Abstract
Effective gene therapy for diseases of the circulation requires vectors capable of systemic delivery. The molecular weight of poly(l-lysine) (pLL) has a significant effect on the circulation of pLL/DNA complexes in mice, with pLL211/DNA complexes displaying up to 20 times greater levels in the blood after 30 minutes compared with pLL20/DNA. It is shown that pLL20/DNA complexes fix mouse complement C3 in vitro, independent of immunoglobulin binding; are less soluble in the blood in vivo; bind erythrocytes; are rapidly removed by the liver, where they associate predominantly with Kupffer cells; and result in a rapid increase in hepatic leukocytes expressing high levels of complement receptor 3 (CR3). The circulation properties of these complexes are also dependent on the type of DNA used, with circular plasmid DNA complexes exhibiting increased circulation compared with linear DNA. PLL211/DNA complexes bind erythrocytes and associate with Kupffer cells but, in contrast, do not fix mouse complement in vitro and are unaffected by the type of DNA used. In rats, both types of complexes produce hematuria and are rapidly removed from the circulation. Correlation of in vivo and in vitro results suggests that the solubility of complexes in physiological saline and species-matched complement fixation and erythrocyte lysis may correlate with systemic circulation. Analysis using human blood in vitro shows no hemolysis, but both types of complexes fix complement and bind IgG, suggesting that pLL/DNA complexes may be rapidly cleared from the human circulation.
Introduction
Gene therapy promises significant advances in the treatment of a range of diseases, including several genetic and acquired disorders affecting a range of cells and tissues. Certain applications of gene therapy may be addressed on the basis of loco-regional administration of therapeutic material—eg, bronchial aspects of cystic fibrosis1 or genetic diseases manifest in specific organs.2,3 However, for many important applications, including the treatment of blood and circulatory disorders4,5 and disseminated or metastatic cancer,6 7 it is necessary to develop intravenous forms of gene therapy capable of systemic circulation. Nonviral vectors are expected ultimately to be preferred for clinical application, particularly in view of the reluctance of patients to be administered viruses in the treatment of nonacutely life-threatening disorders.
The use of the cationic polymer poly(l-lysine) (pLL) in vectors for gene therapy has been widely described.8-11PLL/DNA complexes typically measure less than 100 nm in diameter and can be targeted to various cell lines in vitro by covalent attachment of targeting ligands to the pLL, resulting in transgene expression.12 Unfortunately, pLL/DNA complexes are of limited use in vivo because of their poor circulatory half-lives, typically shorter than 3 minutes.11,13,14 The reasons for the rapid removal of these complexes from the blood are poorly understood, though they are thought to reflect the in vitro observations of protein binding,13 salt aggregation,15 and reversible polycation/DNA interactions.17 Despite these poor circulatory half-lives, transgene expression has been achieved using such complexes14,16 17; however, expression is usually limited to the liver, where most targeted and untargeted pLL/DNA complexes accumulate.
An understanding of the parameters involved in the removal of pLL/DNA complexes from the circulation is fundamental to the development of vectors capable of prolonged plasma circulation and target cell accumulation. Although it is well known that pLL/DNA complexes activate human complement in vitro,18,19 no studies have correlated immune stimulation by pLL/DNA complexes with circulatory half-lives in rodent models. Indeed, evidence shows that pLL/DNA complexes do not stimulate the immune system in mice,11 thus confusing the factors involved in the removal of complexes from the circulation.
In preliminary experiments we observed that the molecular weight of the pLL and the type of DNA used in pLL/DNA complexes have a significant influence on the circulatory properties of the complexes. Here, we show that factors such as poor solubility in blood, complement activation, stimulation of leukocytes, and macrophage capture may contribute to the differential blood circulation observed.
Materials and methods
All reagents were purchased from Sigma (Dorset, United Kingdom) unless otherwise stated.
Preparation of plasmid DNA and 32P-,35S-labeled DNA
A 6.4-kb plasmid encoding lac Z under transcriptional regulation of the cytomegalovirus immediate early promoter was used throughout these studies. Plasmid DNA was prepared in bulk by growth inEscherichia coli and purified using Qiagen columns (Qiagen, Crawley, United Kingdom). Linear plasmid was obtained byHindIII linearization followed by phenol/chloroform extraction. Purity of the DNA was checked by ethidium bromide visualization of agarose gels. 32P- or35S-labeled expression vector was prepared as follows: linearized plasmid was radiolabeled by template extension with32P dCTP or 35S dCTP (Amersham Pharmacia, Little Chalfont, United Kingdom) using the Ready-to-Go Oligolabeling kit (Amersham Pharmacia). Unincorporated nucleotides were removed using microspin columns (Amersham Pharmacia), and the purity of the labeled DNA was checked after agarose gel electrophoresis and quantitative analysis with a PhosphorImager (Molecular Dynamics, Chesham, United Kingdom). Labeled DNA was diluted with nonlabeled calf thymus DNA (DNAct), closed circular plasmid DNA (DNAcp), or linear plasmid DNA (DNAlp) for experimental use.
Formation of pLL/DNA complexes
All complexes were formed in water at a charge ratio of 2.8:1 and a final DNA concentration of 20 μg/mL; this was followed by dilution in HEPES buffer at pH 7.5 to give a final DNA concentration of 10 μg/mL and a final buffer concentration of 25 mM. PLL 20 kd (pLL20) or 211 kd (pLL211) HBr salt (35.84 μg of a 2.5 mg/mL stock solution in water) was added in a single addition to a solution of DNA (20 μg in 1.0 mL water), mixed, and left to stand for 1 hour at room temperature. HEPES buffer (1.0 mL, 50 mM, pH 7.5) was added to the samples and mixed by inversion before experimental analysis. Studies using radiolabeled DNA were performed using a spike of 32P-labeled DNA (0.6 ng/μg unlabeled DNA) in the DNA solution, and the complexes formed as above.
Immobilization of pLL/DNA complexes on magnetic streptavidin beads
Biotin-pLL was formed by reacting pLL (pLL20 or pLL211; 10 mg/mL in 1 M HEPES buffer, pH 8.5) with biotinamidocaproic acid 3-sulfo-N-hydroxy-succinimide ester (1.34 μg) at room temperature for 3 hours. The resultant mixture was centrifuged to remove particulates, and the supernatant was dialyzed against 20 mM NaCl for 48 hours. PLL concentration was calculated using the TNBS method.37 The number of biotin moieties per pLL chain was normalized for pLL20 and pLL211 using HABA analysis.38 All biotin-pLL conjugates condensed DNA as efficiently as nonconjugated pLL, as determined using the ethidium bromide exclusion assay.20 PLL/DNA complexes were then formed using the method described above. Magnetic streptavidin beads (20 μL, washed in phosphate-buffered solution (PBS) before use; Pierce, Chester, United Kingdom) were added to biotin-pLL/DNA complexes (0.4 mL) and left at room temperature for 30 minutes. After magnetic isolation of the beads, the supernatant was removed, and the beads were washed twice in 25 mM HEPES buffer before experimental analysis.
Identification of proteins bound to pLL/DNA complexes
Radiolabeled biotin-pLL/DNA complexes bound to magnetic streptavidin beads were incubated in normal or heat-inactivated serum (0.4 mL; heat-inactivated at 56°C for 30 minutes, with 10 mM EDTA added before incubation with complexes) for 5 minutes. Magnetic beads were isolated and washed in isotonic buffer (3 washes). Laemmeli buffer (0.1 mL) was added to the immobilized complexes and boiled for 3 minutes. The supernatant was decanted to a fresh Eppendorf, and the radioactivity was assayed in a Packard liquid scintillation analyser (1900TR, Packard, Palo Alto, CA). Dot blots were prepared on nitrocellulose membrane (1-μL aliquots), and proteins were identified by antibody blotting using radiography.
Animal models for analysis of pharmacokinetics
Female Balb/c mice (approximate weight, 20 g) and Wistar rats (approximate weight, 200 g) were obtained in-house and kept according to Home Office guidelines.21 pLL/DNA complexes were administered by intravenous tail vein injection (0.1 mL in mice; 1.0 mL in rats; both corresponding to 0.6 μg DNA/mL blood39,40). Blood was sampled from the end of the tail, smeared on glass slides (Superfrost +; BDH, Lutterworth, United Kingdom), air-dried, and fixed in methanol for 10 minutes. Animals were killed after 30 minutes for pharmacokinetic and liver immunohistochemistry studies and after 1 or 10 minutes for blood cell–binding studies. For body distribution studies, organs and carcasses were dissolved in 10 M NaOH at 75°C for 1 hour and assayed for radioactivity (values have been corrected for the levels of blood within the organs36). For immunohistochemistry analysis, organs were removed, snap frozen in liquid nitrogen, and stored at −80°C. Mouse liver was sectioned on a cryostat (12-μm sections) and placed onto coated glass slides (Superfrost +; BDH). The sections were fixed in acetone for 10 minutes and air-dried. Blood cell–binding experiments were performed by centrifuging blood (without anticlotting agents) at 5000 rpm at 4°C immediately upon sampling. Radioactivity within blood samples and organs/carcass was measured in a liquid scintillation analyser (1900TR; Packard), and within blood smears and liver sections it was measured using quantitative analysis with a PhosphorImager (Molecular Dynamics).
Immunohistochemistry of mouse liver sections
Slides were exposed to either rat antimouse F4/80 antibody (Serotec, Oxford, United Kingdom; 1:50 dilution in 1% bovine serum albumin [BSA]/PBS) or rat antimouse CD11b antibody (Research Diagnostics, Flanders, NJ; 1:50 dilution in 1% BSA/PBS) for 2 hours and washed by immersion in 1% BSA/PBS solution (total, 4 washes). Sheep antirat immunoglobulin antibody (DAKO, Ely, United Kingdom; 1:50 dilution in 1% BSA/PBS) was added for 2 hours and washed by immersion in 1% BSA/PBS solution (4 washes). Rat APAAP antibody (DAKO; 1:50 dilution in 1% BSA/PBS) was added for 1 hour, and the slides were washed by immersion in 1% BSA/PBS solution (2 washes). Slides were placed in Tris-buffered saline (2.5 minutes; pH 7.5, 50 mM Tris base, 140 mM NaCl) followed by Tris buffer (2.5 minutes; pH 8.2, 50 mM Tris base) and then immersed in alkaline phosphate substrate (15 minutes; made in the following order: naphthol-phosphate AS-MX [10 mg]; NN-dimethylformamide [1.0 mL]; Tris buffer [49.0 mL]; levamisole [50 μL of 1 M stock]; Fast Red-TR [50 mg]; filtered before use). The slides were then exposed to Mayer's hematoxylin (filtered) for 3 minutes, washed in tap water, and mounted.
Visualization of 35S-labeled pLL/DNA complexes in liver sections and blood smears
Slides were coated with liquid photographic emulsion (NTB-2; Anachem, Luton, United Kingdom) and stored in the dark at 4°C for various lengths of time (between 1 and 7 days). Slides were developed in D-19 developer (diluted 1:1 with deionized water), fixed in sodium thiosulphate (30% wt/vol), and washed gently in tap water to remove excess emulsion.
Erythrocyte lysis assay
Blood was taken from a healthy female human volunteer or a female Wistar rat and divided into EDTA-containing and empty blood tubes. The serum from the coagulated blood sample was mixed with blood cells from the EDTA-treated sample (cells washed × 2 in PBS to remove EDTA) to form an experimental blood substitute containing active serum. Blood (0.5 mL) was mixed with pLL/DNA complexes (30 μL, corresponding to 0.6 μg DNA/mL blood; formed in water and diluted in HEPES buffer as described above), incubated at 37°C for 1 hour, and centrifuged at 5000 rpm for 1 minute to pellet the blood cells, and then 100 μL supernatant was transferred to a flat-bottomed 96-well plate. Hemoglobin released into the supernatant was measured by absorbance at 550 nm in a microplate autoreader (Bio-Tek Instruments, Winooski, VT).
Photon correlation spectroscopy analysis of pLL/DNA complexes
pLL/DNA complexes were analyzed by light scattering on a Zetasizer (Malvern Instruments, Malvern, United Kingdom) equipped with a 70-mW external laser set at a wavelength of 488 nm.
Results
Effect of pLL molecular weight and DNA type on the blood circulation of 2.8:1 charge ratio pLL/DNA complexes in mice
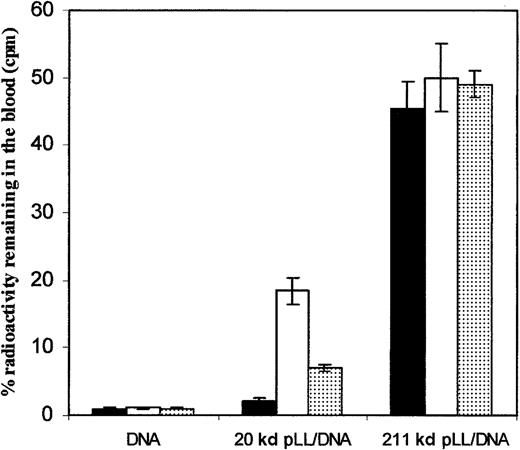
Figure 1 shows the amount of32P-labeled DNA remaining in the blood of female Balb/c mice 30 minutes after intravenous injections of pLL/DNA complexes formed with pLL20 or pLL211 and either DNAct, DNAcp, or DNAlp. A charge ratio of 2.8:1 pLL/DNA was used because previous work has shown these complexes to be most stable to albumin-induced turbidity.22 The amount of pLL211/DNA complexes in the blood after 30 minutes was up to 20 times higher than those of pLL20/DNA complexes and were unaffected by the type of DNA used. In contrast, pLL20/DNA complexes showed significant differences in circulation between the types of DNA. For example, the use of circular plasmid DNA (DNAcp) led to 9-fold greater levels of DNA remaining in the blood after 30 minutes (18.5% ± 2.0%) compared with that obtained for calf thymus DNA (DNAct) (2.1% ± 0.5%). This difference probably resulted from the physical properties of pLL/DNA complexes formed using linear DNA. For instance, when circular plasmid DNA was linearized (DNAlp), a 3-fold decrease in circulation was seen (7.0% ± 0.5%). Proteins present on DNAct were not responsible for the rapid clearance of pLL20/DNAct complexes because complexes formed with DNAct purified by phenol/chloroform extraction exhibited similar blood clearance profiles (data not shown). Administration of either DNAct, DNAcp, or DNAlp alone showed a similar level of32P-labeled DNA remaining in the blood after 30 minutes. Additionally, complexes formed at ± charge ratios of 1:1 and 3:1 showed similar distribution profiles (data not shown).
pLL211/DNA complexes exhibit increased circulation in mice compared to pLL20/DNA complexes and are unaffected by the type of DNA used.
pLL/DNA complexes, formed with pLL20 (20 kd pLL/DNA) or pLL211 (211 kd pLL/DNA), and either DNAct(▪), DNAcp (■), or DNAlp (░) were administered to female Balb/c mice (0.1 mL of 10 μg/mL DNA) through tail vein injection. Administration of free DNA with radioactive probe is also shown (DNA). Mice were killed after 30 minutes, and the blood was assayed for 32P-levels. Results show the mean ± SD of 3 independent experiments.
pLL211/DNA complexes exhibit increased circulation in mice compared to pLL20/DNA complexes and are unaffected by the type of DNA used.
pLL/DNA complexes, formed with pLL20 (20 kd pLL/DNA) or pLL211 (211 kd pLL/DNA), and either DNAct(▪), DNAcp (■), or DNAlp (░) were administered to female Balb/c mice (0.1 mL of 10 μg/mL DNA) through tail vein injection. Administration of free DNA with radioactive probe is also shown (DNA). Mice were killed after 30 minutes, and the blood was assayed for 32P-levels. Results show the mean ± SD of 3 independent experiments.
Body distribution of pLL/DNA complexes
Table 1 shows the body distribution of 32P-labeled DNA in female Balb/c mice 30 minutes after intravenous injection of pLL/DNA complexes formed with pLL20 or pLL211 and either DNAct, DNAcp, or DNAlp. The values have been corrected for the levels of blood within the organs.36 Free DNA was rapidly removed from the blood (Figure 1, Table2) on administration to mice, and, after 30 minutes, it was found primarily within the liver (84.9% ± 2.0%), with small levels within the carcass, the kidneys, and the intestine, and very low levels within the lungs and the spleen. In contrast, most complexes formed using either pLL20 or pLL211 exhibited significantly altered body distributions, with longer circulating vectors (all pLL211/DNA complexes, pLL20/DNAcp and, to a lesser extent, pLL20/DNAlp) exhibiting decreased accumulation within the liver and increased levels within the carcass. PLL20/DNAct complexes showed relatively high levels of accumulation within the liver (78.9 ± 7.3%) and low levels within the carcass (10.8 ± 3.6%), both similar to the levels obtained using free DNA. However, levels of pLL20/DNAct within the kidneys and spleen are significantly different from those obtained with free DNA. Less than 1% of radioactivity in all samples was associated with the urine and the site of injection in the tail (data not shown).
Poly(l-lysine) molecular weight and DNA type affect the body distribution of pLL/DNA complexes in mice
| . | Carcass . | Liver . | Lungs . | Kidneys . | Intestine . | Spleen . |
|---|---|---|---|---|---|---|
| pLL20/DNAct | 10.8 ± 3.6 | 78.9 ± 7.3 | 1.7 ± 0.5 | 0.8 ± 0.1 | 1.7 ± 0.7 | 9.1 ± 4.2 |
| pLL20/DNAcp | 24.5 ± 5.6 | 60.3 ± 8.5 | 2.0 ± 0.4 | 1.9 ± 0.6 | 4.2 ± 1.1 | 6.9 ± 1.2 |
| pLL20/DNAlp | 16.3 ± 2.3 | 71.4 ± 5.2 | 1.0 ± 0.2 | 1.8 ± 0.3 | 4.6 ± 2.3 | 5.0 ± 1.8 |
| pLL211/DNAct | 36.6 ± 6.8 | 42.8 ± 8.8 | 2.8 ± 0.1 | 1.9 ± 1.0 | 4.7 ± 1.2 | 8.8 ± 2.9 |
| pLL211/DNAcp | 33.4 ± 3.2 | 46.9 ± 3.6 | 2.9 ± 0.4 | 1.8 ± 0.4 | 3.6 ± 0.3 | 12.5 ± 1.6 |
| pLL211/DNAlp | 30.9 ± 0.8 | 50.6 ± 2.9 | 2.5 ± 0.8 | 1.6 ± 0.4 | 2.5 ± 0.3 | 12.1 ± 0.8 |
| DNA* | 9.4 ± 2.9 | 84.9 ± 2.0 | 1.1 ± 0.1 | 6.5 ± 1.8 | 5.1 ± 1.3 | 0.4 ± 0.1 |
| . | Carcass . | Liver . | Lungs . | Kidneys . | Intestine . | Spleen . |
|---|---|---|---|---|---|---|
| pLL20/DNAct | 10.8 ± 3.6 | 78.9 ± 7.3 | 1.7 ± 0.5 | 0.8 ± 0.1 | 1.7 ± 0.7 | 9.1 ± 4.2 |
| pLL20/DNAcp | 24.5 ± 5.6 | 60.3 ± 8.5 | 2.0 ± 0.4 | 1.9 ± 0.6 | 4.2 ± 1.1 | 6.9 ± 1.2 |
| pLL20/DNAlp | 16.3 ± 2.3 | 71.4 ± 5.2 | 1.0 ± 0.2 | 1.8 ± 0.3 | 4.6 ± 2.3 | 5.0 ± 1.8 |
| pLL211/DNAct | 36.6 ± 6.8 | 42.8 ± 8.8 | 2.8 ± 0.1 | 1.9 ± 1.0 | 4.7 ± 1.2 | 8.8 ± 2.9 |
| pLL211/DNAcp | 33.4 ± 3.2 | 46.9 ± 3.6 | 2.9 ± 0.4 | 1.8 ± 0.4 | 3.6 ± 0.3 | 12.5 ± 1.6 |
| pLL211/DNAlp | 30.9 ± 0.8 | 50.6 ± 2.9 | 2.5 ± 0.8 | 1.6 ± 0.4 | 2.5 ± 0.3 | 12.1 ± 0.8 |
| DNA* | 9.4 ± 2.9 | 84.9 ± 2.0 | 1.1 ± 0.1 | 6.5 ± 1.8 | 5.1 ± 1.3 | 0.4 ± 0.1 |
pLL/DNA complexes, formed with pLL20 or pLL211, and either DNAct, DNAcp, or DNAlp were administered into female Balb/c mice (0.1 mL of 10 μg/mL DNA) through tail vein injection. Mice were killed after 30 minutes, and the organs and carcasses were dissolved in 10 M NaOH at 75°C for 1 hour. Radioactivity in the samples was measured in a liquid scintillation analyzer (1900TR; Packard). The values are corrected for the amount of blood present in organs of decapitated mice.36
pLL indicates poly(l-lysine); DNAct, calf thymus DNA; DNAcp, closed circular plasmid DNA; DNAlp, linear plasmid DNA.
DNA value is that obtained after the administration of DNAct and the radioactive probe because there was no significant difference using either DNAcp or DNAlp. Results show the mean ± SD of 3 independent experiments.
Poly(l-lysine)/DNAct complexes associate with blood cells in vivo
| Time . | pLL20/DNAct . | pLL211/DNAct . | DNAct . |
|---|---|---|---|
| 1 min | 91.4 ± 5.7 | 92.0 ± 5.3 | 4.6 ± 0.1 |
| (83.4 ± 3.7) | (94.0 ± 2.9) | (75.0 ± 1.2) | |
| 10 min | 85.6 ± 7.2 | 95.5 ± 1.0 | 3.5 ± 0.1 |
| (17.4 ± 5.5) | (66.0 ± 5.3) | (5.0 ± 0.9) |
| Time . | pLL20/DNAct . | pLL211/DNAct . | DNAct . |
|---|---|---|---|
| 1 min | 91.4 ± 5.7 | 92.0 ± 5.3 | 4.6 ± 0.1 |
| (83.4 ± 3.7) | (94.0 ± 2.9) | (75.0 ± 1.2) | |
| 10 min | 85.6 ± 7.2 | 95.5 ± 1.0 | 3.5 ± 0.1 |
| (17.4 ± 5.5) | (66.0 ± 5.3) | (5.0 ± 0.9) |
pLL20/DNAct, pLL211/DNAct complexes or DNActwere administered intravenously to female Balb/c mice (0.1 mL of 10 μg/mL DNA). The mice were killed after 1 and 10 minutes, and the blood was sampled and immediately spun at 5000 rpm at 4°C to pellet the blood cells. Pellet and supernatant were assayed for32P-levels, and the results show the percentage of radioactivity present in the blood associating with the cell pellet. Numbers in brackets indicate the percentage of the original32P dose remaining in the blood at the time of sampling. Results show the mean ± SD of 3 independent experiments. Similar results were obtained using pLL/DNAcp complexes (data not shown).
For abbreviations, see Table 1.
Effect of salt on the size of pLL/DNA complexes
pLL/DNA complexes formed in water at a 2.8:1 ± charge ratio measured 50 to 60 nm in diameter irrespective of pLL molecular weight (pLL20/DNAct = 54.1 ± 1.5 nm; pLL211/DNAct = 61.9 ± 7.2 nm). Figure2 shows that the addition of physiological levels of NaCl (150 mM) to pLL20/DNAcp complexes resulted in significant increases in the diameter of the complexes over time, with sharp increases observed after only 2 minutes. This increase in size was probably due to hydrophobic aggregation of the complexes as a result of increased solvent polarity. In contrast, pLL211/DNAcp complexes displayed significantly smaller increases in diameter on the addition of 150 mM NaCl, suggesting that these complexes may be more stable under physiological conditions. Similar results were obtained using pLL/DNAct complexes.
pLL20/DNA complexes display increased aggregation in salt compared to pLL211/DNA.
pLL/DNAcp complexes were analyzed for light scattering on a Zetasizer photon correlation spectroscope in the presence or absence of 150 mM NaCl. pLL20/DNAcp (●, +NaCl; ♦, −NaCl); pLL211/DNAcp (▴, +NaCl; ▪, −NaCl); note that ♦ and ▪ are superimposed and therefore indistinguishable. Results show the mean ± SD of 3 independent experiments. Similar results were obtained using pLL/DNActcomplexes (data not shown).
pLL20/DNA complexes display increased aggregation in salt compared to pLL211/DNA.
pLL/DNAcp complexes were analyzed for light scattering on a Zetasizer photon correlation spectroscope in the presence or absence of 150 mM NaCl. pLL20/DNAcp (●, +NaCl; ♦, −NaCl); pLL211/DNAcp (▴, +NaCl; ▪, −NaCl); note that ♦ and ▪ are superimposed and therefore indistinguishable. Results show the mean ± SD of 3 independent experiments. Similar results were obtained using pLL/DNActcomplexes (data not shown).
Association of pLL/DNA complexes with mouse blood cells in vivo
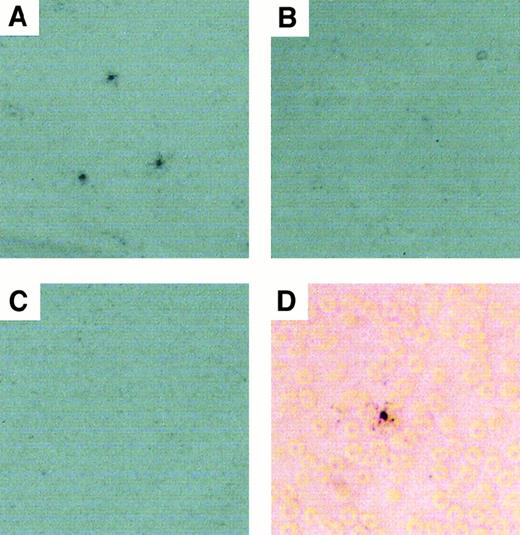
Recent investigations have shown that polycation-DNA complexes bind to erythrocytes in vitro23; therefore, we examined whether the degree of binding of pLL/DNA complexes to blood cells in vivo might have contributed to their differential removal from the blood. The results in Table 2 show that pLL20/DNAct and pLL211/DNAct complexes bind to blood cells in vivo, whereas free DNA does not. Autoradiography of blood smears taken from mice 1 minute after the administration of pLL/DNA complexes show that the pLL20/DNAct complexes (Figure3A) produced a punctate distribution of radioactivity, suggesting that the complexes might have aggregated in the blood. In contrast, neither pLL211/DNActcomplexes (Figure 3B) nor DNA (Figure 3C) displayed punctate distribution of radioactivity, indicating that pLL211/DNAct complexes might have been relatively soluble within the blood. Closer examination of the smears confirms that pLL20/DNAct complexes associated with erythrocytes in vivo (Figure 3D), with no evidence of white blood cell association (data not shown). Radioactive quantification of the blood smears by PhosphorImager (Molecular Dynamics) analysis showed all smears to contain comparable amounts of radioactivity (data not shown). Similar results were obtained using pLL/DNAcpcomplexes.
pLL20/DNA complexes, but not pLL211/DNA, show punctate distribution of radioactivity in blood smears, and both types of complexes bind erythrocytes.
pLL20/DNAct or pLL211/DNAct complexes were administered intravenously to female Balb/c mice (0.1 mL of 10 μg/mL DNA). Blood was sampled from the end of the tail after 1 minute and smeared onto glass slides. Radioactivity in the smear was developed by exposure of the slides to liquid photographic emulsion and viewed by light microscopy. (A) pLL20/DNAct (80 × magnification); (B) pLL211/DNAct (80 × magnification); (C) DNA alone (80 × magnification); (D) pLL20/DNAct (400 × magnification). Similar results were obtained using pLL/DNAcp complexes (data not shown).
pLL20/DNA complexes, but not pLL211/DNA, show punctate distribution of radioactivity in blood smears, and both types of complexes bind erythrocytes.
pLL20/DNAct or pLL211/DNAct complexes were administered intravenously to female Balb/c mice (0.1 mL of 10 μg/mL DNA). Blood was sampled from the end of the tail after 1 minute and smeared onto glass slides. Radioactivity in the smear was developed by exposure of the slides to liquid photographic emulsion and viewed by light microscopy. (A) pLL20/DNAct (80 × magnification); (B) pLL211/DNAct (80 × magnification); (C) DNA alone (80 × magnification); (D) pLL20/DNAct (400 × magnification). Similar results were obtained using pLL/DNAcp complexes (data not shown).
Intrahepatic distribution of pLL/DNA complexes in mice in vivo
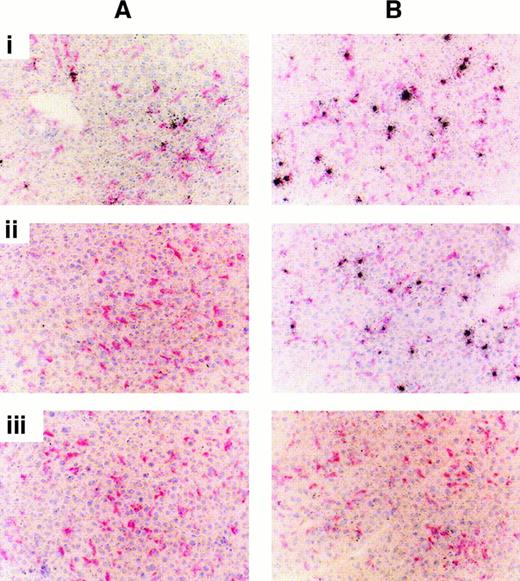
The liver is the main organ of accumulation of pLL/DNA complexes injected intravenously in mice; therefore, we investigated the intrahepatic distribution of complexes to identify any cells that might have been responsible for their capture. Figure4 shows the distribution of pLL/DNAct complexes in mouse liver using autoradiography after 1-day (Figure 4A) or 7-day (Figure 4B) exposure of the slides to the photographic emulsion. PLL20/DNActcomplexes (Figure 4Ai) showed punctate distribution of radioactivity within the liver after 1-day exposure, associating predominantly with Kupffer cells, whereas pLL211/DNAct complexes (Figure 4Aii) did not give a detectable signal. This is unlikely to be due to less radioactivity on slides prepared from livers of mice treated with pLL211/DNAct complexes, because PhosphorImager (Molecular Dynamics) analysis indicates that the levels of radioactivity are approximately 70% of liver slides from mice treated with pLL20/DNAct complexes. The punctate appearance of radioactivity in the liver sections was most likely due to the presence of aggregates leading to “hot spots.” Seven-day exposure of liver sections from mice administered pLL20/DNAct (Figure 4Bi) or pLL211/DNAct (Figure 4Bii) complexes resulted in punctate distribution of the radioactivity and, in both cases, associated predominantly with Kupffer cells. Livers from mice treated with pLL20/DNAct complexes displayed “hot spots” with some high-intensity foci, thought to correspond with those detected after 1 day. The type of DNA used in the pLL/DNA complexes had no effect on their intrahepatic distribution, with both pLL20/DNAcp and pLL211/DNAcp complexes displaying the same radiographic signal intensities as their respective pLL/DNAct ones (data not shown). DNA alone (Figure 4Biii) was washed from the tissue sections during the autoradiographic development process, though 85% of DNA was found in the liver after 30 minutes (Table 1).
pLL20/DNA and pLL211/DNA complexes associate with Kupffer cells in mice.
pLL20/DNAct or pLL211/DNAct complexes were administered intravenously to female Balb/c mice (0.1 mL of 10 μg/mL DNA). Liver sections were stained with hematoxylin (blue/purple nuclei) and the macrophage marker F4/80 (pink/red cytoplasm), and the radioactivity was viewed by exposure of the slides to liquid photographic emulsion for (A) 1 day or (B) 7 days. (i) pLL20/DNAct; (ii) pLL211/DNAct; (iii) DNA alone (note: DNA is washed from the slides during the development process). Similar results were obtained using pLL/DNAcp complexes (data not shown).
pLL20/DNA and pLL211/DNA complexes associate with Kupffer cells in mice.
pLL20/DNAct or pLL211/DNAct complexes were administered intravenously to female Balb/c mice (0.1 mL of 10 μg/mL DNA). Liver sections were stained with hematoxylin (blue/purple nuclei) and the macrophage marker F4/80 (pink/red cytoplasm), and the radioactivity was viewed by exposure of the slides to liquid photographic emulsion for (A) 1 day or (B) 7 days. (i) pLL20/DNAct; (ii) pLL211/DNAct; (iii) DNA alone (note: DNA is washed from the slides during the development process). Similar results were obtained using pLL/DNAcp complexes (data not shown).
Activation of complement by pLL/DNA complexes in mouse serum in vitro
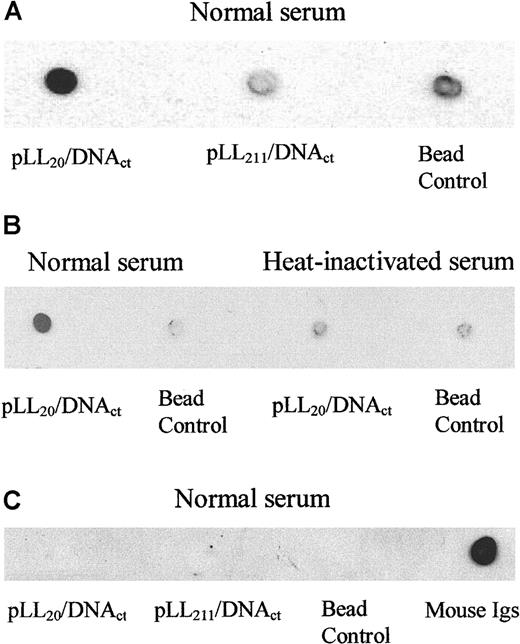
Complement activation by pLL/DNA complexes might have been a contributory factor for their rapid removal from the circulation of mice; therefore, we investigated the binding of complement protein C3 to pLL/DNA complexes incubated in mouse serum in vitro. Figure5A shows, by dot blotting, that pLL20/DNAct complexes bound C3, whereas pLL211/DNAct complexes did not. Binding of C3 to the pLL20/DNAct complexes could be inhibited by heat treatment of the serum (Figure 5B), indicating that C3 binding was due to activation of complement and not nonspecific binding. Association of C3 with pLL20/DNAct complexes might have been due to its interaction with the polycation, because pLL20 alone also binds C3. Similar results were obtained using pLL/DNA complexes formed at 0.8:1, 1:1, 1.2:1, 1.5:1, and 2:1 charge ratios (data not shown), showing that though complement C3 bound to pLL20 alone, it also bound to pLL20/DNA complexes. Both pLL20/DNA and pLL211/DNA complexes were equally bound to the magnetic streptavidin beads, and less than 1% of the radiolabeled probe associated with the serum supernatant and subsequent washes in all samples (data not shown). There is no evidence of immunoglobulin binding to pLL20/DNAct complexes (Figure 5C), suggesting that complement activation was through the alternative complement pathway. Similar results were obtained using pLL/DNAcp complexes.
pLL20/DNAct complexes, but not pLL211/DNA, fix mouse complement C3 independently of immunoglobulins.
Biotin-pLL20/DNAct or biotin-pLL211/DNAct complexes were immobilized on streptavidin magnetic beads and incubated in mouse serum at 37°C for 5 minutes. Beads were isolated, washed in isotonic buffer, and boiled in Laemelli buffer to isolate the bound proteins. Dot blots were prepared on nitrocellulose membrane, and proteins were identified by antibody blotting and radiography. (A) C3 binding to pLL/DNAct complexes in normal mouse serum. (B) C3 binding to pLL20/DNAct complexes in normal or heat-inactivated serum. (C) Total mouse immunoglobulins binding to pLL/DNAct complexes in normal serum. Bead control represents serum-treated beads. Similar results were obtained using pLL/DNAcp complexes (data not shown).
pLL20/DNAct complexes, but not pLL211/DNA, fix mouse complement C3 independently of immunoglobulins.
Biotin-pLL20/DNAct or biotin-pLL211/DNAct complexes were immobilized on streptavidin magnetic beads and incubated in mouse serum at 37°C for 5 minutes. Beads were isolated, washed in isotonic buffer, and boiled in Laemelli buffer to isolate the bound proteins. Dot blots were prepared on nitrocellulose membrane, and proteins were identified by antibody blotting and radiography. (A) C3 binding to pLL/DNAct complexes in normal mouse serum. (B) C3 binding to pLL20/DNAct complexes in normal or heat-inactivated serum. (C) Total mouse immunoglobulins binding to pLL/DNAct complexes in normal serum. Bead control represents serum-treated beads. Similar results were obtained using pLL/DNAcp complexes (data not shown).
Intrahepatic distribution of complement receptor 3 positive cells after administration of pLL/DNA complexes in mice
The complement receptor 3 (CR3) receptor is expressed on leukocytes and is involved in phagocytosis and cell motility. In normal liver, CR3 is differentially expressed on at least 2 subpopulations of liver cells (Figure 6Aiii). The first expresses low levels of CR3 and is normally present throughout the parenchyma; this has been identified as tissue resident macrophages.24 The second cell population expresses high levels of CR3 (hereafter termed CR3h cells) and is present in low numbers within the parenchyma. PLL20/DNAct (Figure 6Ai) and pLL211/DNAct (Figure 6A ii) complexes both associate predominantly with cells expressing low levels of CR3 (ie, Kupffer cells), though there is some association with CR3hcells, and both types of complexes led to higher levels of CR3h cells compared with the control. Quantitative cell analysis in liver sections (Figure 6B) showed that the administration of pLL20/DNAct complexes led to the highest number of CR3h cells. This suggests that the pLL component of these complexes stimulated the immune system because the levels of pLL20/DNAct and free DNA within the liver were comparable (Table 1), yet the number of CR3h cells was 50% lower when DNA alone was administered. Lower levels of CR3hcells were seen in liver sections of mice administered pLL20/DNAcp and pLL211/DNA complexes, possibly due to the reduced amounts of these complexes within the liver compared to pLL20/DNActcomplexes. Mice administered either pLL211/DNAct or pLL211/DNAcp complexes showed no increase in CR3h cells compared to DNA alone (Figure 6B).
pLL20/DNA complexes lead to increased levels of CR3-expressing cells within the liver of mice.
Method as for Figure 4. Briefly, mice were injected intravenously with pLL/DNA complexes, and liver sections were stained using antimouse CR3 antibody (nuclei, blue; CR3, pink). (A) pLL20/DNAct (i); pLL211/DNAct (ii); buffer alone (iii). (B) Average number of cells per field of view (200 × magnification) expressing high levels of CR3 pLL20/DNAct (1), pLL20/DNAcp (2), pLL211/DNAct (3), pLL211/DNAcp (4), DNA alone (5), or buffer alone (6).
pLL20/DNA complexes lead to increased levels of CR3-expressing cells within the liver of mice.
Method as for Figure 4. Briefly, mice were injected intravenously with pLL/DNA complexes, and liver sections were stained using antimouse CR3 antibody (nuclei, blue; CR3, pink). (A) pLL20/DNAct (i); pLL211/DNAct (ii); buffer alone (iii). (B) Average number of cells per field of view (200 × magnification) expressing high levels of CR3 pLL20/DNAct (1), pLL20/DNAcp (2), pLL211/DNAct (3), pLL211/DNAcp (4), DNA alone (5), or buffer alone (6).
pLL/DNA complex behavior in female Wistar rats
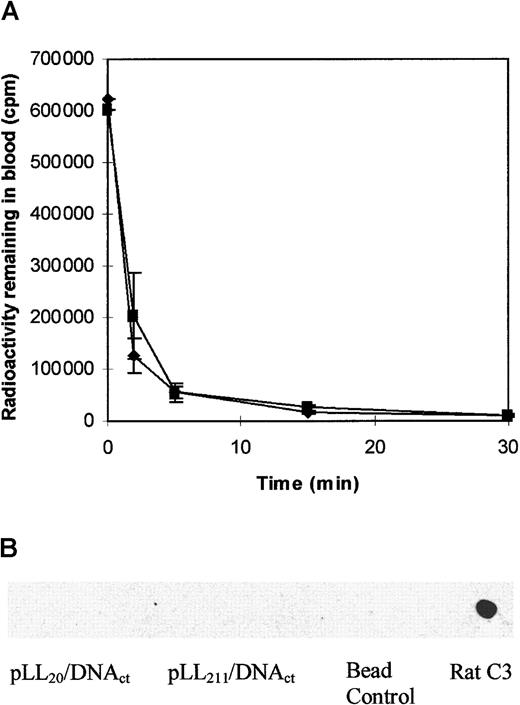
To assess whether the effect of pLL molecular weight on the circulation of pLL/DNA complexes was reproducible between species, we administered pLL20/DNAct or pLL211/DNAct complexes intravenously to female Wistar rats. Figure 7A shows that both pLL20/DNAct and pLL211/DNAct complexes were rapidly cleared from the bloodstream within minutes, and in both cases blood clearance was faster than it was in mice (data not shown). Additionally, erythrocyte toxicity was inferred for both complexes by the presence of hemoglobin in the urine at the time of killing. Rats treated with buffer or DNA alone showed no signs of hematuria. Figure 7B shows that rat complement C3 did not bind to either pLL20/DNAct or pLL211/DNAct complex.
pLL20/DNAct and pLL211/DNAct complexes are rapidly removed from the circulation of female Wistar rats but do not bind complement C3 in vitro.
(A) pLL20/DNAct (♦) or pLL211/DNAct (▪) complexes were administered intravenously to female Wistar rats (1.0 mL of 10 μg/mL DNA), and blood was taken at specified intervals from the end of the tail. Results show the mean ± SD of 3 independent experiments. (B) Rat complement C3 binding to pLL/DNAct complexes incubated in rat serum in vitro. Method as for Figure 5. Bead control represents serum-treated beads.
pLL20/DNAct and pLL211/DNAct complexes are rapidly removed from the circulation of female Wistar rats but do not bind complement C3 in vitro.
(A) pLL20/DNAct (♦) or pLL211/DNAct (▪) complexes were administered intravenously to female Wistar rats (1.0 mL of 10 μg/mL DNA), and blood was taken at specified intervals from the end of the tail. Results show the mean ± SD of 3 independent experiments. (B) Rat complement C3 binding to pLL/DNAct complexes incubated in rat serum in vitro. Method as for Figure 5. Bead control represents serum-treated beads.
Effect of pLL20/DNAct and pLL211/DNAct complexes in human blood in vitro
To predict the behavior of pLL/DNA complexes in humans, we investigated the interactions with human blood of pLL20/DNAct or pLL211/DNAct complexes. Figure8A shows that neither complex caused hemolysis in human blood, whereas it was apparent in rat blood. Figure8Bi shows that human complement C3 bound to both pLL20/DNAct and pLL211/DNAct complexes and that this binding was specific because it could be blocked by heat-inactivation of the serum (Figure 8Bii). Additionally, Figure 8C shows that IgG associated with both types of pLL/DNA complex (IgA, IgD, IgE, and IgM do not bind; data not shown), suggesting that complement activation may be through the classical pathway. Similar results were obtained using pLL/DNAcp complexes.
pLL20/DNAct and pLL211/DNAct complexes fix human complement C3 and bind IgG but are not hematotoxic in vitro.
(A) Hematotoxicity of pLL20/DNAct (■) or pLL211/DNAct (▪) complexes was determined by their incubation in human or rat blood at 37°C for 1 hour. Blood cells were removed by centrifugation, and the presence of hemoglobin in the supernatant was measured by absorbance at 550 nm. (B) Binding of complement protein C3 to pLL/DNA complexes incubated in normal (i) or heat-inactivated human serum (ii). Method as for Figure 5. (C) Binding of IgG to pLL/DNA complexes incubated in normal human serum.
pLL20/DNAct and pLL211/DNAct complexes fix human complement C3 and bind IgG but are not hematotoxic in vitro.
(A) Hematotoxicity of pLL20/DNAct (■) or pLL211/DNAct (▪) complexes was determined by their incubation in human or rat blood at 37°C for 1 hour. Blood cells were removed by centrifugation, and the presence of hemoglobin in the supernatant was measured by absorbance at 550 nm. (B) Binding of complement protein C3 to pLL/DNA complexes incubated in normal (i) or heat-inactivated human serum (ii). Method as for Figure 5. (C) Binding of IgG to pLL/DNA complexes incubated in normal human serum.
Discussion
Understanding the factors regulating differential circulation of polyelectrolyte DNA complexes in the bloodstream may provide insights into clearance mechanisms and possible means to evade them. Complexes formed using high- and low-molecular-weight pLL show quite different circulation properties in mice, and one clear morphologic difference between them is the appearance of a punctate distribution of radioactivity in the blood using pLL20/DNA complexes not apparent using pLL211/DNA. Although the precise reasons for this distribution are unclear, the observation of physiological salt-induced aggregation of pLL20/DNA complexes in vitro suggests that these complexes may have relatively poor solubility in blood. We have also shown that pLL20/DNA complexes bind erythrocytes in vivo, and it is possible that this, along with poor solubility, results in the punctate radioactivity distribution observed in the blood smears.
We have also shown specific binding of complement protein C3 to pLL20/DNA, but not pLL211/DNA, complexes in mouse serum. The influence of molecular weight of polycations on complement activation by pLL and pLL/DNA complexes has been reported previously. Siegal et al19 have shown, in acute-phase human serum in vitro, that low-molecular-weight pLL (4 and 23 kd) activates complement significantly more than high-molecular-weight pLL (70 kd). In contrast, in normal serum, complement is activated independently of the molecular weight of pLL used. Additionally, they showed that in both acute-phase and normal human serum, complement activation is mediated by the acute-phase C-reactive protein (CRP). However, we have found that CRP does not associate with pLL/DNA complexes incubated in normal human serum (data not shown), suggesting that either CRP is not involved in complement activation or that, more likely, CRP does not associate with the pLL/DNA complex after complement activation. Plank et al18have also shown that pLL activates complement in normal human serum in vitro, but, in their study, high-molecular-weight pLL (approximately 50 kd) and corresponding pLL/DNA complexes were shown to activate complement to a greater extent than low-molecular-weight pLL (1, 4, 6, and 25 kd). The apparent contradiction between these 2 studies may reflect the different methods used to determine complement activation, with Siegal et al19 using a direct measure of complement protein depletion and Plank et al18 using an indirect sensitized sheep erythrocyte hemolysis assay. Despite the discrepancies between these 2 studies, which were both performed with human and not mouse serum, they do show that complement is readily activated by pLL and that molecular weight can influence the level of activation achieved. Additionally, we show that complement activation may be through different complement pathways in mouse and human serum. Although the exact mechanisms of complement activation require further investigation, the binding of IgG (but not other immunoglobulins) to pLL/DNA complexes in human (but not mouse serum) suggests the presence of antibody-mediated immunity in normal human serum.
After intravenous injection to mice, pLL20/DNA complexes accumulate within the liver where they associate with macrophages (Kupffer cells). Kupffer cell–associated complement receptor 3 (CR3) is the major pathway for clearance of C3-opsonized particles from the mouse bloodstream,25 and it is possible that C3-opsonized pLL20/DNA complexes may follow this route. Administration of pLL20/DNA complexes also led to a rapid increase in the number of cells within the liver expressing high levels of CR3 (hereafter termed CR3h cells), with fewer CR3hcells being induced by DNA alone or by pLL211/DNA complexes. This is perhaps a reflection of the differential accumulation of polyelectrolyte complexes within the liver (Table 1). However, it is also possible that pLL211/DNA complexes may exhibit reduced interaction with the immune system, exemplified by their inability to mediate complement activation in mouse serum in vitro, resulting in lower numbers of CR3h cells. CR3h cells may be leukocytes infiltrating from the circulation—eg, monocytes24 or natural killer cells26—whose expression of CR3 is associated with phagocytosis and cell motility.26,27 Alternatively, CR3h cells may be stimulated Kupffer cells.28Precise identification of CR3h cells has not been determined; however, the infiltration of CR3-expressing cells and the up-regulation of CR3 on phagocytic cells are associated with immune activation,29 especially by complement-mediated events.30-32 Hence, it is feasible that the increase in CR3h cells within the liver follows the interaction of pLL20/DNA complexes with components of the immune system. Although there is no direct evidence that complement activation by pLL20/DNA complexes leads directly to Kupffer cell capture and increased CR3h cells, it is clear that the immune system becomes provoked by these complexes in some way. PLL211/DNA complexes are eventually captured by Kupffer cells, albeit more slowly than pLL20/DNA complexes, but the mechanisms involved in their removal are unclear.
The properties of polyelectrolyte complexes formed using pLL20 are influenced by the DNA used. For example, pLL20/DNAcp complexes display extended circulation times in mice compared to pLL20/DNAct or pLL20/DNAlp complexes. Using circular dichroism, it has been shown that polyelectrolyte complexes formed using circular or linear plasmid DNA exhibit different topologies, with linear DNA forming less tightly organized structures than circular DNA.33 The greater circulation time of complexes containing the circular DNA may reflect this phenomenon. The same effect is not observed using pLL211, probably because the larger polycation enhances the stability of both types of DNA complex.
The body distribution of pLL/DNA complexes in mice appears to correlate with the circulation half-life of the vector, with pLL211/DNA and pLL20/DNAcpcomplexes exhibiting increased carcass accumulation and decreased liver accumulation compared to DNA alone. Decreased liver accumulation of these complexes may reflect reduced Kupffer cell capture, whereas increased accumulation within the carcass is most likely due to phagocytic capture of the complexes or trapping within capillary beds. All pLL/DNA complexes display decreased accumulation within the kidneys compared to DNA alone, suggesting that DNA within pLL/DNA complexes is protected from degradation and subsequent excretion. It is interesting to note that all pLL/DNA complexes displayed significant levels within the spleen, whereas DNA alone exhibited very low levels (0.4% ± 0.1%). Thus it appears that mechanisms within the spleen may be involved in pLL recognition and capture.
PLL20/DNAct complexes showed high levels of accumulation within the liver and low levels within the carcass, both similar to the levels obtained using free DNA. It is unlikely that the radiolabeled DNA probe within pLL20/DNActcomplexes becomes dissociated from the pLL because more than 99% of the probe was retrieved intact after incubation in blood or serum (data not shown). In addition, pLL20/DNAct complexes bound blood cells in vivo whereas DNA alone did not (see Table 2), and liver-captured radioactivity of pLL20/DNAct(and also pLL211/DNAct complexes) was not eluted during the processing of liver sections, as is the case for free DNA (Figure 3).
Destabilization of pLL20/DNA complexes has been described as a possible mechanism for the removal of polyelectrolyte complexes from the circulation.13 17 Therefore, we hypothesized that incubation of pLL20/DNAct complexes in mouse blood might result in increased degradation of the DNA contained within the complexes compared with pLL211/DNActcomplexes. Incubation of pLL/DNA complexes formed using either pLL20 or pLL211 and either DNAct or DNAcp in 100% serum or blood (mouse, rat, or human) resulted in negligible DNA degradation of the radiolabeled probe in all of the samples, whereas free DNA was substantially degraded (data not shown). This finding suggests that both types of pLL/DNA complexes protect the DNA from degradation by serum nucleases. Therefore, it seems unlikely that destabilization of pLL/DNA complexes, leading to DNA degradation or capture, is an important factor mediating their rapid removal from the blood.
Intravenous administration of pLL/DNAct complexes to Wistar rats resulted in rapid blood clearance, with signs of hematuria. The hematuria may be mediated by direct erythrocyte lysis, demonstrable by both types of complex in rat blood in vitro, and this may also contribute to their rapid clearance in vivo. High doses of DNA vectors have been used in mice to increase transgene expression,34and it is possible that a high dose in rats would lead to considerable hematuria. Neither pLL20/DNAct nor pLL211/DNAct complexes fixed complement in rat serum in vitro. The different behaviors of pLL/DNA complexes in the 2 in vivo rodent models are surprising, though differences in their behavior in vitro are equally striking. Indeed, with careful development, it may be possible to predict behavior in vivo from a relatively small number of in vitro tests. The ability to do this would decrease our reliance on in vivo animal testing8,16,17,23,34 and provide a constructive strategy to address the question of which animal model is the most appropriate for predicting pharmacokinetics of gene delivery vectors in humans.35 At present, in vitro evaluation of polycation-DNA complexes is usually tailored toward optimization of the vector for specific animal models rather than for the prediction of its behavior in humans. Therefore, a range of in vitro tests, including ones described in this and other studies,18 19 may ultimately be preferable to rodent models for predicting the behavior of polyelectrolyte complexes in humans. After an early assessment using currently available techniques, we predict that the administration of either pLL20/DNA or pLL211/DNA complexes to humans may not result in erythrocyte toxicity; however, the complement activation exhibited by these complexes in vitro may also be found in vivo, potentially leading to rapid removal of the complexes from the circulation.
Overall, this study raises important questions for the use of simple polyelectrolyte complexes in vivo and of the use of rodent models in such research. It is clear that liver phagocytes play a major role in the capture of pLL/DNA complexes in mice, and it is likely that such a mechanism exists in humans. We have also shown that complement activation, immune stimulation, or both by pLL20/DNA complexes may be involved in their decreased circulation half-life in mice. Therefore, if pLL/DNA complexes are to be useful as systemic gene therapy vectors in humans, phagocytic capture and immune provocation must be avoided.
We thank Dr Mark Drayson for his advice.
Supported by the Cancer Research Campaign and the Biotechnology and Biological Sciences Research Council.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christopher M. Ward, Paterson Institute for Cancer Research, Christie Hospital NHS Trust, Wilmslow Rd, Manchester, M20 4BX, United Kingdom; e-mail: wardcm@hotmail.com.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal