Abstract
PU.1 is an Ets family transcription factor essential for myelomonocyte and B-cell development. We previously showed that overexpression of PU.1 in murine erythroleukemia (MEL) cells inhibits growth and erythroid differentiation and induces apoptosis of the cells. In an effort to identify target genes of PU.1 concerning these phenomena by using a messenger RNA differential display strategy, we found that some myeloid-specific and lymphoid-specific genes, such as the osteopontin gene, are transcriptionally up-regulated in MEL cells after overexpression of PU.1. We then found that expression of several myelomonocyte-specific genes, including the CAAT-enhancer-binding protein-α and granulocyte-macrophage colony-stimulating factor receptor genes, was induced in MEL cells after overexpression of PU.1. B-cell–specific genes were also examined, and expression of the CD19 gene was found to be induced. Expression of the myelomonocyte-specific proteins CD11b and F4/80 antigen but not the B-cell–specific proteins B220 and CD19 was also induced. After overexpression of PU.1, MEL cells became adherent and phagocytic and showed enhanced nitroblue tetrazolium reduction activity. Expression of myelomonocyte-specific and B-cell–specific genes was not induced when a mutant PU.1 with part of the activation domain deleted (a change found to inhibit erythroid differentiation of MEL cells) was expressed. These results indicate that PU.1 induces a lineage switch in MEL cells toward myelomonocytic cells and that its activation domain is essential for this effect. The results also suggest that the pathway of the lineage switch is distinct from that of inhibition of erythroid differentiation in MEL cells.
Introduction
Hematopoiesis is the process by which progenitor cells acquire characteristics of certain types of hematopoietic cells. It is believed that this process is mediated by hematopoietic cell–specific transcription factors that confer cell specificity by means of regulation of the expression of cell-type–specific genes. Targeted disruption of such transcription-factor genes results in impaired hematopoiesis or defects in the development of particular types of hematopoietic cells.1-6
PU.1, a member of the Ets family of transcription factors, is expressed in hematopoietic cells, predominantly in myelomonocytes and B cells. PU.1 regulates cell-type–specific expression of genes such as the CD18 and granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) genes in myelomonocytes and the immunoglobulin (Ig) heavy- and light-chain genes in B cells. Targeted disruption of thePU.1 gene was found to cause severe defects in the development process in both myeloid and lymphoid cells, characterized by the absence of macrophages and B cells and the delayed appearance of neutrophils.7 8 This finding indicates that PU.1 plays a fundamental role in myelomonocyte and B-cell development.
In Friend virus–induced murine erythroleukemia (MEL), expression of the PU.1 gene is deregulated by proviral integration of spleen focus-forming virus.9 10 This deregulated expression of PU.1 in erythroblasts is thought to be one of the causes of leukemogenesis.
In a previous study, which we conducted to elucidate the functional role of PU.1 in the regulation of growth and differentiation of MEL cells, we introduced an expression plasmid construct of thePU.1 gene into MEL cells. We found that when PU.1 was overexpressed in the cells in the presence of the differentiation-inducing reagent dimethyl sulfoxide (DMSO), growth inhibition and apoptosis were induced but erythroid differentiation was not.11 These results suggest that although PU.1 contributes to the generation of erythroleukemia by inhibiting differentiation of erythroid cells, it induces growth suppression and cell death under special circumstances. Subsequently, we showed that expression of the c-myc and bcl-2 genes is down-regulated in apoptosis12 and DNA-binding activity of GATA-1 transcription factor, which is important for the survival and differentiation of erythroid cells,13,14 is markedly reduced.15 However, the exact mechanism (or mechanisms) by which PU.1 inhibits growth and erythroid differentiation and induces apoptosis has not been determined. Because PU.1 is a transcription factor, it is logical to assume that the cellular effects induced by its overexpression are mediated by means of regulation of expression of its target genes. Therefore, using a messenger RNA (mRNA) differential display strategy, we attempted to isolate and identify the target genes of PU.1 that are transcriptionally up-regulated or down-regulated during induction of the effects of PU.1 in MEL cells.
We observed transcriptional up-regulation of some myeloid-specific and lymphoid-specific genes in addition to up-regulation or down-regulation of genes suggested to be involved in growth inhibition or apoptosis. This finding indicated the possibility that PU.1 not only inhibits erythroid differentiation of MEL cells but also induces expression of myelomonocyte-specific and B-cell–specific genes in the cells. In the current study, we addressed this possibility and found that PU.1 induces expression of several myelomonocyte-specific and B-cell–specific genes as well as morphologic and functional changes in MEL cells.
Materials and methods
Cell culture
Three clones of MEL cells transfected with a zinc-inducible expression plasmid of the wild-type PU.1 gene (PU.1-1, PU.1-2, and PU.1-3 cells) and 2 clones of MEL cells transfected with a zinc-inducible expression plasmid of the mutant PU.1 gene encoding the PU.1 protein with a deletion of the glutamine-rich region of the activation domain (PU.1-ΔA-1 and PU.1-ΔA-2 cells) were used. A clone of MEL cells transfected with a blank plasmid (mock cells) was used as a control. Transfectants were established after electroporation of a plasmid into Friend virus–induced MEL-B8/3 cells as described previously.11 Transfectants and parental MEL-B8/3 cells were maintained in RPMI 1640 medium supplemented with 10% fetal-bovine serum. Differentiation induction of the cells was done by adding 1.5% DMSO to the culture medium. Expression of the exogenousPU.1 genes was induced by adding 100 μM zinc chloride (ZnCl2) to the culture medium.
mRNA differential display
Total RNA samples were extracted from PU.1-1 cells cultured with 1.5% DMSO and 100 μM ZnCl2 for 0, 24, 48, and 72 hours. The reverse transcriptase (RT) reaction of the samples and the following polymerase chain reaction (PCR) amplification were done with the RNAimage mRNA differential display system (Genhunter, Nashville, TN) according to the manufacturer's instructions. For the PCR, 3 kinds of oligo(dT) primers and 80 kinds of arbitrary primers were used. PCR products were resolved in a 6% polyacrylamide DNA sequencing gel. Differentially expressed bands were excised, after which the PCR products were eluted from the gel slices and again amplified by PCR. The final PCR products were subcloned into a pT7 blue T-vector (Novagen, Madison, WI) by using the AT-cloning strategy. The DNA sequence of the PCR products were determined by using a genetic analyzer (310; PE Biosystems, Foster City, CA).
Northern blot analysis
Twenty micrograms of total RNA samples was denatured with formamide and separated in an 1% agarose gel containing formaldehyde. The RNA was transferred to a nylon membrane and then hybridized with DNA probes labeled with phosphorus 32–deoxycytidine triphosphate. The hybridization process was done as described previously.16The complementary DNA (cDNA) fragments used as probes for the osteopontin gene, the eosinophil cationic protein gene, and the B144 gene were obtained by using mRNA differential display. The cDNA fragments used as probes for the CAAT-enhancer-binding protein–α (C/EBP-α) and C/EBP-ε genes were amplified by RT-PCR from HL-60 human myelomonocytic leukemia cells by using the primers shown in Table1. A BamHI fragment of mouse β-actin cDNA was used as the internal control.
Primers used in the reverse transcriptase–polymerase chain reaction analysis
| Gene . | 5′ Primer . | 3′ Primer . |
|---|---|---|
| Human C/EBP-α | 5′-GAATCTCCTAGTCCTGGCTC | 5′-GATGAGAACAGCAACGAGTAC |
| Mouse C/EBP-γ | 5′-CAGAGAGCGGAACAATATGG | 5′-TCAGCTGTTCAGTGTTGAGC |
| Mouse C/EBP-δ | 5′-AGAACGAGAAGCTGCATCAG | 5′-GTCTGAGGTATAGGTCGTTC |
| Human C/EBP-ε | 5′-CAGACAGGAAGGCGCTGGG | 5′-CGGCAGTGGCCAAAGGGGCCTT |
| Mouse GM-CSFR | 5′-CTGATGTCATGAAGCGATGC | 5′-TCACGGTGACATCAATGTCG |
| Mouse G-CSFR | 5′-AAACCTTCCAGTTACCCAGC | 5′-TGGAGTCAGAGCGAATGTAC |
| Mouse M-CSFR | 5′-AACATATGGACTTCGCCCTC | 5′-ACCTTCCAGTCTAGAGTGAC |
| Mouse MPO | 5′-GTCCAGATCATCACATACCG | 5′-CCTATCCTCATGACTTGCTC |
| Mouse CD18 | 5′-TTCAGTGTGCTGGTATGACG | 5′-ACACCAATCAGTACGACACC |
| Mouse neutrophil elastase | 5′-CATGCTACTGGCATTGTTCC | 5′-TCGTTCAGCAGTTGTGATGG |
| Mouse HLS7 | 5′-TGGCTGTTGGTCATCACATC | 5′-TCTTCCACTCTCAATGGCTG |
| Mouse MLL (ALL-1) | 5′-ATTCCTCTTTCAGCTCAGCC | 5′-GGTTACACAGTTGTCACTGC |
| Mouse CD19 | 5′-AATCTGGAGATCCCAGATGG | 5′-TAGCGGCTGGACATATTTGC |
| Mouse Pax-5 | 5′-AGCATAGTGTCTACAGGCTC | 5′-TGGCTTTCATGTCATCCAGG |
| Mouse Igκ chain | 5′-CAAAATGGCGTCCTGAACAG | 5′-TTGATGTCTTGTGAGTGGCC |
| Mouse Igμ chain | 5′-CTGGTCTCAAACCTGGCAAC | 5′-CTGTCACAGTCAGGATGCTG |
| Mouse β-actin | 5′-CACCCTGTGCTGCTCACCGAGGCC | 5′-CCACACAGATGACTTGCGCTCAGG |
| Gene . | 5′ Primer . | 3′ Primer . |
|---|---|---|
| Human C/EBP-α | 5′-GAATCTCCTAGTCCTGGCTC | 5′-GATGAGAACAGCAACGAGTAC |
| Mouse C/EBP-γ | 5′-CAGAGAGCGGAACAATATGG | 5′-TCAGCTGTTCAGTGTTGAGC |
| Mouse C/EBP-δ | 5′-AGAACGAGAAGCTGCATCAG | 5′-GTCTGAGGTATAGGTCGTTC |
| Human C/EBP-ε | 5′-CAGACAGGAAGGCGCTGGG | 5′-CGGCAGTGGCCAAAGGGGCCTT |
| Mouse GM-CSFR | 5′-CTGATGTCATGAAGCGATGC | 5′-TCACGGTGACATCAATGTCG |
| Mouse G-CSFR | 5′-AAACCTTCCAGTTACCCAGC | 5′-TGGAGTCAGAGCGAATGTAC |
| Mouse M-CSFR | 5′-AACATATGGACTTCGCCCTC | 5′-ACCTTCCAGTCTAGAGTGAC |
| Mouse MPO | 5′-GTCCAGATCATCACATACCG | 5′-CCTATCCTCATGACTTGCTC |
| Mouse CD18 | 5′-TTCAGTGTGCTGGTATGACG | 5′-ACACCAATCAGTACGACACC |
| Mouse neutrophil elastase | 5′-CATGCTACTGGCATTGTTCC | 5′-TCGTTCAGCAGTTGTGATGG |
| Mouse HLS7 | 5′-TGGCTGTTGGTCATCACATC | 5′-TCTTCCACTCTCAATGGCTG |
| Mouse MLL (ALL-1) | 5′-ATTCCTCTTTCAGCTCAGCC | 5′-GGTTACACAGTTGTCACTGC |
| Mouse CD19 | 5′-AATCTGGAGATCCCAGATGG | 5′-TAGCGGCTGGACATATTTGC |
| Mouse Pax-5 | 5′-AGCATAGTGTCTACAGGCTC | 5′-TGGCTTTCATGTCATCCAGG |
| Mouse Igκ chain | 5′-CAAAATGGCGTCCTGAACAG | 5′-TTGATGTCTTGTGAGTGGCC |
| Mouse Igμ chain | 5′-CTGGTCTCAAACCTGGCAAC | 5′-CTGTCACAGTCAGGATGCTG |
| Mouse β-actin | 5′-CACCCTGTGCTGCTCACCGAGGCC | 5′-CCACACAGATGACTTGCGCTCAGG |
C/EBP indicates CAAT-enhancer-binding protein; GM-CSFR, granulocyte-macrophage colony-stimulating factor receptor; G-CSFR, granulocyte colony-stimulating factor receptor; M-CSFR, macrophage colony-stimulating factor receptor; MPO, myeloperoxidase; HLS7, hematopoietic lineage switch 7; MLL, mixed lineage leukemia; ALL, acute lymphoblastic leukemia; Ig, immunoglobulin.
RT-PCR analysis
Preparation of first-strand cDNA and PCR amplification were done as described previously.12 PCR amplification consisted of 1 minute at 94°C, 2 minutes at 55°C, and 3 minutes at 72°C for 25 to 35 cycles. PCR products were resolved in a 1% agarose gel containing ethidium bromide. The intensity of the bands was quantified by using the National Institutes of Health Image computer program. The primers used are listed in Table 1.
Western blot analysis
Fifty micrograms of total protein samples was subjected to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The membrane was then probed with an antimouse PU.1 antibody (T-21; Santa Cruz Biotechnology, Santa Cruz, CA) as described previously.11 Bound antibody was detected by using an electrogenerated chemiluminescence system (Amersham, United Kingdom).
Flow cytometric analysis
Flow cytometric analysis was done with a fluorescence-activated cell-sorter scanner (Becton Dickinson, Mountain View, CA), and data were processed with the Cell Quest program (Becton Dickinson). Cells (5 × 105) were incubated first with 2.4G2 monoclonal antibody (mAb) (Pharmingen, San Diego, CA) to block the Fc receptor and were then incubated with mAbs. Dead cells were stained with 0.05 μM YOYO-3 (Molecular Probes, Eugene, OR). The mAbs used were phycoerythrin-conjugated rat antimouse CD11b, biotin-conjugated rat antimouse F4/80 and B220, and fluorescein isothiocyanate–conjugated rat antimouse CD19 (Pharmingen).
Immunohistochemical analysis
Cells cytocentrifuged onto a slide were fixed in acetone and blocked with normal rabbit serum. The cells were incubated with rat antimouse CD3, CD4, CD5, CD8, B220, CD19, CD11b, Ter119 (Pharmingen), or F4/80 (Caltag, Burlingame, CA) mAb and then with a biotin-conjugated rabbit antirat Ig antibody (Dako, Denmark). Subsequently, the cells were incubated with streptavidin–horseradish peroxidase conjugate. Bound peroxidase was detected by using diaminobenzidine as a substrate.
Nitroblue tetrazolium reduction assay
Cells were incubated with 1 mg/mL nitroblue tetrazolium (NBT) and 100 ng/mL 12-O-tetradecanoyl phorbol 13-acetate for 1 hour at 37°C. After incubation, the proportion of formazan-positive cells was determined by examining the cells under a microscope.
Measurement of phagocytic activity
Cells were incubated with latex beads (diameter, 1.09 μm) for 2.5 hours at 37°C. After 4 washes with phosphate-buffered saline, the proportion of cells taking up the beads was determined by examining the cells under a microscope.
Results
Identification of genes in MEL cells whose expression changed with PU.1-mediated growth inhibition, differentiation inhibition, and apoptosis
Using an mRNA differential display strategy, we isolated 251 bands whose intensity was changed in one clone of MEL-B8/3 cells transfected with a zinc-inducible expression plasmid of the wild-typePU.1 gene (PU.1-1 cells) after overexpression of PU.1. Sequencing and a search of the Basic Local Alignment Search Tool database (National Center for Biotechnology Information) revealed that 100 of the 251 PCR products originated from known genes. Of these 100 genes, expression of 37 was up-regulated and that of 63 was down-regulated.
Representative genes are listed in Table2. The transcriptionally up-regulated genes were, for instance, the CDC10 gene involved in cytokinesis17 and hematopoietic cell–specific genes that are not normally expressed in the erythroid cell lineage. The transcriptionally down-regulated genes included, for instance, the adenosine triphosphate synthase α-subunit gene encoded by mitochondrial DNA,18 some ribosomal protein genes, and the nucleolin gene involved in cell proliferation and ribosomal RNA processing.19 20 Some histone genes and neural cell–specific genes were also included in this category.
Representative genes identified in the messenger RNA differential display analysis
| Up-regulated genes . | Down-regulated genes . |
|---|---|
| Cytokinesis-related genes | Mitochondria-related genes |
| CDC10 | ATP synthase α subunit |
| Hematopoietic cell-specific genes | Cytochrome C oxidase 6B |
| Osteopontin | Lon protease-like protein |
| Eosinophil cationic protein | Ribosome-related genes |
| B144 | Nucleolin |
| Other Breast heat shock protein 73 | v-fos Transformation effector protein/ribosomal protein S3a |
| Histone genes | |
| Histone H2A F/Z variant | |
| Histone H3.3 | |
| Cell cycle-related and cell growth-related genes | |
| Ran/TC4 binding protein | |
| Gas5 | |
| Inosine-5′-monophosphate dehydrogenase | |
| Neural cell-specific genes | |
| Monocarboxylate transporter | |
| p205 | |
| NEDD-6 |
| Up-regulated genes . | Down-regulated genes . |
|---|---|
| Cytokinesis-related genes | Mitochondria-related genes |
| CDC10 | ATP synthase α subunit |
| Hematopoietic cell-specific genes | Cytochrome C oxidase 6B |
| Osteopontin | Lon protease-like protein |
| Eosinophil cationic protein | Ribosome-related genes |
| B144 | Nucleolin |
| Other Breast heat shock protein 73 | v-fos Transformation effector protein/ribosomal protein S3a |
| Histone genes | |
| Histone H2A F/Z variant | |
| Histone H3.3 | |
| Cell cycle-related and cell growth-related genes | |
| Ran/TC4 binding protein | |
| Gas5 | |
| Inosine-5′-monophosphate dehydrogenase | |
| Neural cell-specific genes | |
| Monocarboxylate transporter | |
| p205 | |
| NEDD-6 |
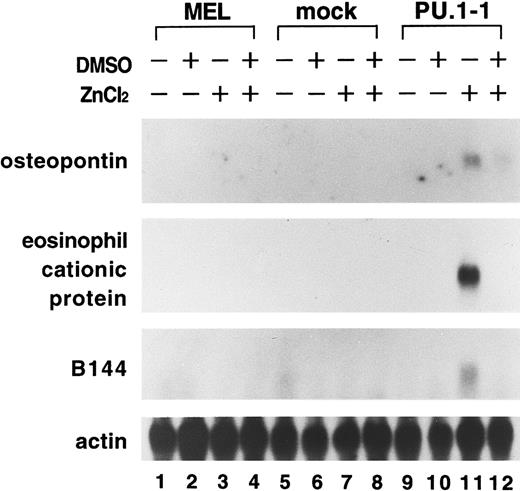
Of particular interest was the up-regulation of expression of hematopoietic cell–specific genes, including the osteopontin gene, encoding for an extracellular adhesion protein thought to be involved in monocyte maturation21,22; the eosinophil cationic protein gene, encoding for a toxic protein in the granules of eosinophils23; and the B144 gene, which is located in the major histocompatibility complex class III region and abundantly expressed in hematopoietic cells, macrophages, and B cells.24,25 Northern blot analysis confirmed that expression of these genes was induced in PU.1-1 cells treated with ZnCl2 for 48 hours (Figure 1, lane 11). In cells treated with both DMSO and ZnCl2, levels of expression of these genes were lower than in cells treated with ZnCl2 alone (Figure 1, lanes 11 and 12). This may have been because of the reduction in cell viability resulting from apoptosis induced under these conditions. Alternatively, it may be possible that stimulation of DMSO, which triggers erythroid differentiation of MEL cells, affected expression of these genes, although we previously showed that erythroid differentiation of MEL cells induced by DMSO is inhibited by overexpression of PU.1.11 These results suggested the interesting possibility that PU.1 could induce expression of genes specific for myeloid and lymphoid lineages in erythroid cells. We therefore focused our study on assessing this possibility.
Induction of expression of the osteopontin, eosinophil cationic protein, and B144 genes in MEL cells after overexpression of PU.1.
Total RNA was extracted from parental MEL-B8/3 cells (lanes 1-4), mock cells (lanes 5-8), and PU.1-1 cells (lanes 9-12) cultured for 48 hours without reagents (lanes 1, 5, and 9), with DMSO (lanes 2, 6, and 10), with ZnCl2 (lanes 3, 7, and 11), or with DMSO and ZnCl2 (lanes 4, 8, and 12). Expression of the osteopontin, eosinophil cationic protein, and B144 genes was examined by Northern blot analysis.
Induction of expression of the osteopontin, eosinophil cationic protein, and B144 genes in MEL cells after overexpression of PU.1.
Total RNA was extracted from parental MEL-B8/3 cells (lanes 1-4), mock cells (lanes 5-8), and PU.1-1 cells (lanes 9-12) cultured for 48 hours without reagents (lanes 1, 5, and 9), with DMSO (lanes 2, 6, and 10), with ZnCl2 (lanes 3, 7, and 11), or with DMSO and ZnCl2 (lanes 4, 8, and 12). Expression of the osteopontin, eosinophil cationic protein, and B144 genes was examined by Northern blot analysis.
Induction of myelomonocyte-specific and B-cell–specific gene expression in MEL cells by overexpression of PU.1
Expression of several genes specific for myelomonocyte and B-cell lineages was examined by Northern blot and RT-PCR analyses in PU.1-1 cells treated with and without ZnCl2. As shown in Figure2A, expression of the genes encoding for C/EBP-α, C/EBP-ε, and C/EBP-δ transcription factors, which play important roles in the development and maturation of myelomonocytes, was up-regulated in the cells cultured for 48 hours with ZnCl2 (lane 11). Similarly, levels of expression of the genes encoding for receptors of myelomonocyte-specific growth factors (the GM-CSFR, granulocyte colony-stimulating factor receptor [G-CSFR], and macrophage colony-stimulating factor receptor [M-CSFR] genes), the myeloperoxidase (MPO) gene, and the CD18 gene encoding for a β chain of β2 family of integrins were higher in PU.1-1 cells treated with ZnCl2 than in those of parental MEL and mock cells treated with ZnCl2 (Figure 2A, lanes 3, 7, and 11).
Induction of expression of myelomonocyte-specific and B-cell–-specific genes in MEL cells after overexpression of PU.1.
(A) Total RNA was extracted from parental MEL-B8/3 cells (lanes 1-4), mock cells (lanes 5-8), and PU.1-1 cells (lanes 9-12) cultured for 48 hours without reagents (lanes 1, 5, and 9), with DMSO (lanes 2, 6, and 10), with ZnCl2 (lanes 3, 7, and 11), or with DMSO and ZnCl2 (lanes 4, 8, and 12). Expression of myelomonocyte-specific and B-cell–specific genes was examined by Northern blot (the C/EBP genes) or RT-PCR analysis. (B) The intensity of the bands obtained in the RT-PCR analysis was quantified, and ratios of the values for parental MEL-B8/3 (■), mock (░), and PU.1-1 (▨) cells treated with ZnCl2 are shown in the graphs. The calculated ratio of the value for PU.1 cells to the average value for parental and mock cells is shown in each graph. (C) Total RNA was extracted from PU.1-2 cells (lanes 1 and 2) and PU.1-3 cells (lanes 3 and 4) cultured for 48 hours without reagents (lanes 1 and 3) or with ZnCl2 (lanes 2 and 4). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. (D) Total protein was extracted from PU.1-1 cells (lanes 1 and 2), PU.1-2 cells (lanes 3 and 4), and PU.1-3 cells (lanes 5 and 6) cultured for 8 hours without reagents (lanes 1, 3, and 5) or with ZnCl2 (lanes 2, 4, and 6). Expression of PU.1 protein was examined by Western blot analysis.
Induction of expression of myelomonocyte-specific and B-cell–-specific genes in MEL cells after overexpression of PU.1.
(A) Total RNA was extracted from parental MEL-B8/3 cells (lanes 1-4), mock cells (lanes 5-8), and PU.1-1 cells (lanes 9-12) cultured for 48 hours without reagents (lanes 1, 5, and 9), with DMSO (lanes 2, 6, and 10), with ZnCl2 (lanes 3, 7, and 11), or with DMSO and ZnCl2 (lanes 4, 8, and 12). Expression of myelomonocyte-specific and B-cell–specific genes was examined by Northern blot (the C/EBP genes) or RT-PCR analysis. (B) The intensity of the bands obtained in the RT-PCR analysis was quantified, and ratios of the values for parental MEL-B8/3 (■), mock (░), and PU.1-1 (▨) cells treated with ZnCl2 are shown in the graphs. The calculated ratio of the value for PU.1 cells to the average value for parental and mock cells is shown in each graph. (C) Total RNA was extracted from PU.1-2 cells (lanes 1 and 2) and PU.1-3 cells (lanes 3 and 4) cultured for 48 hours without reagents (lanes 1 and 3) or with ZnCl2 (lanes 2 and 4). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. (D) Total protein was extracted from PU.1-1 cells (lanes 1 and 2), PU.1-2 cells (lanes 3 and 4), and PU.1-3 cells (lanes 5 and 6) cultured for 8 hours without reagents (lanes 1, 3, and 5) or with ZnCl2 (lanes 2, 4, and 6). Expression of PU.1 protein was examined by Western blot analysis.
The intensity of the bands obtained in the RT-PCR analysis was quantified, and the ratio of the value for PU.1-1 cells treated with ZnCl2 to the average value for parental and mock cells treated with ZnCl2 was calculated for each gene. According to the calculation, expression of the myelomonocyte-specific genes was up-regulated 1.7- to 22.4-fold in PU.1-1 cells compared with parental and mock cells (Figure 2B).
The degree of expression of some genes was weaker in the cells cultured with DMSO and ZnCl2 (Figure 2A, lane 12) than in the cells cultured with ZnCl2 alone (Figure 2A, lane 11), possibly because of the reasons described above. The presence of detectable levels of expression of some genes in the parental and mock cells, as well as in the PU.1-1 cells cultured without ZnCl2, may have been due to the function of steady-state levels of PU.1 originally expressed in MEL cells. Changes in levels of expression of some genes in the parental and mock cells cultured in the presence of DMSO may have reflected erythroid differentiation of these cells.
On the other hand, expression of the neutrophil elastase gene was not detected in the RT-PCR analysis (data not shown). Expression of the hematopoietic lineage switch 7 (HLS7) gene, which has been shown to be involved in cell-lineage conversion from erythroid to myeloid cells,26,27 and the mixed lineage leukemia (MLL, also known as acute lymphoblastic leukemia 1) gene, which is thought to be involved in monocyte differentiation,28 was not induced in PU.1-1 cells treated with ZnCl2 (lane 11).
Expression of the gene encoding for the B-cell marker CD19 was up-regulated in PU.1-1 cells cultured with ZnCl2 (Figure2A, lane 11, and Figure 2B). Basal levels of expression of the C/EBP-γ gene, which is highly expressed in early B cells,29 were observed in all cells in the RT-PCR analysis, but expression of the gene was not up-regulated after overexpression of PU.1 (data not shown). Expression of the Pax-5 gene, encoding for a B-cell–specific transcription factor, and the Igκ-chain and Igμ-chain genes was not detected in the RT-PCR analysis in any cells cultured under any conditions (data not shown).
Induction of expression of myelomonocyte-specific and B-cell–specific genes is not peculiar to PU.1-1 cells because similar results were obtained in 2 additional independent clones of the transfectants, PU.1-2 and PU.1-3 cells, after overexpression of PU.1 (Figure 2C). Western blot analysis showed that the degree of induction of expression of the PU.1 protein was comparable in these clones after treatment with ZnCl2 (Figure 2D). Taken together, these results indicate that expression of a considerable number of myelomonocyte-specific and B-cell–specific genes is up-regulated in MEL cells after overexpression of PU.1.
Induction of expression of myelomonocyte-specific proteins in MEL cells by overexpression of PU.1
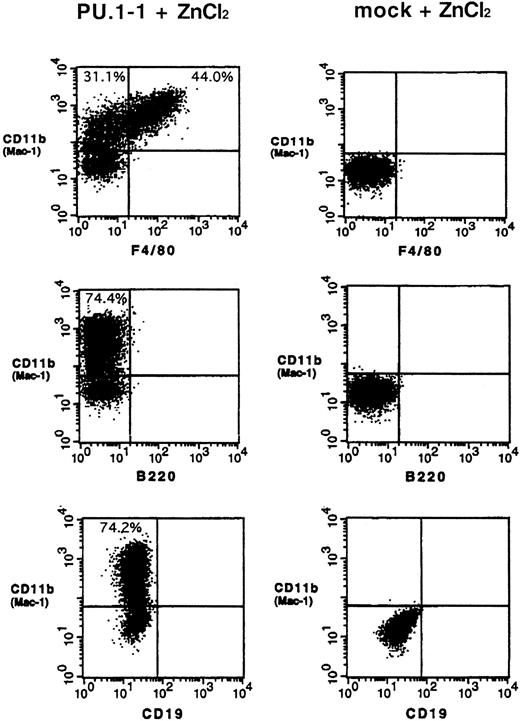
The results described above prompted us to examine whether myelomonocyte-specific and B-cell–specific proteins are expressed in MEL cells after overexpression of PU.1. Flow cytometric analysis found that 70% to 80% of PU.1-1 cells treated with ZnCl2expressed myelomonocyte-specific protein CD11b (Mac-1) on their surface. Moreover, 40% to 50% of CD11b-positive cells were also positive for another myelomonocyte-specific protein, F4/80 antigen (Figure 3, left). Similar results were obtained in studies using PU.1-2 cells; that is, about 50% of cells were positive for CD11b and 30% of CD11b-positive cells were also positive for F4/80 antigen (data not shown). These proteins were not detected on the surface of PU.1-1 and PU.1-2 cells cultured without ZnCl2 (data not shown) or mock cells cultured with or without ZnCl2 (Figure 3, right, and data not shown).
Induction of expression of the myelomonocyte-specific proteins CD11b (Mac-1) and F4/80 antigen on the surface of MEL cells after overexpression of PU.1.
PU.1-1 cells (left lane) and mock cells (right lane) were cultured for 48 hours with ZnCl2 and subjected to flow cytometric analysis. Results of a representative experiment are shown. Similar results were obtained in a duplicate experiment (data not shown).
Induction of expression of the myelomonocyte-specific proteins CD11b (Mac-1) and F4/80 antigen on the surface of MEL cells after overexpression of PU.1.
PU.1-1 cells (left lane) and mock cells (right lane) were cultured for 48 hours with ZnCl2 and subjected to flow cytometric analysis. Results of a representative experiment are shown. Similar results were obtained in a duplicate experiment (data not shown).
In contrast to the results regarding myelomonocyte-specific proteins, expression of the B-cell–specific proteins B220 and CD19 was not detected on the surface of PU.1-1 and PU.1-2 cells or mock cells cultured with or without ZnCl2 (Figure 3 and data not shown). Because expression of the CD19 gene was up-regulated in PU.1-1 cells after overexpression of PU.1, we used immunohistochemical analysis to assess whether B220 and CD19 proteins were present in the cytoplasm. Although CD11b and F4/80 antigen were detected in cells cultured with ZnCl2, neither of the B-cell–specific proteins was present (data not shown). These findings suggest that even though expression of B-cell–specific genes is induced by overexpression of PU.1 in MEL cells, this does not necessarily result in expression of the proteins.
Morphologic and functional changes in MEL cells after overexpression of PU.1
We next examined the effects of overexpression of PU.1 on morphologic features and function of MEL cells by using PU.1-1 and PU.1-2 cells. These cells were originally spherical and grew in suspension. However, when treated with ZnCl2, some cells of both clones became spindle shaped and adherent by 48 hours (Figure4A and data not shown). Such changes were not observed in the parental and mock cells treated with ZnCl2 (data not shown). Because expression levels of the genes encoding for some myelomonocyte-specific growth factor receptors were up-regulated in these cells treated with ZnCl2, we attempted to determine the effects of these factors on morphologic changes in the cells. As shown in Figure 4A, the number of adherent cells increased when granulocyte-macrophage colony-stimulating factor (GM-CSF; 1 ng/mL) was added to the culture along with ZnCl2. Similar results were obtained when macrophage colony-stimulating factor (M-CSF; 6 ng/mL) was added instead of GM-CSF (data not shown). These results suggest that the receptors are expressed on the cell surfaces and that they are functional.
Induction of morphologic and functional changes in MEL cells after overexpression of PU.1.
(A) PU.1-1 cells cultured for 48 hours without reagents (upper panel), with ZnCl2 (middle panel), or with ZnCl2 and GM-CSF (1 ng/mL; lower panel) are shown. Arrowheads indicate the adherent cells (spindle shape). (B) NBT reduction activity was measured in PU.1-1 cells, PU.1-2 cells, and mock cells cultured for 48 hours with (▨) or without (■) ZnCl2. Data represent the mean ± SD values from 3 independent experiments. (C) PU.1-1 cells were cultured for 48 hours with ZnCl2, and this was followed by incubation with latex beads for 2.5 hours. Experiments were performed in triplicate. Representative cells taking up latex beads after treatment with ZnCl2 are shown.
Induction of morphologic and functional changes in MEL cells after overexpression of PU.1.
(A) PU.1-1 cells cultured for 48 hours without reagents (upper panel), with ZnCl2 (middle panel), or with ZnCl2 and GM-CSF (1 ng/mL; lower panel) are shown. Arrowheads indicate the adherent cells (spindle shape). (B) NBT reduction activity was measured in PU.1-1 cells, PU.1-2 cells, and mock cells cultured for 48 hours with (▨) or without (■) ZnCl2. Data represent the mean ± SD values from 3 independent experiments. (C) PU.1-1 cells were cultured for 48 hours with ZnCl2, and this was followed by incubation with latex beads for 2.5 hours. Experiments were performed in triplicate. Representative cells taking up latex beads after treatment with ZnCl2 are shown.
Levels of NBT reduction activity were markedly increased in both PU.1-1 and PU.1-2 cells cultured with ZnCl2 for 48 hours compared with mock cells cultured under the same conditions (Figure 4B). Moreover, 11% to 14% of cells of both clones took up latex beads after treatment with ZnCl2 for 48 hours (Figure 4C and data not shown). No such activity was observed in cells cultured without ZnCl2 or mock cells cultured with or without ZnCl2 (data not shown). These results indicate that morphologic and functional changes are provoked in MEL cells as a result of the alteration in gene-expression profile after overexpression of PU.1. In summary, we found that PU.1 can reprogram MEL cells that have myelomonocytic characteristics.
Activation domain of PU.1 is required for induction of myelomonocyte-specific and B-cell–specific gene expression in MEL cells
In a previous study,11 we established a transfectant by introducing a zinc-inducible expression plasmid of a mutant version of PU.1 with a deletion of the glutamine-rich region of the activation domain (referred to as PU.1-ΔA mutant) into MEL-B8/3 cells. Using this transfectant, we showed that induced expression of this mutant PU.1 in MEL cells inhibits erythroid differentiation of the cells, although it does not induce growth inhibition and apoptosis.11 We therefore investigated whether expression of myelomonocyte-specific and B-cell–specific genes is induced in the transfectant when PU.1-ΔA mutant is expressed.
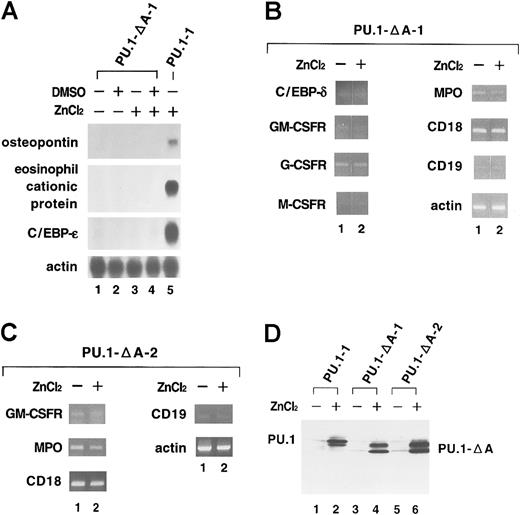
As shown in Figure 5A and 5B, no induction of expression was observed in any of the genes examined (the osteopontin, eosinophil cationic protein, C/EBP-ε, C/EBP-δ, GM-CSFR, G-CSFR, M-CSFR, MPO, CD18, and CD19 genes) in one clone of the transfectant (PU.1-ΔA-1 cells) treated with ZnCl2 for 48 hours (Figure 5A, lane 4, and Figure 5B, lane 2). We obtained similar results in an additional independent clone of the transfectant (PU.1-ΔA-2 cells; Figure 5C). Moreover, no adherent cells were detected in either clone after expression of PU.1-ΔA mutant (data not shown). Western blot analysis showed that the levels of expression of induced mutant PU.1 protein in these cells were comparable to those of induced wild-type PU.1 protein in PU.1-1 cells (Figure 5D). These results indicate that the glutamine-rich subdomain of the activation domain is essential for induction of expression of myelomonocyte-specific and B-cell–specific genes in MEL cells and suggest that intrinsic transcriptional activity of PU.1 is necessary for reprogramming MEL cells.
No induction of expression of myelomonocyte-specific and B-cell–specific genes in MEL cells after overexpression of a mutant PU.1 with part of the activation domain deleted (PU.1-ΔA).
(A) Total RNA was extracted from PU.1-ΔA-1 cells cultured for 48 hours without reagents (lane 1), with DMSO (lane 2), with ZnCl2 (lane 3), or with DMSO and ZnCl2 (lane 4). Expression of myelomonocyte-specific and B-cell–specific genes was examined by Northern blot analysis. Total RNA from PU.1 cells cultured for 48 hours with ZnCl2 was used as the positive control (lane 5). (B) Total RNA was extracted from PU.1-ΔA-1 cells cultured for 48 hours without ZnCl2 (lane 1) or with ZnCl2 (lane 2). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. Although the photograph is separated between lanes 1 and 2, PCR products of each gene were run in the same gel. (C) Total RNA was extracted from PU.1-ΔA-2 cells cultured for 48 hours without ZnCl2 (lane 1) or with ZnCl2 (lane 2). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. (D) Total protein was extracted from PU.1-1 cells (lanes 1 and 2), PU.1-ΔA-1 cells (lanes 3 and 4), and PU.1-ΔA-2 cells (lanes 5 and 6) cultured for 8 hours without reagents (lanes 1, 3, and 5) or with ZnCl2 (lanes 2, 4, and 6). Expression of wild-type and mutant PU.1 proteins was examined by Western blot analysis.
No induction of expression of myelomonocyte-specific and B-cell–specific genes in MEL cells after overexpression of a mutant PU.1 with part of the activation domain deleted (PU.1-ΔA).
(A) Total RNA was extracted from PU.1-ΔA-1 cells cultured for 48 hours without reagents (lane 1), with DMSO (lane 2), with ZnCl2 (lane 3), or with DMSO and ZnCl2 (lane 4). Expression of myelomonocyte-specific and B-cell–specific genes was examined by Northern blot analysis. Total RNA from PU.1 cells cultured for 48 hours with ZnCl2 was used as the positive control (lane 5). (B) Total RNA was extracted from PU.1-ΔA-1 cells cultured for 48 hours without ZnCl2 (lane 1) or with ZnCl2 (lane 2). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. Although the photograph is separated between lanes 1 and 2, PCR products of each gene were run in the same gel. (C) Total RNA was extracted from PU.1-ΔA-2 cells cultured for 48 hours without ZnCl2 (lane 1) or with ZnCl2 (lane 2). Expression of myelomonocyte-specific and B-cell–specific genes was examined by RT-PCR analysis. (D) Total protein was extracted from PU.1-1 cells (lanes 1 and 2), PU.1-ΔA-1 cells (lanes 3 and 4), and PU.1-ΔA-2 cells (lanes 5 and 6) cultured for 8 hours without reagents (lanes 1, 3, and 5) or with ZnCl2 (lanes 2, 4, and 6). Expression of wild-type and mutant PU.1 proteins was examined by Western blot analysis.
Discussion
In this study, we used an mRNA differential display strategy to identify PU.1 target genes involved in the inhibition of growth and erythroid differentiation and the induction of apoptosis in MEL cells. Some of the genes we identified are known to be involved in cell division and regulation of mitochondrial and ribosomal functions that might participate in growth inhibition or apoptosis.
In addition, expression of 3 genes—the osteopontin, eosinophil cationic protein, and B144—that are usually expressed in the myelomonocyte and B-cell lineages but not in the erythroid lineage was up-regulated in MEL cells after overexpression of PU.1. Because of this finding, we extended our study and showed that expression of many genes and proteins specific for these lineages is induced in MEL cells after overexpression of PU.1 (Table 3) and that, consequently, morphologic and functional changes are also induced in the cells. These results indicate that overexpression of PU.1 not only inhibits erythroid differentiation but also induces lineage switch in MEL cells.
Genes and proteins up-regulated by overexpression of PU.1 in murine erythroleukemia cells
| Myelomonocyte specific . | B-cell specific . | ||
|---|---|---|---|
| Messenger RNA | Messenger RNA | ||
| Osteopontin | + | CD19 | + |
| Eosinophil cationic protein | + | C/EBP-γ | − |
| B144 | + | Pax-5 | − |
| C/EBP-α | + | Igκ | − |
| C/EBP-δ | + | Igμ | − |
| C/EBP-ε | + | Protein | |
| GM-CSFR | + | CD19 | − |
| G-CSFR | + | B220 | − |
| M-CSFR | + | ||
| MPO | + | ||
| CD18 | + | ||
| Neutrophil elastase | − | ||
| HLS7 | − | ||
| MLL1 | − | ||
| Protein | |||
| CD11b (Mac-1) | + | ||
| F4/80 antigen | + | ||
| Myelomonocyte specific . | B-cell specific . | ||
|---|---|---|---|
| Messenger RNA | Messenger RNA | ||
| Osteopontin | + | CD19 | + |
| Eosinophil cationic protein | + | C/EBP-γ | − |
| B144 | + | Pax-5 | − |
| C/EBP-α | + | Igκ | − |
| C/EBP-δ | + | Igμ | − |
| C/EBP-ε | + | Protein | |
| GM-CSFR | + | CD19 | − |
| G-CSFR | + | B220 | − |
| M-CSFR | + | ||
| MPO | + | ||
| CD18 | + | ||
| Neutrophil elastase | − | ||
| HLS7 | − | ||
| MLL1 | − | ||
| Protein | |||
| CD11b (Mac-1) | + | ||
| F4/80 antigen | + | ||
Plus signs indicate gene is up-regulated; minus signs indicate gene is not up-regulated.
For abbreviations, see Table 1.
PU.1 is known to bind to and activate the promoter of genes such as the GM-CSFR,30 M-CSFR,31 G-CSFR,32CD18,33 and CD11b.34 Ets-binding sites were reported to be present in the promoter regions of the osteopontin35 and C/EBP-ε36 genes. PU.1 may be able to induce expression of these endogenous genes directly through binding to their promoter regions in MEL cells. Binding of PU.1 to these regions may change chromatin conformation by means of histone modifications. In this context, we previously showed that PU.1 physically and functionally interacts with CREB-binding protein (CBP), which has intrinsic histone acetyltransferase activity.37
It is remarkable that expression of the genes encoding for some C/EBP transcription factors was up-regulated because C/EBP-α, C/EBP-β, and C/EBP-ε, and PU.1 are critical for myeloid differentiation.38,39 The C/EBP family was shown to regulate expression of, for instance, the GM-CSFR and M-CSFR genes synergistically with some other transcription factors, including PU.1 and AML1.30 40 PU.1 may activate expression of C/EBP transcription factors hierarchically, consequently activating expression of target genes in a synergistic manner. Our results also suggest that expression of all C/EBP genes is not regulated in a similar fashion, since we found that expression of the C/EBP-γ gene was not up-regulated by PU.1 overexpression (data not shown).
The HLS7 gene was isolated from Raf-transformed and Myc-transformed erythroid cells that spontaneously acquired myeloid characteristics.26,27 The HLS7 gene was shown to be homologous to the human myeloid leukemia factor 1 gene and to inhibit differentiation of erythroid cells and induce differentiation of myeloid cells.27 The MLLgene is often rearranged by chromosomal translocations in human lymphocytic and myelomonocytic leukemias and is thought to play a role in the regulation of myelomonocyte differentiation.28However, no increased expression was observed in these genes after overexpression of PU.1 in MEL cells. The results of this study suggest that the pathway through which PU.1 induces lineage switch in MEL cells is distinct from pathways used by HLS7 or MLL or that these 2 genes are not downstream from PU.1, even if they are in the same pathway as PU.1.
Reflecting the induction of the gene-expression profile for myelomonocytes, morphologic and functional changes associated with macrophage-like phenotypes were produced in MEL cells. Induction of these changes appeared to depend on the levels of expression of PU.1, since no such changes were observed in PU.1 cell clones before induction of PU.1 overexpression or in mock cells, both of which originally expressed steady-state levels of PU.1 and detectable levels of some myelomonocyte-specific genes (Figure 2). Addition of GM-CSF or M-CSF to the culture medium increased the number of adherent cells. We also observed that addition of these growth factors at least partly, if not completely, rescued PU.1-1 cells from apoptosis induced by overexpression of PU.1 in the presence of DMSO (data not shown). These results suggest that the growth factor receptors induced to be expressed by overexpression of PU.1 are functional in MEL cells. In contrast to the morphologic findings, no apparent increase in NBT reduction activity or the proportion of phagocytic cells taking up latex beads was observed when GM-CSF or M-CSF was added (data not shown). These growth factors might not have any effects on these functions in MEL cells.
In the flow cytometric and immunohistochemical analyses, induction of expression of the myelomonocyte-specific proteins CD11b (Mac-1) and F4/80 antigen was observed in PU.1 cell clones after overexpression of PU.1, but there were no detectable levels of expression of B-cell–specific B220 and CD19 proteins. Because induction of gene expression was observed in the case of CD19, we suggest that posttranscriptional modifications, including translation or posttranslational modifications (or both) in these B-cell–specific proteins may not occur completely in the erythroid background. Expression of the Igκ-chain and Igμ-chain genes was not also induced after overexpression of PU.1. This may have been because of lack of a mechanism in MEL cells to manage the gene rearrangement necessary before gene expressions. We suggest that a threshold on the pathway to B cells might be higher than that on the pathway to myeloid cells. In addition, our immunohistochemical analysis did not detect T-cell–specific proteins (CD3, CD4, CD5, or CD8) in MEL cells after overexpression of PU.1 (data not shown)—results consistent with the idea that PU.1 is not expressed or involved in the development of this cell lineage.41 42
To address the possibility of whether loss of erythroid characteristics was induced by the overexpression of PU.1 concomitant with the lineage switch in MEL cells, we determined levels of expression of the β-globin gene and genes encoding for hematopoietic transcription factors that play important roles in erythropoiesis (GATA-1, SCL, and LMO2) by using RT-PCR analysis. We found that expression of these genes was not down-regulated by overexpression of PU.1 (data not shown), which is consistent with our previous finding that expression of genes encoding for erythrocyte-specific transcription factors, such as GATA-1, NF-E2 (p45 and p18), and SCL, was not reduced in MEL cells, even after PU.1 overexpression.15 We also examined expression of the erythroid marker protein Ter119 by using immunohistochemical analysis and found no expression of this protein in MEL cells before or after overexpression of PU.1 (data not shown). Moreover, no erythroid cell–specific genes were detected in our mRNA differential display analysis (data not shown).
The experiments using PU.1-ΔA cell clones demonstrated that the activation domain of PU.1 is essential for induction of expression of myelomonocyte-specific and B-cell–specific genes in MEL cells. It is thought that not all the transcription factors that function synergistically with PU.1 to induce expression of these genes are provided in the erythroid background. Intrinsic transcriptional activity of PU.1 may be essential in the presence of such a shortage of cooperators. In PU.1-ΔA cell clones, in addition to impairment of the transactivation domain of PU.1-ΔA, there is no induction of expression of C/EBP transcription factors. This may be one of the reasons for the lack of induction of expression of the myelomonocyte-specific and B-cell–specific genes in the cells. Another possible reason is the inability of PU.1-ΔA mutant to bind to CBP.37 As discussed above, the participation of histone or chromatin modifications is considered to be necessary to induce expression of myelomonocyte-specific and B-cell–specific genes in MEL cells. The dominant negative effect of this mutant PU.1, if any, appears to be mild because basal levels of expression of myelomonocyte-specific and B-cell–specific genes were preserved after induction of expression of the mutant (Figure 5B and 5C, lanes 1 and 2). We previously found that PU.1-ΔA mutant inhibited erythroid differentiation of MEL cells.11 Because of this finding and the results of the current study, we suggest that the mechanism that induces lineage switch is distinct from that inducing differentiation inhibition of MEL cells; that is, PU.1-mediated inhibition of erythroid differentiation of MEL cells does not result from lineage switch in the cells.
This study provides evidence that PU.1 induces a lineage switch in MEL cells, which have already been committed to the erythroid lineage, toward myelomonocytes. In line with this idea, Nerlov and Graf43 demonstrated that PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitor cells (MEPs) established by infecting chicken blastoderm cells with the Myb-Ets–encoding E26 leukemia virus. Taken together, these results indicate that PU.1 can impose myelomonocytic characteristics not only on hematopoietic progenitor cells but also on cells already committed to other lineages, thus suggesting that PU.1 acts as a master regulator to mediate lineage commitment of progenitors in hematopoiesis. Additionally, consistent with our observations, Nerlov and Graf43 also showed that the activation domain of PU.1 is required for myeloid lineage commitment of MEPs. Moreover, DeKoter and Singh44 found that a low concentration of PU.1 promotes B-cell differentiation, whereas a high concentration promotes macrophage differentiation in hematopoietic progenitors. Apart from PU.1, GATA-1 was shown to induce megakaryocyte and erythrocyte traits in M1 myeloid cells,45 and E2A was found to induce expression of B-cell–specific genes in macrophage-like cells.46
The findings of this study, along with the examples of lineage switch of hematopoietic cells induced by hematopoietic cell–specific transcription factors, provide evidence that transcription factors play fundamental roles in the commitment of hematopoietic cells and offer new insight into hematopoiesis in general.
We thank Dr Y. Hashimoto, the director of Sasaki Institute, for critical reading of the manuscript.
Supported by grants-in-aid for Scientific Research on Priority Areas and for Scientific Research (C) from the Ministry of Education, Science and Culture of Japan, and by the Uehara Memorial Foundation, Tokyo, Japan, and Obstetrics-Gynecology Akiyama Memorial Hospital, Hakodate, Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Tsuneyuki Oikawa, Department of Cell Genetics, Sasaki Institute, 2-2, Kanda-Surugadai, Chiyoda-ku, Tokyo, 101-0062, Japan; e-mail: oikawa@sasaki.or.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal