Abstract
Patient–tumor-specific oligonucleotides were generated for the detection of minimal residual disease (MRD) in a highly specific and sensitive clonotypic polymerase chain reaction (cPCR). The clone-specific region of highest diversity, CDR-III, was PCR amplified and sequenced. Nested CDR-III clonotypic primers were used in a semi-nested cPCR with a sensitivity of at least 1 in 105cells. Patients with protocol-eligible Rai intermediate or high-risk chronic lymphocytic leukemia (CLL) received induction with fludarabine 25 mg/m2 per day for 5 days every 4 weeks for 6 cycles, followed by consolidative high-dose cyclophosphamide (1.5, 2.25, or 3g/m2). cPCR was performed on peripheral blood and bone marrow mononuclear cells. All 5 patients achieving a clinical partial remission (PR) studied by cPCR were positive. Five patients achieved nodular PR (nPR) (residual nodules or suspicious lymphocytic infiltrates in a bone marrow biopsy as the sole suggestion of residual disease). Five of 5 patients with nPR were cPCR positive. In contrast, flow cytometry for CD5–CD19 dual staining and κ–λ clonal excess detected MRD in only 3 of the same 5 nPR patients, all of whom were cPCR positive, and immunohistochemistry detected MRD in only 1 of 4 assessable patients. Three of 7 CR patients evaluable by cPCR had MRD. Only 1 CR patient had MRD by flow cytometry; that patient was also cPCR positive. These data support the conclusions that nodular PR in CLL represents MRD and that clonotypic PCR detects MRD in CLL more frequently than flow cytometry or immunohistochemistry.
Introduction
Cancer patients with minimal residual disease (MRD) are in clinical complete remission but still have occult disease whose regrowth can result in relapse. During the occult phase of the illness, detection may be possible by molecular testing, which is far more sensitive than physical examination, radiologic study, or conventional histologic examination. Detecting MRD affords an opportunity to identify patients who will have relapses despite their apparent cancer-free state. These patients can be offered further potentially curative therapy, while those who are truly cured can be spared the toxicity associated with additional treatment.
Tumor-characteristic chromosomal breakpoints can be used as highly sensitive and specific markers of MRD, though these approaches have limited applicability for lymphoid neoplasms. Traditional Southern blot analysis used in most diagnostic laboratories detects a malignant population only when it exceeds 1% to 5% of the background nonmalignant cells.1 Although the polymerase chain reaction (PCR) is at least 1000-fold more sensitive, not all lymphoid tumors have recurrent amplifiable translocations. Even for tumors with well-characterized recurrent translocations such as follicular lymphoma, only 70% of t(14;18) translocations identified by cytogenetics can be detected by PCR using oligonucleotides spanning the translocation breakpoint cluster regions.2-4 In the remaining cases the translocation breakpoints lie outside the amplifiable regions. Moreover, in some tumors, such as diffuse large cell lymphoma in which the t(14;18) translocation is present in only 25% of patients, restricting analysis to amplifiable cases introduces potential biologic bias. A better assay would be directed at a tumor marker universally present in the lymphomas.
The B and T cell malignancies are unusual cancers in that the clonal population can be identified by the rearrangement of the immunoglobulin genes (reviewed in Sklar and Longtime5) or T-cell receptor unique to that clone. In pre-B cells the immunoglobulin heavy chain (IgH) locus undergoes sequential rearrangement of diversity (D) and joining (J) genes followed by rearrangement of the variable (V) and D genes, which results in an intact IgH variable segment. This is a direct result of the recombination event occurring in the pre-B cell that brings together distant parts of the genome, markedly increasing the diversity of the antigen recognition regions. When a malignant cell expands, all the progeny carry and express the same immunoglobulin or T cell receptor as the parent cell, with minor, if any, mutations. (An exception occurs in acute lymphoblastic leukemia, in which further V-D or D-J recombination may occur.)6-10 Because the same clone can be demonstrated at both diagnosis and relapse of recurrent low- and intermediate-grade lymphoma,2 11-13 the malignant clone persists after treatment during the phase of occult disease.
PCR strategies for B and T cell neoplasms depend on this genomic recombination event to bring together the target sequences within sufficient proximity, allowing successful amplification of the region defined by the primer pair. For B cells this involves the amplification of the V-D-J rearrangement using consensus V region primers and a downstream primer in the JH region.4Amplification and isolation of the clonal V-D-J rearrangement allows for sequencing of the clone-specific region composed of nontemplated N sequences and the DH segment (N-DH-N), referred to as complementarity determining region-III (CDR-III).5This region is ideally suited for strategies that detect a specific clone of B cells because the nontemplated N sequences inserted by terminal deoxytidyl-transferase are random and increase the diversity of the area above that of the recombination event alone. We have designed a clonotypic PCR using nested 5′ patient specific primers and consensus JH primers to uniquely amplify the malignant clone for the detection of MRD.
In the current analysis, clonotypic PCR of peripheral blood (PB) and bone marrow (BM) mononuclear cells was used to assess patients with intermediate or high-risk chronic lymphocytic leukemia (CLL) treated on an ongoing clinical trial to determine whether molecular evidence of disease can be used as a surrogate marker. In 1988 the National Cancer Institute-sponsored Working Group (NCI-WG) on CLL specified that patients in complete remission (CR) had to have less than 30% lymphocytes on bone marrow aspiration, and it recommended that the clinical significance of lymphoid nodules in CR patients be assessed prospectively.14 Subsequently, it was demonstrated that patients who were in CR according to aspirate criteria and physical examination but who were found on bone marrow biopsy to have either interstitial involvement or lymphoid nodules had inferior progression-free survival compared to those in CR.15 In 1996 a revision of the NCI-WG guidelines defined this subgroup as having nodular partial remission (nPR), stating “It is, unfortunately, difficult with current techniques to determine the clonality of these nodules.”16 In our trial, a number of patients achieved nPR. Clonotypic PCR demonstrated that these patients have MRD.
Patients, materials, and methods
Patient characteristics
Between August 1992 and April 1996, 35 patients with intermediate- and advanced-stage CLL, as defined by the 3-stage Rai system, were enrolled in an institutional review board–approved Memorial Sloan-Kettering Cancer Center protocol number 92-78.17 Eligibility, treatment plans, and clinical results have been described elsewhere. Eligible patients had lymphadenopathy, hepatomegaly or splenomegaly, anemia or thrombocytopenia. A circulating lymphocytosis level of at least 5000 cells/μL and a bone marrow aspirate with lymphoid cells constituting at least 30% of all nucleated cells were required. Seven patients were previously treated, and the remaining patients were untreated. Study therapy consisted of induction with fludarabine 25 mg/m2per day for 5 days every 4 weeks for 6 cycles followed by consolidation with high-dose cyclophosphamide at one of 3 dose levels (1.5, 2.25, or 3 g/m2). Complete remission (CR) was defined by normal results of physical examination, normal complete blood count, bone marrow aspirate with less than 30% lymphocytes, and morphologically normal bone marrow biopsy. Nodular PR was defined by a bone marrow biopsy with residual nodules in the absence of other disease. Patients were prospectively studied for MRD as part of the study protocol. The study was approved by the institutional review board of the Memorial Sloan-Kettering Cancer Center, and all patients gave informed consent.
Clinical specimens
DNA extraction for PCR amplification of IgH genes was performed on pretreatment specimens that consisted of diagnostic lymph node biopsies, involved bone marrow, or peripheral blood with a circulating lymphocytosis. All lymph node and bone marrow biopsy specimens were reviewed by MSKCC hematopathologists. During protocol treatment, peripheral blood and bone marrow were obtained for analysis after 3 and 6 cycles of fludarabine and after 3 cycles of high-dose cyclophosphamide. Bone marrow aspirates were collected in EDTA for DNA extraction for PCR and in heparin sulfate for flow cytometry. When available, bone marrow was preferentially used in the analysis.
Mononuclear cells were isolated by Ficoll gradient and stored at −140°C in liquid nitrogen. DNA extraction was performed with proteinase K and was purified using a QIAamp spin column (QIAGEN, Chatsworth, CA). In 2 instances, follow-up samples of fresh bone marrow aspirates were unavailable, and DNA was extracted from bone marrow aspirates or paraffin section using octane for paraffin removal. The minute amount of material obtained precluded accurate determination of its concentration without losing the DNA. Therefore, results from these reactions were strictly qualitative.
Polymerase chain reaction amplification of rearranged IgH genes
The experimental design is illustrated in Figure1. The rearranged VH gene from B cell tumors was amplified, as previously described, using upstream primers based on a set of 6 family-specific VHleader primers18 in conjunction with a consensus JH primer.4 The latter was modified by the 5′ addition of a T7 sequence, facilitating sequencing and destabilizing a 5′-end loop that could interfere with the primer's performance. PCR reactions were carried out in 50-μL volumes containing 100 ng template DNA, 0.5 μM each of the 5′ and 3′ primers, 200 μM each dNTP, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, and 0.625 U Amplitaq (Perkin-Elmer Cetus, Norwalk, CT). The samples were overlain with 100 μL mineral oil and subjected to 37 cycles of denaturation (15 seconds at 95°C; first cycle, 3 minutes), annealing (15 seconds at 55°C; 30 seconds, first cycle), and elongation (15 seconds at 72°C; 30 seconds, first cycle) using the Perkin-Elmer thermal cycler. After the last cycle, a final elongation step for 7 minutes at 72°C was performed. PCR fragments (10 μL) were separated on 1.2% agarose gels in 90 mM Tris-borate–2 mM EDTA (TBE buffer) and visualized with ethidium bromide staining.
Clonotypic PCR technique.
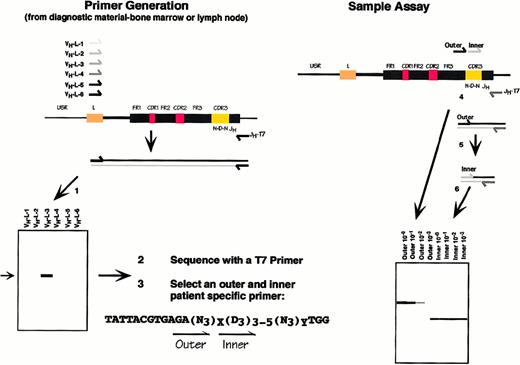
The CDR-III region is formed by the sequential D-JH and V-JH rearrangements in the pre-B cell. Both joins involve the addition of nontemplated “N” sequences and are therefore clone specific. (1) The V-D-J rearrangement is amplified by PCR using the family-specific VHL primers. A 550-bp product represents the rearranged VH allele. (2) The PCR product is sequenced directly, and primers are selected (3) from within the N-D-N region that are clonotypic. (4, 5) Samples are amplified in a semi-nested reaction and (6) are displayed on a high-resolution agarose gel.
Clonotypic PCR technique.
The CDR-III region is formed by the sequential D-JH and V-JH rearrangements in the pre-B cell. Both joins involve the addition of nontemplated “N” sequences and are therefore clone specific. (1) The V-D-J rearrangement is amplified by PCR using the family-specific VHL primers. A 550-bp product represents the rearranged VH allele. (2) The PCR product is sequenced directly, and primers are selected (3) from within the N-D-N region that are clonotypic. (4, 5) Samples are amplified in a semi-nested reaction and (6) are displayed on a high-resolution agarose gel.
In the event that this approach failed to reveal a monoclonal band, an alternative set of primers based on Fr2a and Fr3a consensus sequences were used with a downstream semi-nested pair of JH primers (LJH and VLJH), as previously described, with minor modifications.19-21 PCR conditions were identical to those above with the exception of the cycling conditions, as follows: an initial denaturation step at 95°C for 7 minutes, followed by 3 cycles of denaturation (45 seconds at 93°C), annealing (45 seconds at 50°C), and elongation (110 seconds at 72°C). After the last cycle, a final elongation step for 7 minutes at 72°C was performed. The second round of amplification with the semi-nested 3′ was carried out under the same conditions.
All experiments were run with a negative and a positive control. The former consisted of all the PCR reagents in the absence of a DNA template. The latter used alternative primer pairs for the bcl-2untranslated region (bcl-2 UT 5′ TCAGCCTTGAAACATTGATG and bcl-2 UT 3′ CAAGGTCAAAGGGACAACAG), which amplified under the same conditions as the immunoglobulin VH-family specific leader primers, resulting in a 450-bp fragment. The positive control verified the integrity of the DNA template. To prevent contamination, simultaneous amplification of a positive control template was not performed. PCR reactions were carried out in a dedicated hood into which amplified PCR products were never introduced.
Clone-specific primer design
Nested primer pairs unique to the malignant clone for the detection of MRD were created based on the sequence of the CDR-III region of the malignant clone (Figure 1). The initial PCR product reflecting the monoclonal immunoglobulin rearrangement was gel purified and directly sequenced using a Taq DNA polymerase-based sequencing strategy (Dye Terminator Cycle Sequencing Reaction Kit; Perkin Elmer, Foster City, CA) on an automated sequencer (ABI Prism, Foster City, CA). Bidirectional sequencing was performed for verification. The compatibility and expected performance characteristics of these primers with the appropriate downstream T7JH or VLJH primer were tested using a software package, Oligo 5.0 (LifeBiotechnologies, Plymouth, MN). Thermodynamic properties, including annealing temperatures, were assessed before implementation, increasing the likelihood that PCR using these primers would be successful.
Clonotypic polymerase chain reaction
Conditions for clonotypic PCR were identical to those used for isolating the initial V-D-J sequence with the exception of the annealing temperatures, which were modified for each primer pair based on the PCR predicted by Oligo 5.0. Primer pairs were first tested on 100 ng of the diagnostic sample from which the initial V-D-J was amplified. Ten-fold serial dilutions of a 100-ng template were made to estimate disease burden. In the first round of amplification, the outer 5′ CDR-III clone-specific primer was used. In the second round of amplification, 1 μL of a 10−3 dilution of the first-round PCR product served as a template with the nested CDR-III clone-specific primer. PCR products were run on a 4% Metaphor gel (FMC BioProducts, Rockland, ME) with a 10-bp ladder (Gibco BRL, Gaithersburg, MD). The predicted sizes of both products served to verify the specificity of the reaction products. Negative controls included a no DNA template and a polyclonal template reaction. The latter also served to test the specificity of the primers. Positive control reactions used the bcl-2 primers described above.
Flow cytometry
Surface membrane antigens were detected by standard direct immunofluorescence using the fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated monoclonal antibodies (mAbs) CD5-FITC and CD19-PE. Antibodies were purchased from Becton Dickinson (Mountain View, CA) and Coulter-Immunotech Immunology (Miami, FL). Dual-color staining was performed by incubating cells simultaneously with optimal concentrations of the 2 monoclonal antibodies of interest. Negative controls using irrelevant monoclonal immunoglobulins of the same isotype were analyzed concurrently. Surface immunoglobulin-bearing lymphocytes (sIg) were assayed using mouse F (ab)2anti-human CD19-PE/goat F(ab)2 anti-kappa FITC and mouse F (ab)2 anti-human CD19-PE/goat F(ab)2 anti-λ FITC (Biosource International, Camarillo, CA). Flow cytometry of the stained cells was performed on an EPICS Profile (Coulter) equipped with a 488-nm argon laser. The target lymphocyte population was gated according to forward and side scatter characteristics of the cells. Multiparameter Data Acquisition and Display System (Coulter Electronics) was used to acquire, display, and store output from the EPICS.
Immunologic detection of MRD was accomplished by dual-color staining of CD5–CD19 or clonal excess in defined CD19+populations.22 23 CD5–CD19 positivity in more than 5% of the cells (representing the nonspecific staining background) was considered positive for MRD. Clonal excess was defined as a κ–λ ratio greater than 5.0 or a λ–κ ratio greater than 3.0 in the CD19+ lymphocyte gate. Only one of the immunologic criteria was required to consider a sample positive for MRD.
Bone marrow aspirates and peripheral blood were assayed by flow cytometry. Cases reported as negative were negative in both bone marrow and peripheral blood. Cases reported as positive had positive bone marrow aspirates with either positive or negative peripheral blood flow cytometry. No cases were positive in the peripheral blood and negative in the marrow.
Immunohistochemistry
Five-micrometer sections from paraffin-embedded tissues were stained with hematoxylin and eosin. Immunoperoxidase studies were performed on paraffin sections using a biotin–avidin peroxidase complex method.24 The mAbs used were CD20, CD5, CD3, CD10, and Bcl-2.
Results
Study population and clinical results
The results of this recently completed trial have been reported elsewhere.25 Briefly, 35 evaluable patients with intermediate- and advanced-stage CLL were enrolled between April 1992 and August 1996, and 30 were evaluable for response. Five patients were inevaluable and were removed from the study because of protocol violations. Clinical evaluation at the point of maximal response included 3 treatment failures, 13 PRs, 5 nPRs, and 9 CRs. Analysis of the subgroup of previously untreated patients revealed that consolidation with high-dose cyclophosphamide improved the quality of response in 9 patients and generally decreasing tumor burden and converting PR to CR in 5 patients.17
Sensitivity of clonotypic polymerase chain reaction
Semi-nested PCR could identify at least 1 malignant cell in a background of 105 polyclonal cells (Figure2A). Serial 10-fold dilutions of 100 ng patient DNA derived from BM (approximately 104 cells) were mixed into 1 μg polyclonal DNA (approximately 105 cells) extracted from normal PB mononuclear cells. The primary clonotypic PCR using the outer 5′ patient-specific primer and a JHconsensus primer detected 1 cell in 102 polyclonal cells. After a second round of amplification substituting the inner 5′ patient-specific primer, one cell in 105 polyclonal cells was detected. This compared favorably to an alternative technique in which the primary-round PCR product was electrophoresed, transferred, and probed with an inner CDR-III primer end labeled with32P (Figure 2B). This detected one malignant cell of 103 polyclonal cells.
Sensitivity of PCR versus hybridization of an internal probe to the first round product.
(A) Primary and semi-nested PCR demonstrating a sensitivity of one cell in 102 polyclonal cells after the primary round of amplification and one cell in 105 after the semi-nested second round. Primary PCR: serial 10-fold dilutions of 100 ng patient DNA derived from BM (approximately 104 cells) were mixed into 1μg polyclonal DNA (approximately 105 cells) extracted from normal PB mononuclear cells. Lane 1: positive control with 100 ng patient DNA, no polyclonal DNA. Lanes 2 through 8: log concentration of polyclonal versus patient DNA (eg, lane 2: polyclonal vs patient DNA is 100:1). Lane 9: negative control, 1 μg polyclonal DNA, no patient DNA. M: 100-bp markers. Semi-nested PCR: 1 μL 10−3 dilution of the first-round product serves as a template in the second round of amplification with the inner CDR-III clone-specific primer. Each lane corresponds to a dilution of the primary PCR product directly above it on the gel. Lane 10: no template from primary PCR. Anticipated product lengths were 101 bp and 75 bp for first- and second-round products, respectively. Lanes with an effective patient–polyclonal concentration of 10−6–10−8 are not expected to contain any malignant cells. The 350-bp product seen in lanes 6 to 9 reflects a nonspecific product amplified from polyclonal DNA with the clonotypic primers. (B) Demonstration of a sensitivity of 1 cell in 10−3 polyclonal cells after hybridization to a32P-labeled CDR-III inner primer. Hybridization of the probe to the semi-nested product serves as a positive control. The gel from Figure 3A was transferred by vacuum Southern blot and hybridized at 42°C to a 19-mer oligonucleotide in 6 × SSPE/1% sodium dodecyl sulfate (SDS). The filter was washed (6 × SSPE/1%SDS × 10 minutes × 3 at room temperature, followed by 1 × SSPE/1% SDS × 3 minutes at 42°C) and exposed overnight.
Sensitivity of PCR versus hybridization of an internal probe to the first round product.
(A) Primary and semi-nested PCR demonstrating a sensitivity of one cell in 102 polyclonal cells after the primary round of amplification and one cell in 105 after the semi-nested second round. Primary PCR: serial 10-fold dilutions of 100 ng patient DNA derived from BM (approximately 104 cells) were mixed into 1μg polyclonal DNA (approximately 105 cells) extracted from normal PB mononuclear cells. Lane 1: positive control with 100 ng patient DNA, no polyclonal DNA. Lanes 2 through 8: log concentration of polyclonal versus patient DNA (eg, lane 2: polyclonal vs patient DNA is 100:1). Lane 9: negative control, 1 μg polyclonal DNA, no patient DNA. M: 100-bp markers. Semi-nested PCR: 1 μL 10−3 dilution of the first-round product serves as a template in the second round of amplification with the inner CDR-III clone-specific primer. Each lane corresponds to a dilution of the primary PCR product directly above it on the gel. Lane 10: no template from primary PCR. Anticipated product lengths were 101 bp and 75 bp for first- and second-round products, respectively. Lanes with an effective patient–polyclonal concentration of 10−6–10−8 are not expected to contain any malignant cells. The 350-bp product seen in lanes 6 to 9 reflects a nonspecific product amplified from polyclonal DNA with the clonotypic primers. (B) Demonstration of a sensitivity of 1 cell in 10−3 polyclonal cells after hybridization to a32P-labeled CDR-III inner primer. Hybridization of the probe to the semi-nested product serves as a positive control. The gel from Figure 3A was transferred by vacuum Southern blot and hybridized at 42°C to a 19-mer oligonucleotide in 6 × SSPE/1% sodium dodecyl sulfate (SDS). The filter was washed (6 × SSPE/1%SDS × 10 minutes × 3 at room temperature, followed by 1 × SSPE/1% SDS × 3 minutes at 42°C) and exposed overnight.
Isolation of clonotypic sequence
Using 100 ng DNA from marrow or lymph node obtained at diagnosis, a monoclonal band was identified in all 19 patients evaluated. The VH leader primers were used successfully in 12 patients (63%). In 5 additional patients, the Fr2 or Fr3 primers were successful when used with the 3′ nested combination LJH followed by VLJH. In 2 patients, monoclonal bands identified by either set of primers provided sequence that subsequently led to unsuccessful primers (see below). Thus, the combination of the 2 primer sets was able to amplify the CDR-III of the malignant clone and lead to the design of successful clonotypic PCR primers in 89% of the patients.
The failure of the VH leader primers to consistently amplify the monoclonal product may be particular to CLL. Bahler et al26 used these primers to determine VH gene usage successfully in 36 of 36 patients with follicular lymphoma. For the 12 cases amplified by VH leader primers, the following breakdown was noted: 6 patients with VH1, 5 patients with VH4, and 1 patient with both VH1 and VH3. This is an unusual distribution for CLL, which typically is predominantly VH3 more than VH1 more than VH4 more than others.27 28
Creation of clone-specific primer
Clone-specific primers were selected from within the CDR-III region. Careful attention was given to the thermodynamics, as directed by the software package Oligo 5.0 (LifeBiotechnologies). Primers were chosen that were predicted to have adequate PCR performance. This limited the potential primers that could be created from the region of interest. After sequencing and primer design, the clone-specific primers were tested on the initial diagnostic material to confirm that a band of the appropriate size was obtained.
As evaluated by end-point dilution analysis of the pretreatment sample, the clone-specific primers were able to detect a single cell in 14 of 17 evaluable patients (Table 1)—that is, the PCR remained positive after a 10−4 dilution of a specimen with 104 cells. In patient 26, it was necessary to assay 10 cells to detect a malignant cell; in patient 30, it was 100 cells. In patient 28, the malignant clone could only be detected in the pretreatment DNA if 600 ng was assayed. Finally, in 2 patients, the predicted size band could not be amplified from the pretreatment despite primer modification. These 2 patients were considered inevaluable by clonotypic PCR. In one of these patients, the only pretreatment DNA available was extracted from a dried bone marrow aspirate slide demonstrating 73% lymphoid cells.
Clonotypic PCR correlated with clinical status at maximal response
| Patient . | Relative disease burden per 100 ng DNA . | |||
|---|---|---|---|---|
| Pretreatment/source of DNA assayed . | Post-treatment/source of DNA assayed . | Flow cytometry (PB and BM) . | Treatment response . | |
| 6 | 10−4/PB, BM | 10−4/PB, BM | + | PR |
| 11 | 10−5/PB, BM | 10−4/BM | + | PR |
| 12 | 10−5/PB, BM | 10−5/BM | + | PR |
| 19 | 10−4/PB, BM | 10−7/BM | Not done | PR |
| 22 | 10−3/PB, 10−5/BM | 10−5/BM | + | PR |
| 2 | 10−5/PB | 100/BM | − | nPR |
| 4 | 10−4/BM | 10−2/BM | − | nPR |
| 7 | 10−5/PB | 10−5/BM | − | nPR |
| 10 | 10−4/BM | Positive/BM* | + | nPR |
| 13 | 10−4/LN | Positive/BM† | + | nPR |
| 26 | 10−3/BM‡ | 10−2/BM | + | CR |
| 28 | >100 (600 ng)/BM | Negative/BM | − | CR |
| 1 | 10−5/PB | 10−1/BM | − | CR |
| 16 | Inevaluable/PB1-153 | Inevaluable1-153 | − | CR |
| 17 | Inevaluable1-153 | Inevaluable1-153 | − | CR |
| 23 | 10−3/BM 10−5/PB | 100/BM 10−3/PB | − | CR |
| 25 | 10−4/BM | Negative | − | CR |
| 27 | 10−4/BM | Negative/BM | − | CR |
| 30 | 10−2/BM | Negative/BM | − | CR |
| Patient . | Relative disease burden per 100 ng DNA . | |||
|---|---|---|---|---|
| Pretreatment/source of DNA assayed . | Post-treatment/source of DNA assayed . | Flow cytometry (PB and BM) . | Treatment response . | |
| 6 | 10−4/PB, BM | 10−4/PB, BM | + | PR |
| 11 | 10−5/PB, BM | 10−4/BM | + | PR |
| 12 | 10−5/PB, BM | 10−5/BM | + | PR |
| 19 | 10−4/PB, BM | 10−7/BM | Not done | PR |
| 22 | 10−3/PB, 10−5/BM | 10−5/BM | + | PR |
| 2 | 10−5/PB | 100/BM | − | nPR |
| 4 | 10−4/BM | 10−2/BM | − | nPR |
| 7 | 10−5/PB | 10−5/BM | − | nPR |
| 10 | 10−4/BM | Positive/BM* | + | nPR |
| 13 | 10−4/LN | Positive/BM† | + | nPR |
| 26 | 10−3/BM‡ | 10−2/BM | + | CR |
| 28 | >100 (600 ng)/BM | Negative/BM | − | CR |
| 1 | 10−5/PB | 10−1/BM | − | CR |
| 16 | Inevaluable/PB1-153 | Inevaluable1-153 | − | CR |
| 17 | Inevaluable1-153 | Inevaluable1-153 | − | CR |
| 23 | 10−3/BM 10−5/PB | 100/BM 10−3/PB | − | CR |
| 25 | 10−4/BM | Negative | − | CR |
| 27 | 10−4/BM | Negative/BM | − | CR |
| 30 | 10−2/BM | Negative/BM | − | CR |
Results of semiquantitative clonotypic PCR at pretreatment and post-treatment time-points are presented along with clinical response. Results of PCR are reported as the 10-fold serial end-point dilution of 100 ng (roughly 104 cell equivalents), which still yielded a positive PCR result on the semi-nested round of PCR. Hence, there is an inverse relation between the disease burden and the serial dilution reported positive. The more dilute the specimen recorded as positive, the greater the disease burden.
Paraffin section used as source of DNA; only qualitative result available.
Bone marrow aspirate slide used as a source of DNA; only qualitative result available.
s/p 3 cycles fludarabine; no pretreatment available.
Primers problematic (see text).
Illustration of technique
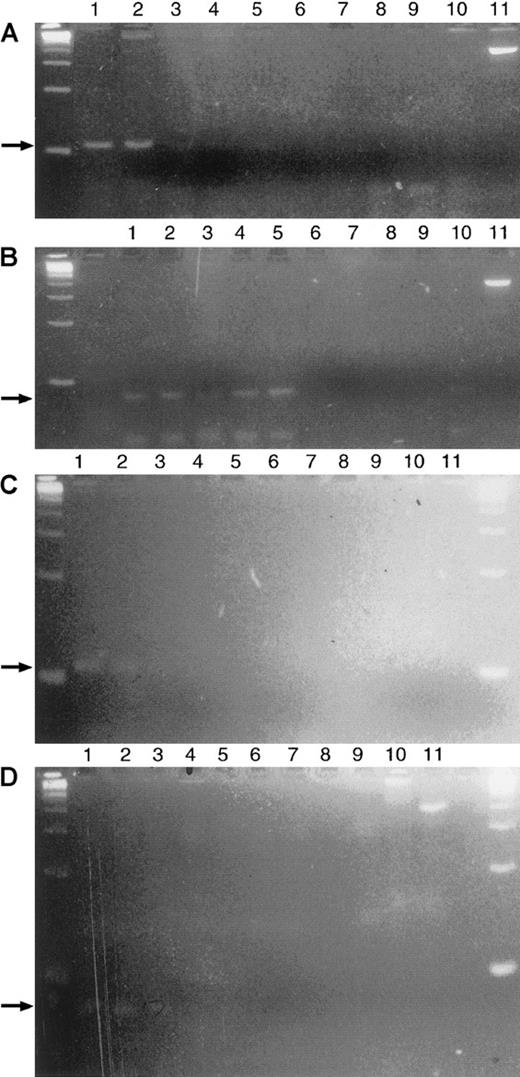
A representative clonotypic PCR is illustrated in Figure3 for a patient who achieved nPR after treatment. At nPR, mature lymphoid cells comprised 18% of the bone marrow aspirate. Before treatment the malignant clone could be detected at a 10−4 dilution (Figure 3A-B); after chemotherapy, it could be detected at a 10−2 dilution (Figure 3C-D). The relative abundance of the malignant clone was reduced by a factor of 2 logs in the bone marrow.
Demonstration of clonotypic PCR before and after treatment in an nPR patient.
(A) Clonotypic PCR product in the initial pretreatment samples of this patient at a 10−4 dilution in the BM after primary PCR. (B) After seminested PCR, the clonotype is detected at a 10−4 dilution with greater intensity confirming the previous primary PCR result. (C) Persistent clonotype PCR after treatment from a nodular PR BM specimen for this same patient. Primary PCR detects a clone at a 10−2 dilution. (D) Seminested PCR confirms the primary PCR finding, detecting a clone at a 10−2 dilution. DNA was extracted from an aspirate corresponding to the bone marrow biopsy shown in Figure 4B. (A-D) Lanes 1-8, serial 10-fold dilutions of patient DNA starting with a 100 ng template; lane 9, negative control with no template DNA; lane 10, 1 ug polyclonal DNA; lane 11, positive control with bcl-2 primers and 100 ng of template.
Demonstration of clonotypic PCR before and after treatment in an nPR patient.
(A) Clonotypic PCR product in the initial pretreatment samples of this patient at a 10−4 dilution in the BM after primary PCR. (B) After seminested PCR, the clonotype is detected at a 10−4 dilution with greater intensity confirming the previous primary PCR result. (C) Persistent clonotype PCR after treatment from a nodular PR BM specimen for this same patient. Primary PCR detects a clone at a 10−2 dilution. (D) Seminested PCR confirms the primary PCR finding, detecting a clone at a 10−2 dilution. DNA was extracted from an aspirate corresponding to the bone marrow biopsy shown in Figure 4B. (A-D) Lanes 1-8, serial 10-fold dilutions of patient DNA starting with a 100 ng template; lane 9, negative control with no template DNA; lane 10, 1 ug polyclonal DNA; lane 11, positive control with bcl-2 primers and 100 ng of template.
Clonotypic polymerase chain reaction
The results of clonotypic PCR, correlated with clinical response, are presented in Table 1. To confirm the validity of the clonotypic PCR technique, the first 5 patients who achieved PR were studied. All 5 patients had disease detectable histologically in the core BM biopsy. For these patients, there was no decrement in disease burden after treatment, as judged by end-point dilution analysis.
Clonotypic PCR analysis detected disease in all 5 patients who achieved nPR. Of these 5 patients, 3 were evaluable by limiting-dilution analysis at both the pretreatment and the nPR time-point. A decrement in disease burden was seen in 2 of 3 patients.
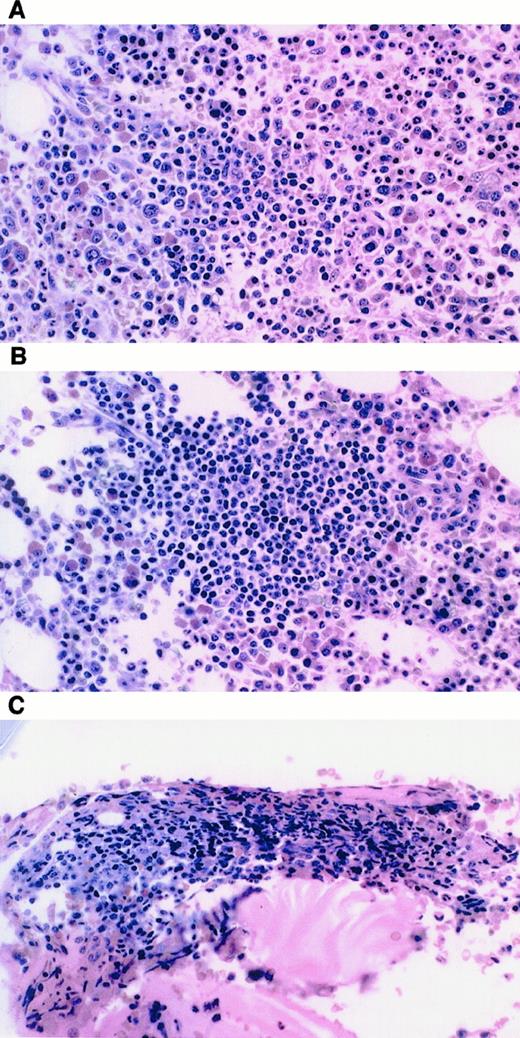
The nPR patients had a variety of bone marrow findings, as demonstrated in Figure 4 and as correlated with PCR results in Table 2. By definition, all patients with nPR had less than 30% mature lymphocytes on bone marrow aspiration. Noting that biopsy findings were not included in the definition of response, biopsy specimens ranged from minute clusters of lymphocytes to dense clusters more readily identifiable as consistent with CLL. Immunohistochemistry staining identified disease in only 1 of 4 evaluable patients. This patient (patient 4) was only weakly CD20+, CD5−, CD10−, rare CD3+ and was strongly bcl-2+.
Examples of bone marrow biopsies from patients achieving nPR.
(A) Patient 2: few irregular clusters of small lymphocytes. (B) Patient 4: several aggregates of small, round lymphocytes consistent with CLL. (C) Patient 7: 1-minute cluster of small lymphocytes, no identifiable lymphoma.
Examples of bone marrow biopsies from patients achieving nPR.
(A) Patient 2: few irregular clusters of small lymphocytes. (B) Patient 4: several aggregates of small, round lymphocytes consistent with CLL. (C) Patient 7: 1-minute cluster of small lymphocytes, no identifiable lymphoma.
Correlation of clonotypic PCR results with bone marrow biopsy results at nPR
| Patient . | PCR result . | Disease assessment on bone marrow biopsy . |
|---|---|---|
| 2 | 100 | Few irregular clusters of small lymphocytes |
| 4 | 10−2/PB | Several aggregates of small, round lymphocytes |
| 7 | 10−5 | Single, minute cluster of small lymphocytes |
| 10 | Positive* | Two lymphoid follicles |
| 13 | Positive† | Irregular, dense clusters of small lymphoid cells |
| Patient . | PCR result . | Disease assessment on bone marrow biopsy . |
|---|---|---|
| 2 | 100 | Few irregular clusters of small lymphocytes |
| 4 | 10−2/PB | Several aggregates of small, round lymphocytes |
| 7 | 10−5 | Single, minute cluster of small lymphocytes |
| 10 | Positive* | Two lymphoid follicles |
| 13 | Positive† | Irregular, dense clusters of small lymphoid cells |
Results of PCR are presented as the minimum number of cells assayed by limiting-dilution analysis, which still yielded a positive PCR result on the semi-nested round of PCR.
Paraffin section used as source of DNA; only qualitative result available.
Bone marrow aspirate slide used as source of DNA; only qualitative result available.
The disease burden, as estimated by end-point dilution analysis, did not correlate with the findings on biopsy. For example, in patient 7, though there was only a single minute cluster of lymphocytes on biopsy, PCR of a single cell by end-point dilution was still positive. In contrast, in patient 4, the marrow had several aggregates of small, round lymphocytes consistent with CLL and positive immunohistochemistry staining, yet 100 cells were assayed before a malignant clone could be detected. Clonotypic PCR results were compared with flow cytometric assessments of residual disease using dual staining for CD5–CD19 and κ–λ clonal excess. Flow cytometry detected disease in only 3 of 5 patients positive by clonotypic PCR.
Clonotypic PCR yielded variable results in the 7 evaluable patients who had CR. PCR findings were positive for 3 patients and negative for 4. Flow cytometry findings were negative in all but 1 patient whose PCR findings were positive. As judged by end-point dilution, disease burden was reduced in the 2 assessable patients after treatment.
Post-response follow-up
Four patients have had serial PCR determinations during follow-up after maximal response. In 3 of these 4, increasing disease burdens have been demonstrated, as judged by end-point dilution. None have had clinical relapse.
Patient 26 achieved CR. Clonotypic PCR of the BM was positive at a 10−2 dilution, a log reduction from pretreatment. On repeat aspirate 2 months later, the bone marrow aspirate was suggestive of malignant cells, but BM biopsy findings were negative. This material was not available for PCR analysis. Seven months after treatment, the BM aspirate was negative, the biopsy findings were suspicious, and clonotypic PCR was positive at a 10−3 dilution.
Patient 27 became PCR negative after treatment. Pretreatment BM tested positive at 10−4 dilution. Four months after CR, the patient remained PCR negative, but findings were positive 1 year after follow-up at a 100 dilution.
At CR the undiluted PB and BM DNA of patient 23 remained PCR positive. This was a 4-log improvement from pre-treatment PCR. Eight months into follow-up, the BM biopsy specimen had a few clusters of small lymphocytes, consistent with lymphoma, with only 20% lymphocytes on aspirate. PCR of the PB demonstrated the malignant clone at a 10−3 dilution. BM was unavailable for analysis. After additional observation at 5 and 13 months, the BM biopsy and aspirate showed no evidence of disease, and clonotypic PCR of the BM remained positive at a 10−1 dilution. Finally patient 30, initially positive at a 10−2 dilution in the BM, achieved PCR negativity after treatment and remained negative after 1 year of follow-up.
Discussion
We have used clonotypic PCR to detect MRD in CLL after high-dose, non-myeloablative chemotherapy. Disease was detectable in all patients at nPR and in some patients at CR. A clone-specific technique was chosen because, unlike chronic myelocytic leukemia, acute promyelocytic leukemia, or follicular lymphoma, CLL is not associated with a recurrent chromosomal translocation that could serve as a convenient marker of MRD. CLL is analogous to acute lymphocytic leukemia (ALL), in which molecular probes for clone-specific immunoglobulin rearrangements have been used for the detection of MRD.
Several PCR strategies for B cell neoplasms have been devised and applied primarily to the assessment of ALL. In this report we describe a technique of 2 oligonucleotide primers within the CDR-III region that can be used with a consensus JH primer. This enables semi-nested PCR to be performed, increasing both sensitivity and specificity. Although further cycles of amplification increase sensitivity, the requirement that the first round product be made amplifiable by the nested second primer verifies specificity. High-resolution electrophoresis with a 10-bp DNA ladder allowed for relatively precise size determination of the products, again demonstrating specificity. Additionally, the 2-step PCR technique allows a sample to be readily assayed in 2 days from the time the sample arrives in the laboratory. It is unlikely that conventional primers alone are as sensitive for MRD. In 2 patients who had nPR and for whom material was available, the VH leader primers or the framework primers demonstrated a smear with a dominant band or with multiple bands (data not shown).
In the cases evaluated by VH leader primers, we saw a skewing of the repertoire, with 50% VH1 and 42% VH4. This is surprising for 2 reasons. First, because VH3 is the largest group of VH genes, it is the one most often used by randomly rearranged genes.5Moreover, in recent larger series of randomly collected CLL cases,27-29 VH3 is usually the predominant subtype, with more VH3 than VH1, more VH1 than VH4, and more VH4 than others. Most likely this discrepancy reflects our small sample size. VH typing restriction is believed to reflect an antigen selection process in the development of CLL. Although earlier authors have not reported somatic mutations in CLL,30,31 others more recently have demonstrated equal expression of mutated and unmutated VH genes, with inferior clinical outcome in the latter subgroup.27 32 Additional follow-up will be necessary to confirm clinical significance in our study.
This clonotypic PCR technique demonstrated MRD in all 5 patients who achieved nPR. Traditionally, response assessment in CLL has relied on bone aspirate differentials, independent of biopsy findings.14 As a consequence, in the absence of other evidence of disease, patients with suspicious BM biopsy specimens were still considered to be in CR. We have demonstrated that patients with the recently defined category of nPR16 can have a dramatic range of findings, from lymphoid clusters to nodules, consistent with involvement by CLL. Moreover, immunohistochemistry fails to demonstrated MRD in most patients. Our PCR results are the first molecular evidence that these lymphoid nodules represent disease. This conclusion is supported by our observation that patients who achieved nPR in this clinical trial had remission lengths essentially identical to those of patients who achieved PR but markedly shorter than those of patients who achieved CR.25 Robertson et al15report that patients who achieve nPR after treatment with fludarabine plus prednisone have an overall survival rate between those who achieve CR and those who achieve PR.
Clonotypic PCR detected MRD in 3 of 6 evaluable patients in CR. These patients will be followed up to determine whether PCR positivity can predict relapse. Using single-round amplification techniques, which are expected to be less sensitive than the methodology used in our study, others have reported molecular CR in patients with CLL. Maloum et al33 demonstrated that 1 of 7 patients became PCR negative after fludarabine treatment alone, whereas 3 of 3 attained molecular remission after autologous transplantation. Esteve et al34studied 12 patients with low-grade lymphoproliferative disorders treated with fludarabine, 3 of whom had CLL. In this small study, all 3 patients with CLL achieved molecular CR. Analyzed without regard to disease type, patients who were in molecular CR had longer relapse-free survival.
Tracking MRD in CLL by clonal excess or dual staining on flow cytometry is attractive because of its simplicity, sensitivity, and rapid turnaround. In numerous studies of CLL, investigators have sought to detect minimal residual disease using flow cytometric techniques. Despite the sensitivity of this technique, no standards defining what constitutes a “positive” result have been established. In this study, we have chosen to define the presence of flow cytometric residual disease as either the ability to detect a κ–λ clonal excess ratio of greater than 5:1 (3:1 if λ clonal excess is detected) or a CD5–CD19 dual-staining population exceeding 5%. This level represents a value approximately 2 SD above the nonspecific staining for this dual-marker combination measured in normal controls (data not shown) and is comparable to results described elsewhere.15,22,23,35 Flow cytometry proved to be less sensitive than our molecular approach. Flow cytometry detected MRD in only 2 of 5 nPR patients and in 1 of 3 CR patients who tested positive by clonotypic PCR. Additionally, flow cytometry did not detect disease in any patient with negative findings on PCR. These flow cytometric results are consistent with a previous report15 that 2-parameter flow cytometry demonstrated residual disease in 11% and 49% of patients in CR and nPR, respectively.
Southern blot analysis is an alternative method for monitoring MRD. The technique is at least 100 times less sensitive than PCR for CLL and results depend on the demonstration of rearranged immunoglobulin genes. Robertson et al15 treated CLL patients with fludarabine alone. After evaluating 20% of them by Southern blotting, they detected a residual clonal population in 5 of 7 CR patients and in 6 of 8 nPR patients. Selection bias might have played a role in determining on whom to perform this test.
The limiting-dilution technique used here is only semiquantitative. By this method, disease burden did not decrease in the PR patients but did decrease with treatment in 2 of 3 nPR analyzable patients. We were unable to correlate the extent of disease on biopsy and estimated disease burden by end-point dilution analysis. This disparity may be explained by sampling error—the biopsy section and the aspirate were not identical samples, and the involvement of the bone marrow might have been patchy. Alternatively, end-point dilution may be a poor measure of assessing disease burden. Because the primer pairs differ in sequence, they have different performance characteristics and sensitivity. However, in this series, clone-specific primers were able to detect a single malignant cell in 14 of 17 evaluable patients. In only 1 patient were the primers truly problematic; a malignant clone was detected only if 600 ng pretreatment DNA was assayed. Despite these caveats, clonotypic PCR with end-point dilution detected increasing disease burdens in 3 of 4 patients who remain in clinical remission under observation. Current research efforts include refining the quantitative aspects of clonotypic PCR36 and prospectively evaluating the meaning of the increasing molecular burden of disease.
In summary, our results support the hypothesis that nodular PR in CLL represents MRD. Additionally, clonotypic PCR appears more sensitive than flow cytometry for the detection of MRD. Our findings also demonstrate the wide heterogeneity of bone marrow biopsy findings in patients achieving nPR. As the follow-up increases, we will assess the value of clonotypic PCR in predicting clinical outcome. If PCR proves to be a reliable indicator of clinical outcome, it could be used as a surrogate end-point in clinical trials. In addition, it may prove to be useful in predicting which patients are at the greatest risk for relapse, targeting them for additional treatment while sparing those with the least risk from associated morbidity.
Supported in part by the Norman and Rosita Winston Foundation; the Horace W. Goldsmith Foundation; the Mortimer J. Lacher, MD Lymphoma Foundation Fund; the Leukemia Research Foundation of America; and National Institutes of Health grants K12-CA01712-03, K12-CA09512, 1-K08CA73825-01A1, and R01 CA67823-02 USPHS (NCI CA05826-34).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ariela Noy, Dept of Medicine/Division of Hematologic Oncology, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021; e-mail: noya@mskcc.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal