | M | 68 | 36 | 97 | 166 | −/− | −/− | +/− | ||
| V-93-150 | M | 70 | 74 | 310 | 159 | −/− | −/− | +/− | ||
| VI-1 | F | 59 | 27 | 79 | 142 | +/− | ||||
| 3-151 | F | 17 | 38 | 47 | 155 | +/− | ||||
| 3-151,3-152 | M | 13 | 25 | 58 | 132 | +/− | ||||
| 3-151 | M | 7 | 34 | 22 | 145 | +/− | ||||
| Family 3 | ||||||||||
| V-4 | F | 78 | ND | 32 | ND | −/− | −/− | +/− | ||
| VI-1 | F | 50 | 33 | 19 | ND | −/− | −/− | +/− | ||
| VI-2 | F | 46 | 15 | 30 | 128 | −/− | −/− | +/− | ||
| VI-6 | M | 34 | 26 | 183 | 156 | −/− | −/− | +/− | ||
| VII-3 | M | 27 | 49 | 84 | 147 | −/− | −/− | +/− | ||
| VII-4 | M | 19 | 35 | 33 | ND | −/− | −/− | +/− | ||
| Family 4 | ||||||||||
| I-23-153 | F | 72 | 39 | 180 | 112 | −/− | +/− | +/− | ||
| III-13-153 | M | 18 | 40 | 75 | 132 | −/− | −/− | +/− | ||
| III-2 | M | 16 | 30 | 44 | 140 | −/− | −/− | +/− | ||
| . | Sex . | Age . | TS (%) . | SF (μg/L) . | Hb (g/L) . | HFEmutations . | TFR2 mutations . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| C282Y . | H63D . | Y250X . | E60X . | M172K . | ||||||
| Family 1 | ||||||||||
| V-2 | M | 68 | 36 | 97 | 166 | −/− | −/− | +/− | ||
| V-93-150 | M | 70 | 74 | 310 | 159 | −/− | −/− | +/− | ||
| VI-1 | F | 59 | 27 | 79 | 142 | +/− | ||||
| 3-151 | F | 17 | 38 | 47 | 155 | +/− | ||||
| 3-151,3-152 | M | 13 | 25 | 58 | 132 | +/− | ||||
| 3-151 | M | 7 | 34 | 22 | 145 | +/− | ||||
| Family 3 | ||||||||||
| V-4 | F | 78 | ND | 32 | ND | −/− | −/− | +/− | ||
| VI-1 | F | 50 | 33 | 19 | ND | −/− | −/− | +/− | ||
| VI-2 | F | 46 | 15 | 30 | 128 | −/− | −/− | +/− | ||
| VI-6 | M | 34 | 26 | 183 | 156 | −/− | −/− | +/− | ||
| VII-3 | M | 27 | 49 | 84 | 147 | −/− | −/− | +/− | ||
| VII-4 | M | 19 | 35 | 33 | ND | −/− | −/− | +/− | ||
| Family 4 | ||||||||||
| I-23-153 | F | 72 | 39 | 180 | 112 | −/− | +/− | +/− | ||
| III-13-153 | M | 18 | 40 | 75 | 132 | −/− | −/− | +/− | ||
| III-2 | M | 16 | 30 | 44 | 140 | −/− | −/− | +/− | ||
For abbreviations, see Table 2.
HCV chronic hepatis.
Children of patients VII-1 and VII-2.
δβ-thalassemia.
β-thalassemia trait.
Patient II-2 of family 4 had a severe disease. HFE2 was excluded by studying the intrafamily segregation of marker alleles of chromosome 1q because the patient was haploidentical to a healthy sister (not shown). Consanguinity was not reported in this family. However, a homozygous T→A transversion was identified in exon 4 at position 515 ofTFR2 cDNA (Figure 2B). This nucleotide change resulted in a lysine for methionine substitution at position 172 of the protein (M172K). T515A segregation within the family was studied by direct sequencing. I-2, III-1, and III-2 were heterozygous for the mutation, and II-1 had the normal genotype. The same mutation was not found by RNA-SSCP among 50 healthy controls.
RT-PCR was performed on total RNA obtained from peripheral blood buffy coats and lymphoblastoid cell lines (LCL) of patients with differentTFR2 substitutions and in a healthy control. As expected on the bases of the α-transcript restricted pattern of expression, attempts at amplifying the α-transcripts by RT-PCR using RNA from buffy coats or LCL were unsuccessful. Fragments corresponding to the β-transcript were obtained in the healthy control and in all patients, except in Y250X homozygotes (not shown).
Clinical findings in HFE3 patients
Clinical data from all HFE3 patients with a defined mutation inTFR2 are summarized in Table 2. A partial description of the clinical phenotype in index cases was previously reported.18,22,23 No patient was C282Y homozygous or heterozygous. Patient II-3 of family 2 was H63D homozygous. In family 3, a rather variable phenotype was observed. The 2 probands (VI-7 and VI-8) had a typical disorder, with a more severe phenotype in the younger patient.20 VI-5 had modest liver fibrosis, a remarkable degree of steatosis, and increased liver iron staining. The hepatic iron index was borderline (1.99), and approximately 5 g iron had to be removed to attain iron depletion. VI-3, a 45-year-old woman, showed only increased transferrin saturation and serum ferritin. VI-4 and V-2 were undiagnosed, and their identification as E60X homozygotes was obtained through family studies (Table 2). VI-4, a premenopausal woman, had remarkably low transferrin saturation and serum ferritin. The finding of iron deficiency without anemia was unexpected in a homozygous mutant subject. Low intake of dietary iron and long-lasting blood loss through menses without iron supplementation were identified as the causes of iron deficiency. No history of other blood loss or evidence of a hemostasis defect was recorded, but the patient refused a thorough gastrointestinal endoscopic investigation. V-2 had altered iron parameters, abnormal liver function test findings, and severe arthritis but never had phlebotomies. His older brother (V-1) was not available for the study but was reported to be under regular phlebotomy treatment.
II-2 of family 4 had a severe disease (cirrhosis, hypogonadism, and cardiac disease). As shown in Figure 1B the patient had inherited β-thalassemia at the heterozygous state from his mother.
Clinical findings in HFE3 carriers
HFE-3 carriers were parents or children of the patients or siblings with a documented TFR2 mutation. Clinical data and iron parameters of 15 HFE3 heterozygotes are shown in Table 3. All carriers had normal transferrin saturation and serum ferritin levels, except V-9 of family 1. This subject, who showed increased transferrin saturation and serum ferritin, was affected by HCV chronic hepatitis and underwent occasional phlebotomy. In all the other subjects, the condition of HFE3 heterozygosity, even in combination with H63D at the heterozygous state (1 subject) or with β-thalassemia trait (3 subjects), was not associated with iron overload.
Discussion
In this paper we describe 2 new mutations of TFR2 in patients with hemochromatosis with wild-type HFE. This finding confirms the role of TFR2 as the HFE3 gene. Scanning for mutations, the coding sequence of TFR2, a C insertion (84-88 insC) that causes a premature stop codon (E60X) in exon 2, was found at the homozygous state in 5 affected subjects and in 1 nonexpressing, iron-deficient woman from an inbred family. A T515A change in exon 4 that causes a missense (M172K) in the protein was found at the homozygous state in a single nonfamilial case. The presence of HFE3 homozygotes in 2 consecutive generations in family 3 witnesses the high degree of inbreeding. Homozygote-heterozygote mating (between V-2 and V-4) explains the absence of wild-type TFR2in the next-to-last generation and the seemingly dominant inheritance in this branch of the family.
The identification of 13 subjects with homozygous mutations inTFR2 allows a tentative genotype-phenotype correlation. All classic complications of hemochromatosis were observed among patients with HFE3, though the phenotype severity was variable, especially in family 3. As in HFE-associated disease, the intrafamilial variability of the clinical phenotype might be ascribed to environmental factors and modifier genes. Surprisingly VI-4 of family 3 had iron deficiency without anemia that was related to heavy menstrual losses and low dietary iron intake. A consistent proportion of C282Y-homozygous women are reported not to express the disorder during the fertile age,27 28 though iron deficiency is extremely rare in the literature and in our experience.
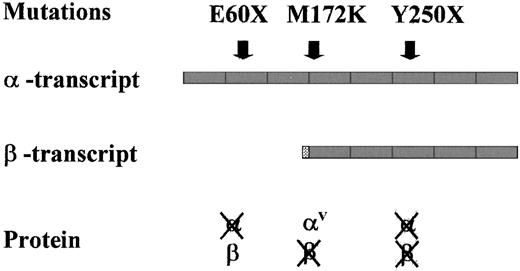
We hypothesize that the different effects of the mutations on the encoded protein may influence the phenotype severity. In theory, as shown in Figure 3, the effect of the reported mutations on TFR2 transcripts is different. E60X occurs in exon 2 and disrupts the predicted α-TFR2transcript but, at variance with Y250X, does not interfere with the β-transcript. T515A causes a missense in the α-protein (M172K). The methionine at position 172 is conserved in the mouse29 and the substitution of a basic for a neutral amino acid might change the α-protein properties. However, the same nucleotide substitution also affects the putative initiation codon of the β-variant, preventing its translation. We speculate that the absence of β-TFR2might be associated with a severe phenotype in Y250X and M172K homozygotes, whereas its persistence might be responsible for a partial function of the protein in E60X homozygotes. However, to establish whether β-TFR2 has any physiological significance requires further investigation. At present, antibodies against TFR2, which could help to confirm our hypothesis, are not available. Preliminary results of RT-PCR on RNA from peripheral blood or LCL showed the absence of β-TFR2 only in Y250X homozygotes, a result that is in keeping with a rapid degradation of a nonsense transcript.30 Such a degradation is not expected in M172K, which affects the β-TFR2 initiation codon, or in E60X, which does not affect the β-transcript.
Schematic representation of
TFR2 structure in the 5′ region of the gene. Two transcripts, possibly resulting from the alternative splicing according to Kawabata et al,19 are shown. β-Transcript lacks exons 1 to 3 and has additional nucleotides at the 5′ end (dotted box) compared to the α-transcript. Position of the mutations is identified, and their possible effects on the proteins are shown. αV, variant with the amino acid change M172K.
Schematic representation of
TFR2 structure in the 5′ region of the gene. Two transcripts, possibly resulting from the alternative splicing according to Kawabata et al,19 are shown. β-Transcript lacks exons 1 to 3 and has additional nucleotides at the 5′ end (dotted box) compared to the α-transcript. Position of the mutations is identified, and their possible effects on the proteins are shown. αV, variant with the amino acid change M172K.
A contribution of the β-thalassemia trait to the phenotype severity cannot be ruled out in II-2 of family 4 because we have observed that the co-inheritance of β-thalassemia trait aggravates the phenotype expression in C282Y HFE homozygotes.23 Mutations ofTFR2 at the heterozygous state were usually well tolerated. As observed in C282Y HFE heterozygous mutants,23 persons with heterozygous mutations of TFR2 in association with the β-thalassemia trait do not develop iron overload (Table 3).
Our results may have practical implications for the molecular diagnosis of hemochromatosis. Genotyping the HFE gene is included in the disease diagnostic protocols. However, in all the reported series, a minority of patients have wild-type or incomplete HFE genotypes (C282Y at the heterozygous or H63D at the heterozygous or homozygous state).5-10,31,32 They are considered to be affected by atypical forms of hemochromatosis or by secondary iron overload.32 Our data show that a different non-HFE determinant may be present in these patients, as exemplified by patient II-3 of family 2. Therefore, screening for mutations of TFR2is a new diagnostic tool that can be offered to patients who do not have HFE mutations or who have incomplete HFE genotypes.
Finally, the identification of TFR2 as the HFE3 gene is of relevance regarding modifier genes in hemochromatosis. The presence of modifier genes that may modulate (either ameliorate or worsen) the phenotype has been demonstrated in mice and hypothesized in humans.33TFR2 is the first obvious modifier to be investigated in C282Y homozygotes.
Supported in part by Telethon, E.U. (contract QLK6-1999-02237), the Italian Ministry of University and Technologic Research, the Italian Ministry of Health, CNR-PF Biotecnologie, and IRCC Pavia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Clara Camaschella, Dipartimento di Scienze Cliniche e Biologiche, Università di Torino, Azienda Ospedaliera San Luigi, 10043-Orbassano, Turin, Italy; e-mail: camaschella@ope.net.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal